Function and Benefits of Natural Competence in Cyanobacteria: From Ecology to Targeted Manipulation
Abstract
1. Introduction
2. The Molecular Basis of Natural Competence
2.1. T4P Are Crucial for Natural Competence in Cyanobacteria
2.2. Com Proteins Mediate DNA Uptake and Processing
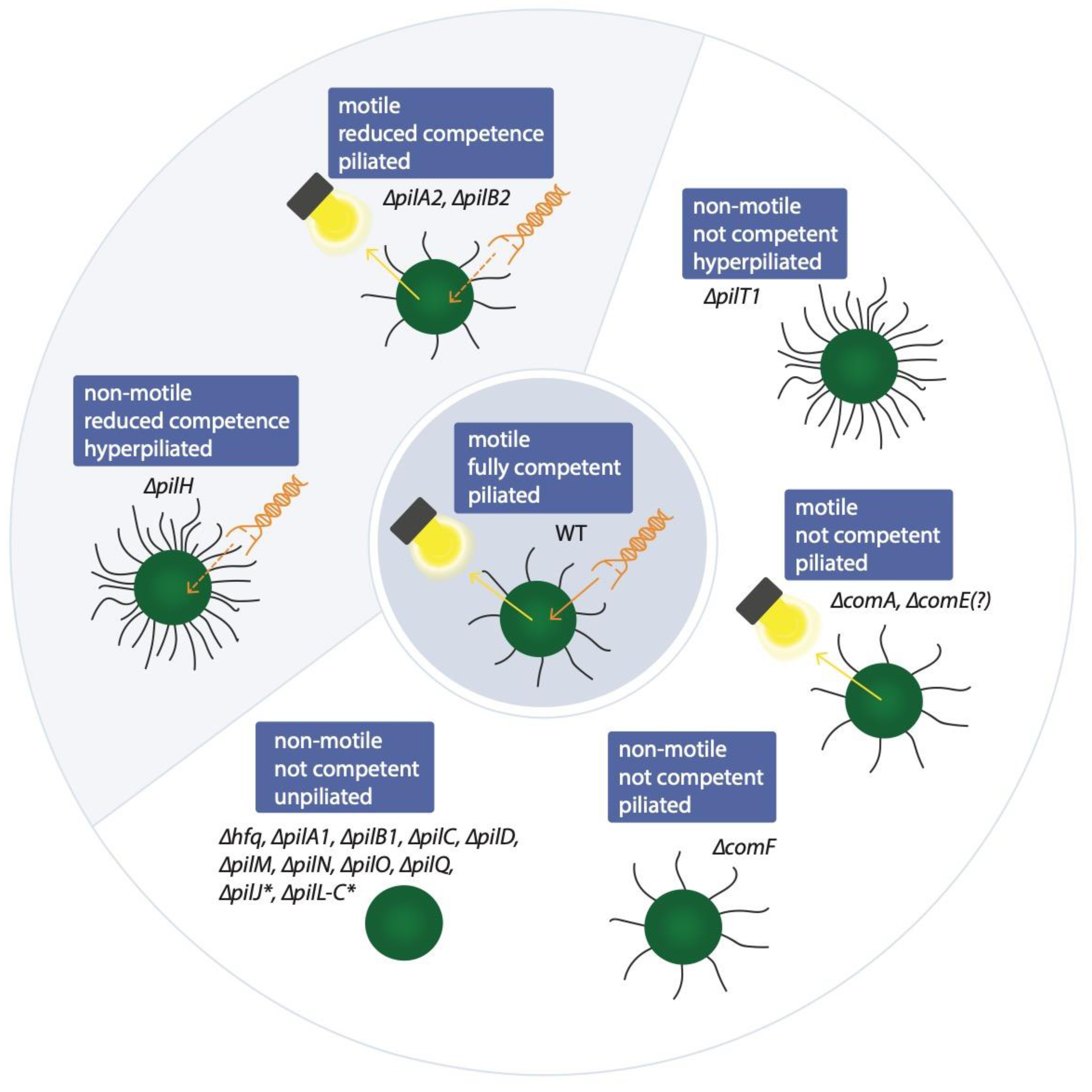
3. Versatile Factors Control Natural Competence
4. Natural Competence Might Be More Frequent among Cyanobacteria than Initially Anticipated
4.1. Experimental Evidence of Natural Competence in Cyanobacteria
4.2. Genomics Give Insights into the Prevalence of Cyanobacterial Natural Competence
5. Benefits and Drawbacks of Natural Competence in an Ecological Context
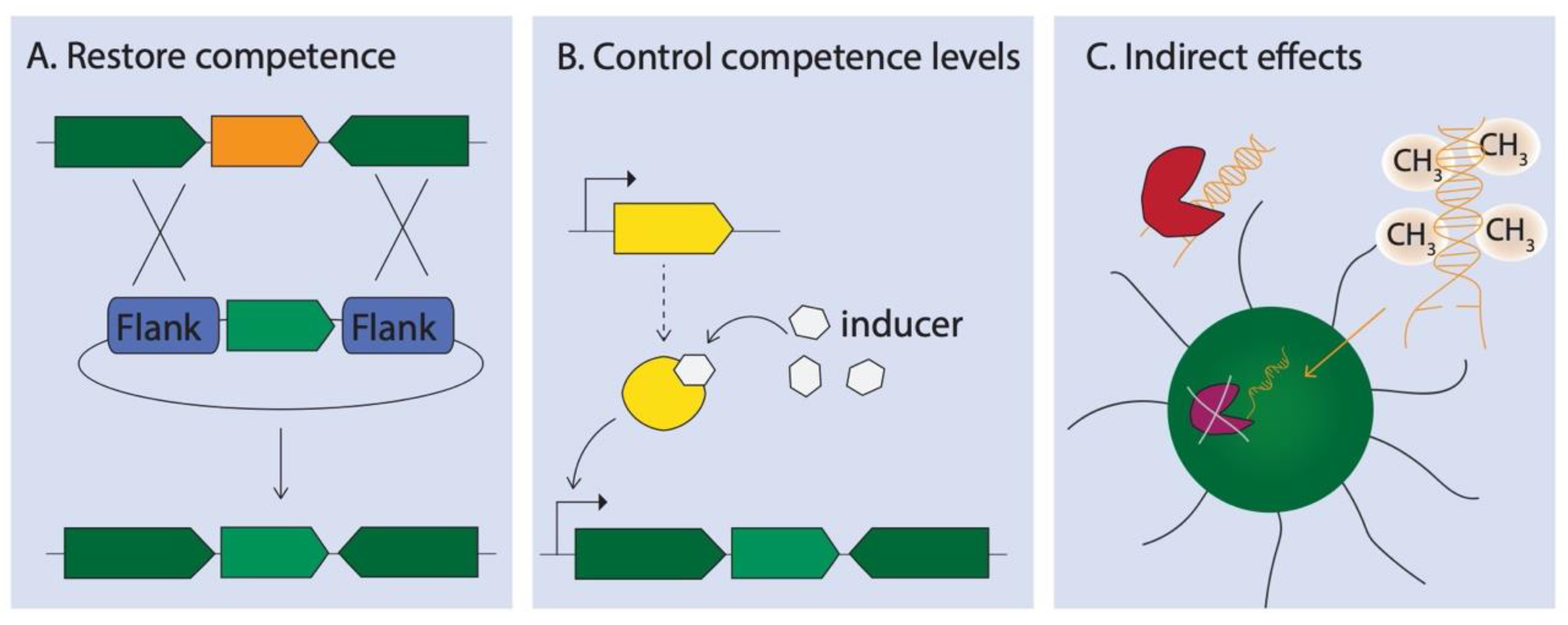
6. Targeted Manipulation of Natural Competence
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Johnsborg, O.; Eldholm, V.; Håvarstein, L.S. Natural genetic transformation: Prevalence, mechanisms and function. Res. Microbiol. 2007, 158, 767–778. [Google Scholar] [CrossRef] [PubMed]
- Dubnau, D.; Blokesch, M. Mechanisms of DNA Uptake by Naturally Competent Bacteria. Annu. Rev. Genet. 2019, 53, 217–237. [Google Scholar] [CrossRef] [PubMed]
- Johnston, C.; Martin, B.; Fichant, G.; Polard, P.; Claverys, J.-P. Bacterial transformation: Distribution, shared mechanisms and divergent control. Nat. Rev. Microbiol. 2014, 12, 181–196. [Google Scholar] [CrossRef] [PubMed]
- Haas, R.; Meyer, T.F.; Putten, J.P.M. Aflagellated mutants of Helicobacter pylori generated by genetic transformation of naturally competent strains using transposon shuttle mutagenesis. Mol. Microbiol. 1993, 8, 753–760. [Google Scholar] [CrossRef]
- Bhaya, D.; Bianco, N.R.; Bryant, D.; Grossman, A. Type IV pilus biogenesis and motility in the cyanobacterium Synechocystis sp. PCC6803. Mol. Microbiol. 2000, 37, 941–951. [Google Scholar] [CrossRef]
- Wolfgang, M.; Lauer, P.; Park, H.; Brossay, L.; Hébert, J.; Koomey, M. PilT mutations lead to simultaneous defects in competence for natural transformation and twitching motility in piliated Neisseria gonorrhoeae. Mol. Microbiol. 1998, 29, 321–330. [Google Scholar] [CrossRef]
- Evans, K.J.; Lambert, C.; Sockett, R.E. Predation by Bdellovibrio bacteriovorus HD100 Requires Type IV Pili. J. Bacteriol. 2007, 189, 4850–4859. [Google Scholar] [CrossRef] [PubMed]
- Pujol, C.; Eugene, E.; Marceau, M.; Nassif, X. The meningococcal PilT protein is required for induction of intimate attachment to epithelial cells following pilus-mediated adhesion. Proc. Natl. Acad. Sci. USA 1999, 96, 4017–4022. [Google Scholar] [CrossRef]
- Klausen, M.; Aaes-Jørgensen, A.; Molin, S.; Tolker-Nielsen, T. Involvement of bacterial migration in the development of complex multicellular structures in Pseudomonas aeruginosa biofilms. Mol. Microbiol. 2003, 50, 61–68. [Google Scholar] [CrossRef]
- Klausen, M.; Heydorn, A.; Ragas, P.; Lambertsen, L.; Aaes-Jørgensen, A.; Molin, S.; Tolker-Nielsen, T. Biofilm formation by Pseudomonas aeruginosa wild type, flagella and type IV pili mutants. Mol. Microbiol. 2003, 48, 1511–1524. [Google Scholar] [CrossRef]
- O’Toole, G.A.; Kolter, R. Flagellar and twitching motility are necessary for Pseudomonas aeruginosa biofilm development. Mol. Microbiol. 1998, 30, 295–304. [Google Scholar] [CrossRef] [PubMed]
- Bieber, D.; Ramer, S.W.; Wu, C.-Y.; Murray, W.J.; Tobe, T.; Fernandez, R.; Schoolnik, G.K. Type IV Pili, Transient Bacterial Aggregates, and Virulence of Enteropathogenic Escherichia coli. Science 1998, 280, 2114–2118. [Google Scholar] [CrossRef] [PubMed]
- Lu, H.-M.; Motley, S.T.; Lory, S. Interactions of the components of the general secretion pathway: Role of Pseudomonas aeruginosa type IV pilin subunits in complex formation and extracellular protein secretion. Mol. Microbiol. 1997, 25, 247–259. [Google Scholar] [CrossRef]
- Shestakov, S.V.; Khyen, N.T. Evidence for genetic transformation in blue-green alga Anacystis nidulans. Mol. Gen. Genet. 1970, 107, 372–375. [Google Scholar] [CrossRef] [PubMed]
- Jacobsen, T.; Bardiaux, B.; Francetic, O.; Izadi-Pruneyre, N.; Nilges, M. Structure and function of minor pilins of type IV pili. Med. Microbiol. Immunol. 2020, 209, 301–308. [Google Scholar] [CrossRef]
- Sun, D. Pull in and Push Out: Mechanisms of Horizontal Gene Transfer in Bacteria. Front. Microbiol. 2018, 9, 2154. [Google Scholar] [CrossRef]
- Yoshihara, S.; Geng, X.X.; Okamoto, S.; Yura, K.; Murata, T.; Go, M.; Ohmori, M.; Ikeuchi, M. Mutational Analysis of Genes Involved in Pilus Structure, Motility and Transformation Competency in the Unicellular Motile Cyanobacterium Synechocystis sp. PCC 6803. Plant Cell Physiol. 2001, 42, 63–73. [Google Scholar] [CrossRef]
- Yura, K.; Toh, H.; Go, M. Putative Mechanism of Natural Transformation as Deduced from Genome Data. DNA Res. 1999, 6, 75–82. [Google Scholar] [CrossRef][Green Version]
- Nakasugi, K.; Svenson, C.J.; Neilan, B.A. The competence gene, comF, from Synechocystis sp. strain PCC 6803 is involved in natural transformation, phototactic motility and piliation. Microbiology 2006, 152, 3623–3631. [Google Scholar] [CrossRef]
- Bergé, M.; Mortier-Barrière, I.; Martin, B.; Claverys, J.-P. Transformation of Streptococcus pneumoniae relies on DprA- and RecA-dependent protection of incoming DNA single strands. Mol. Microbiol. 2003, 50, 527–536. [Google Scholar] [CrossRef]
- Mortier-Barrière, I.; Velten, M.; Dupaigne, P.; Mirouze, N.; Piétrement, O.; McGovern, S.; Fichant, G.; Martin, B.; Noirot, P.; Le Cam, E.; et al. A Key Presynaptic Role in Transformation for a Widespread Bacterial Protein: DprA Conveys Incoming ssDNA to RecA. Cell 2007, 130, 824–836. [Google Scholar] [CrossRef] [PubMed]
- Taton, A.; Erikson, C.; Yang, Y.; Rubin, B.E.; Rifkin, S.A.; Golden, J.W.; Golden, S.S. The circadian clock and darkness control natural competence in cyanobacteria. Nat. Commun. 2020, 11, 1688. [Google Scholar] [CrossRef] [PubMed]
- Dienst, D.; Dühring, U.; Mollenkopf, H.-J.; Vogel, J.; Golecki, J.; Hess, W.R.; Wilde, A. The cyanobacterial homologue of the RNA chaperone Hfq is essential for motility of Synechocystis sp. PCC 6803. Microbiology 2008, 154, 3134–3143. [Google Scholar] [CrossRef] [PubMed]
- Schuergers, N.; Ruppert, U.; Watanabe, S.; Nürnberg, D.J.; Lochnit, G.; Dienst, D.; Mullineaux, C.W.; Wilde, A. Binding of the RNA chaperone Hfq to the type IV pilus base is crucial for its function in Synechocystis sp. PCC 6803. Mol. Microbiol. 2014, 92, 840–852. [Google Scholar] [CrossRef]
- Sergeyenko, T.V.; Los, D.A. Identification of secreted proteins of the cyanobacterium Synechocystis sp. strain PCC 6803. FEMS Microbiol. Lett. 2000, 193, 213–216. [Google Scholar] [CrossRef]
- Yoshihara, S.; Geng, X.; Ikeuchi, M. pilG Gene Cluster and Split pilL Genes Involved in Pilus Biogenesis, Motility and Genetic Transformation in the Cyanobacterium Synechocystis sp. PCC 6803. Plant Cell Physiol. 2002, 43, 513–521. [Google Scholar] [CrossRef]
- Okamoto, S.; Ohmori, M. The Cyanobacterial PilT Protein Responsible for Cell Motility and Transformation Hydrolyzes ATP. Plant Cell Physiol. 2002, 43, 1127–1136. [Google Scholar] [CrossRef]
- Murphy, R.C.; Bryant, D.A.; Porter, R.D.; de Marsac, N.T. Molecular cloning and characterization of the recA gene from the cyanobacterium Synechococcus sp. strain PCC 7002. J. Bacteriol. 1987, 169, 2739–2747. [Google Scholar] [CrossRef]
- Bhaya, D.; Watanabe, N.; Ogawa, T.; Grossman, A.R. The role of an alternative sigma factor in motility and pilus formation in the cyanobacterium Synechocystis sp. strain PCC6803. Proc. Natl. Acad. Sci. USA 1999, 96, 3188–3193. [Google Scholar] [CrossRef]
- Lamb, J.J.; Hohmann-Marriott, M.F. Manganese acquisition is facilitated by PilA in the cyanobacterium Synechocystis sp. PCC 6803. PLoS ONE 2017, 12, e0184685. [Google Scholar] [CrossRef]
- Lamb, J.J.; Hill, R.E.; Eaton-Rye, J.J.; Hohmann-Marriott, M.F. Functional Role of PilA in Iron Acquisition in the Cyanobacterium Synechocystis sp. PCC 6803. PLoS ONE 2014, 9, e105761. [Google Scholar] [CrossRef] [PubMed]
- Allen, R.; Rittmann, B.E.; Curtiss, R. Axenic Biofilm Formation and Aggregation by Synechocystis sp. Strain PCC 6803 Are Induced by Changes in Nutrient Concentration and Require Cell Surface Structures. Appl. Environ. Microbiol. 2019, 85, e02192-18. [Google Scholar] [CrossRef]
- Conradi, F.D.; Zhou, R.-Q.; Oeser, S.; Schuergers, N.; Wilde, A.; Mullineaux, C.W. Factors Controlling Floc Formation and Structure in the Cyanobacterium Synechocystis sp. Strain PCC 6803. J. Bacteriol. 2019, 201, e00344-19. [Google Scholar] [CrossRef] [PubMed]
- Grigorieva, G.; Shestakov, S. Transformation in the cyanobacterium Synechocystis sp. 6803. FEMS Microbiol. Lett. 1982, 13, 367–370. [Google Scholar] [CrossRef]
- Piepenbrink, K.H. DNA Uptake by Type IV Filaments. Front. Mol. Biosci. 2019, 6, 1. [Google Scholar] [CrossRef] [PubMed]
- Pelicic, V. Monoderm bacteria: The new frontier for type IV pilus biology. Mol. Microbiol. 2019, 112, 1674–1683. [Google Scholar] [CrossRef]
- Linhartová, M.; Bučinská, L.; Halada, P.; Ječmen, T.; Šetlík, J.; Komenda, J.; Sobotka, R. Accumulation of the Type IV prepilin triggers degradation of SecY and YidC and inhibits synthesis of Photosystem II proteins in the cyanobacterium Synechocystis PCC 6803. Mol. Microbiol. 2014, 93, 1207–1223. [Google Scholar] [CrossRef]
- Nunn, D.; Bergman, S.; Lory, S. Products of three accessory genes, pilB, pilC, and pilD, are required for biogenesis of Pseudomonas aeruginosa pili. J. Bacteriol. 1990, 172, 2911–2919. [Google Scholar] [CrossRef]
- Nunn, D.N.; Lory, S. Product of the Pseudomonas aeruginosa gene pilD is a prepilin leader peptidase. Proc. Natl. Acad. Sci. USA 1991, 88, 3281–3285. [Google Scholar] [CrossRef]
- Strom, M.S.; Nunn, D.N.; Lory, S. A single bifunctional enzyme, PilD, catalyzes cleavage and N-methylation of proteins belonging to the type IV pilin family. Proc. Natl. Acad. Sci. USA 1993, 90, 2404–2408. [Google Scholar] [CrossRef]
- Bhaya, D.; Takahashi, A.; Shahi, P.; Grossman, A.R. Novel Motility Mutants of Synechocystis Strain PCC 6803 Generated by In Vitro Transposon Mutagenesis. J. Bacteriol. 2001, 183, 6140–6143. [Google Scholar] [CrossRef]
- Yoshihara, S.; Ikeuchi, M. Phototactic motility in the unicellular cyanobacterium Synechocystis sp. PCC 6803. Photochem. Photobiol. Sci. 2004, 3, 512–518. [Google Scholar] [CrossRef] [PubMed]
- Ellison, C.K.; Dalia, T.N.; Ceballos, A.V.; Wang, J.C.-Y.; Biais, N.; Brun, Y.V.; Dalia, A.B. Retraction of DNA-bound type IV competence pili initiates DNA uptake during natural transformation in Vibrio cholerae. Nat. Microbiol. 2018, 3, 773–780. [Google Scholar] [CrossRef] [PubMed]
- Chen, Z.; Li, X.; Tan, X.; Zhang, Y.; Wang, B. Recent Advances in Biological Functions of Thick Pili in the Cyanobacterium Synechocystis sp. PCC 6803. Front. Plant Sci. 2020, 11, 241. [Google Scholar] [CrossRef] [PubMed]
- Diallo, A.; Foster, H.R.; Gromek, K.A.; Perry, T.N.; Dujeancourt, A.; Krasteva, P.V.; Gubellini, F.; Falbel, T.G.; Burton, B.M.; Fronzes, R. Bacterial transformation: ComFA is a DNA-dependent ATPase that forms complexes with ComFC and DprA. Mol. Microbiol. 2017, 105, 741–754. [Google Scholar] [CrossRef]
- Johnsborg, O.; Håvarstein, L.S. Regulation of natural genetic transformation and acquisition of transforming DNA in Streptococcus pneumoniae. FEMS Microbiol. Rev. 2009, 33, 627–642. [Google Scholar] [CrossRef] [PubMed]
- Meibom, K.L.; Blokesch, M.; Dolganov, N.A.; Wu, C.-Y.; Schoolnik, G.K. Chitin Induces Natural Competence in Vibrio cholerae. Science 2005, 310, 1824–1827. [Google Scholar] [CrossRef]
- Lorenz, M.G.; Wackernagel, W. Natural genetic transformation of Pseudomonas stutzeri by sand-adsorbed DNA. Arch. Microbiol. 1990, 154, 380–385. [Google Scholar] [CrossRef]
- Havarstein, L.S.; Coomaraswamy, G.; Morrison, D.A. An unmodified heptadecapeptide pheromone induces competence for genetic transformation in Streptococcus pneumoniae. Proc. Natl. Acad. Sci. USA 1995, 92, 11140–11144. [Google Scholar] [CrossRef]
- Li, Y.-H.; Lau, P.C.Y.; Lee, J.H.; Ellen, R.P.; Cvitkovitch, D.G. Natural Genetic Transformation of Streptococcus mutans Growing in Biofilms. J. Bacteriol. 2001, 183, 897–908. [Google Scholar] [CrossRef]
- Magnuson, R.; Solomon, J.; Grossman, A.D. Biochemical and genetic characterization of a competence pheromone from B. subtilis. Cell 1994, 77, 207–216. [Google Scholar] [CrossRef]
- Kandel, P.P.; Lopez, S.M.; Almeida, R.P.P.; De La Fuente, L. Natural Competence of Xylella fastidiosa Occurs at a High Frequency Inside Microfluidic Chambers Mimicking the Bacterium’s Natural Habitats. Appl. Environ. Microbiol. 2016, 82, 5269–5277. [Google Scholar] [CrossRef] [PubMed]
- Merod, R.T.; Wuertz, S. Extracellular Polymeric Substance Architecture Influences Natural Genetic Transformation of Acinetobacter baylyi in Biofilms. Appl. Environ. Microbiol. 2014, 80, 7752–7757. [Google Scholar] [CrossRef] [PubMed]
- Lorenz, M.G.; Wackernagel, W. High Frequency of Natural Genetic Transformation of Pseudomonas stutzeri in Soil Extract Supplemented with a Carbon/Energy and Phosphorus Source. Appl. Environ. Microbiol. 1991, 57, 1246–1251. [Google Scholar] [CrossRef] [PubMed]
- Dorer, M.S.; Fero, J.; Salama, N.R. DNA Damage Triggers Genetic Exchange in Helicobacter pylori. PLoS Pathog. 2010, 6, e1001026. [Google Scholar] [CrossRef]
- Charpentier, X.; Kay, E.; Schneider, D.; Shuman, H.A. Antibiotics and UV Radiation Induce Competence for Natural Transformation in Legionella pneumophila. J. Bacteriol. 2011, 193, 1114–1121. [Google Scholar] [CrossRef]
- Prudhomme, M.; Attaiech, L.; Sanchez, G.; Martin, B.; Claverys, J.-P. Antibiotic Stress Induces Genetic Transformability in the Human Pathogen Streptococcus pneumoniae. Science 2006, 313, 89–92. [Google Scholar] [CrossRef]
- Herriott, R.M.; Meyer, E.M.; Vogt, M. Defined Nongrowth Media for Stage II Development of Competence in Haemophilus influenzae. J. Bacteriol. 1970, 101, 517–524. [Google Scholar] [CrossRef]
- Nielsen, K.M.; Bones, A.M.; Van Elsas, J.D. Induced Natural Transformation of Acinetobacter calcoaceticus in Soil Microcosms. Appl. Environ. Microbiol. 1997, 63, 3972–3977. [Google Scholar] [CrossRef]
- Traglia, G.M.; Quinn, B.; Schramm, S.T.J.; Soler-Bistue, A.; Ramirez, M.S. Serum Albumin and Ca2+ Are Natural Competence Inducers in the Human Pathogen Acinetobacter baumannii. Antimicrob. Agents Chemother. 2016, 60, 4920–4929. [Google Scholar] [CrossRef]
- Mell, J.C.; Redfield, R.J. Natural Competence and the Evolution of DNA Uptake Specificity. J. Bacteriol. 2014, 196, 1471–1483. [Google Scholar] [CrossRef] [PubMed]
- Seitz, P.; Blokesch, M. Cues and regulatory pathways involved in natural competence and transformation in pathogenic and environmental Gram-negative bacteria. FEMS Microbiol. Rev. 2013, 37, 336–363. [Google Scholar] [CrossRef] [PubMed]
- Porter, R.D. Transformation in Cyanobacteria. CRC Crit. Rev. Microbiol. 1986, 13, 111–132. [Google Scholar] [CrossRef] [PubMed]
- Zang, X.; Liu, B.; Liu, S.; Arunakumara, K.K.I.U.; Zhang, X. Optimum conditions for transformation of Synechocystis sp. PCC 6803. J. Microbiol. 2007, 45, 241–245. [Google Scholar]
- Golden, S.S.; Sherman, L.A. Optimal conditions for genetic transformation of the cyanobacterium Anacystis nidulans R2. J. Bacteriol. 1984, 158, 36–42. [Google Scholar] [CrossRef] [PubMed]
- Wolk, C.P.; Kraus, J. Two approaches to obtaining low, extracellular deoxyribonuclease activity in cultures of heterocyst-forming cyanobacteria. Arch. Microbiol. 1982, 131, 302–307. [Google Scholar] [CrossRef]
- Schuergers, N.; Wilde, A. Appendages of the Cyanobacterial Cell. Life 2015, 5, 700–715. [Google Scholar] [CrossRef]
- Williams, J.G.K.; Szalay, A.A. Stable integration of foreign DNA into the chromosome of the cyanobacterium Synechococcus R2. Gene 1983, 24, 37–51. [Google Scholar] [CrossRef]
- Jaiswal, D.; Sengupta, A.; Sohoni, S.; Sengupta, S.; Phadnavis, A.G.; Pakrasi, H.B.; Wangikar, P.P. Genome Features and Biochemical Characteristics of a Robust, Fast Growing and Naturally Transformable Cyanobacterium Synechococcus elongatus PCC 11801 Isolated from India. Sci. Rep. 2018, 8, 16632. [Google Scholar] [CrossRef]
- Stevens, S.E.; Porter, R.D. Transformation in Agmenellum quadruplicatum. Proc. Natl. Acad. Sci. USA 1980, 77, 6052–6056. [Google Scholar] [CrossRef]
- Herdman, M. Mutations arising during transformation in the blue-green alga Anacystis nidulans. Mol. Gen. Genet. MGG 1973, 120, 369–378. [Google Scholar] [CrossRef] [PubMed]
- Orkwiszewski, K.G.; Kaney, A.R. Genetic transformation of the blue-green bacterium, Anacystis nidulans. Arch. Microbiol. 1974, 98, 31–37. [Google Scholar] [CrossRef] [PubMed]
- Lightfoot, D.A.; Walters, D.E.; Wootton, J.C. Transformation of the Cyanobacterium Synechococcus PCC 6301 Using Cloned DNA. J. Gen. Microbiol. 1988, 134, 1509–1514. [Google Scholar] [CrossRef][Green Version]
- Takeshima, Y.; Takatsugu, N.; Sugiura, M.; Hagiwara, H. High-level expression of human superoxide dismutase in the cyanobacterium Anacystis nidulans 6301. Proc. Natl. Acad. Sci. USA 1994, 91, 9685–9689. [Google Scholar] [CrossRef] [PubMed]
- Takeshima, Y.; Sugiura, M.; Hagiwara, H. A Novel Expression Vector for the Cyanobacterium, Synechococcus PCC 6301. DNA Res. 1994, 1, 181–189. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Sugita, C.; Ogata, K.; Shikata, M.; Jikuya, H.; Takano, J.; Furumichi, M.; Kanehisa, M.; Omata, T.; Sugiura, M.; Sugita, M. Complete nucleotide sequence of the freshwater unicellular cyanobacterium Synechococcus elongatus PCC 6301 chromosome: Gene content and organization. Photosynth. Res. 2007, 93, 55–67. [Google Scholar] [CrossRef] [PubMed]
- Tsinoremas, N.F.; Kutach, A.K.; Strayer, C.A.; Golden, S.S. Efficient Gene Transfer in Synechococcus sp. Strains PCC 7942 and PCC 6301 by Interspecies Conjugation and Chromosomal Recombination. J. Bacteriol. 1994, 176, 6764–6768. [Google Scholar] [CrossRef]
- Onai, K.; Morishita, M.; Kaneko, T.; Tabata, S.; Ishiura, M. Natural transformation of the thermophilic cyanobacterium Thermosynechococcus elongatus BP-1: A simple and efficient method for gene transfer. Mol. Genet. Genom. 2004, 271, 50–59. [Google Scholar] [CrossRef]
- Dittmann, E.; Neilan, B.A.; Erhard, M.; Von Döhren, H.; Börner, T. Insertional mutagenesis of a peptide synthetase gene that is responsible for hepatotoxin production in the cyanobacterium Microcystis aeruginosa PCC 7806. Mol. Microbiol. 1997, 26, 779–787. [Google Scholar] [CrossRef]
- Koksharova, O.A.; Wolk, C.P. Genetic tools for cyanobacteria. Appl. Microbiol. Biotechnol. 2002, 58, 123–137. [Google Scholar] [CrossRef]
- Al-Haj, L.; Lui, Y.; Abed, R.; Gomaa, M.; Purton, S. Cyanobacteria as Chassis for Industrial Biotechnology: Progress and Prospects. Life 2016, 6, 42. [Google Scholar] [CrossRef]
- Stucken, K.; Ilhan, J.; Roettger, M.; Dagan, T.; Martin, W.F. Transformation and Conjugal Transfer of Foreign Genes into the Filamentous Multicellular Cyanobacteria (Subsection V) Fischerella and Chlorogloeopsis. Curr. Microbiol. 2012, 65, 552–560. [Google Scholar] [CrossRef]
- Trehan, K.; Sinha, U. Genetic Transfer in a Nitrogen-fixing Filamentous Cyanobacterium. Microbiology 1981, 124, 349–352. [Google Scholar] [CrossRef]
- Singh, D.T.; Bagchi, S.N.; Modi, D.R.; Singh, H.N. Evidence for Intergenetic Transformation in Filamentous, Diazotrophic Cyanobacteria. New Phytol. 1987, 107, 347–356. [Google Scholar] [CrossRef]
- Verma, S.K.; Singh, A.K.; Katiyar, S.; Singh, H.N. Genetic transformation of glutamine auxotrophy to prototrophy in the cyanobacterium Nostoc muscorum. Arch. Microbiol. 1990, 154, 414–416. [Google Scholar] [CrossRef]
- Nies, F.; Mielke, M.; Pochert, J.; Lamparter, T. Natural transformation of the filamentous cyanobacterium Phormidium lacuna. PLoS ONE 2020, 15, e0234440. [Google Scholar] [CrossRef]
- Springstein, B.L.; Nies, F.; Dagan, T. Natural competence in Chlorogloeopsis fritschii PCC 6912 and other ramified cyanobacteria. bioRxiv 2020, 2020. [Google Scholar] [CrossRef]
- Cassier-Chauvat, C.; Veaudor, T.; Chauvat, F. Comparative Genomics of DNA Recombination and Repair in Cyanobacteria: Biotechnological Implications. Front. Microbiol. 2016, 7, 1809. [Google Scholar] [CrossRef]
- Wendt, K.E.; Pakrasi, H.B. Genomics Approaches to Deciphering Natural Transformation in Cyanobacteria. Front. Microbiol. 2019, 10, 1259. [Google Scholar] [CrossRef]
- Ponce-Toledo, R.I.; Deschamps, P.; López-García, P.; Zivanovic, Y.; Benzerara, K.; Moreira, D. An Early-Branching Freshwater Cyanobacterium at the Origin of Plastids. Curr. Biol. 2017, 27, 386–391. [Google Scholar] [CrossRef]
- Partensky, F.; Garczarek, L. Prochlorococcus: Advantages and Limits of Minimalism. Ann. Rev. Mar. Sci. 2010, 2, 305–331. [Google Scholar] [CrossRef]
- Zerulla, K.; Ludt, K.; Soppa, J. The ploidy level of Synechocystis sp. PCC 6803 is highly variable and is influenced by growth phase and by chemical and physical external parameters. Microbiology 2016, 162, 730–739. [Google Scholar] [CrossRef]
- Koyama, Y.; Hoshino, T.; Tomizuka, N.; Furukawa, K. Genetic transformation of the extreme thermophile Thermus thermophilus and of other Thermus spp. J. Bacteriol. 1986, 166, 338–340. [Google Scholar] [CrossRef]
- Friedrich, A.; Hartsch, T.; Averhoff, B. Natural Transformation in Mesophilic and Thermophilic Bacteria: Identification and Characterization of Novel, Closely Related Competence Genes in Acinetobacter sp. Strain BD413 and Thermus thermophilus HB27. Appl. Environ. Microbiol. 2001, 67, 3140–3148. [Google Scholar] [CrossRef]
- Friedrich, A.; Rumszauer, J.; Henne, A.; Averhoff, B. Pilin-Like Proteins in the Extremely Thermophilic Bacterium Thermus thermophilus HB27: Implication in Competence for Natural Transformation and Links to Type IV Pilus Biogenesis. Appl. Environ. Microbiol. 2003, 69, 3695–3700. [Google Scholar] [CrossRef]
- Vos, M.; Buckling, A.; Kuijper, B. Sexual Selection in Bacteria? Trends Microbiol. 2019, 27, 972–981. [Google Scholar] [CrossRef]
- Szöllősi, G.J.; Derényi, I.; Vellai, T. The Maintenance of Sex in Bacteria Is Ensured by Its Potential to Reload Genes. Genetics 2006, 174, 2173–2180. [Google Scholar] [CrossRef]
- Russo, D.A.; Zedler, J.A.Z. Genomic insights into cyanobacterial protein translocation systems. Biol. Chem. 2020, 2020. [Google Scholar] [CrossRef]
- Watanabe, S. Cyanobacterial multi-copy chromosomes and their replication. Biosci. Biotechnol. Biochem. 2020, 84, 1309–1321. [Google Scholar] [CrossRef]
- Stucken, K.; Koch, R.; Dagan, T. Cyanobacterial defense mechanisms against foreign DNA transfer and their impact on genetic engineering. Biol. Res. 2013, 46, 373–382. [Google Scholar] [CrossRef]
- Cai, F.; Axen, S.D.; Kerfeld, C.A. Evidence for the widespread distribution of CRISPR-Cas system in the Phylum Cyanobacteria. RNA Biol. 2013, 10, 687–693. [Google Scholar] [CrossRef] [PubMed]
- Hou, S.; Brenes-Álvarez, M.; Reimann, V.; Alkhnbashi, O.S.; Backofen, R.; Muro-Pastor, A.M.; Hess, W.R. CRISPR-Cas systems in multicellular cyanobacteria. RNA Biol. 2019, 16, 518–529. [Google Scholar] [CrossRef] [PubMed]
- Hitchcock, A.; Hunter, C.N.; Canniffe, D.P. Progress and challenges in engineering cyanobacteria as chassis for light-driven biotechnology. Microb. Biotechnol. 2020, 13, 363–367. [Google Scholar] [CrossRef] [PubMed]
- Santos-Merino, M.; Singh, A.K.; Ducat, D.C. New Applications of Synthetic Biology Tools for Cyanobacterial Metabolic Engineering. Front. Bioeng. Biotechnol. 2019, 7, 33. [Google Scholar] [CrossRef]
- Klemenčič, M.; Nielsen, A.Z.; Sakuragi, Y.; Frigaard, N.-U.; Čelešnik, H.; Jensen, P.E.; Dolinar, M. Synthetic biology of cyanobacteria for production of biofuels and high-value products. In Microalgae-Based Biofuels and Bioproducts; Gonzalez-Fernandez, C., Muñoz, R., Eds.; Woodhead Publishing: Sawston, UK, 2017; pp. 305–325. [Google Scholar]
- Russo, D.A.; Zedler, J.A.Z.; Jensen, P.E. A force awakens: Exploiting solar energy beyond photosynthesis. J. Exp. Bot. 2019, 70, 1703–1710. [Google Scholar] [CrossRef]
- Yu, J.; Liberton, M.; Cliften, P.F.; Head, R.D.; Jacobs, J.M.; Smith, R.D.; Koppenaal, D.W.; Brand, J.J.; Pakrasi, H.B. Synechococcus elongatus UTEX 2973, a fast growing cyanobacterial chassis for biosynthesis using light and CO2. Sci. Rep. 2015, 5, 8132. [Google Scholar] [CrossRef]
- Li, S.; Sun, T.; Xu, C.; Chen, L.; Zhang, W. Development and optimization of genetic toolboxes for a fast-growing cyanobacterium Synechococcus elongatus UTEX 2973. Metab. Eng. 2018, 48, 163–174. [Google Scholar] [CrossRef]
- Ayers, M.; Sampaleanu, L.M.; Tammam, S.; Koo, J.; Harvey, H.; Howell, P.L.; Burrows, L.L. PilM/N/O/P Proteins Form an Inner Membrane Complex That Affects the Stability of the Pseudomonas aeruginosa Type IV Pilus Secretin. J. Mol. Biol. 2009, 394, 128–142. [Google Scholar] [CrossRef]
- Behle, A.; Saake, P.; Axmann, I.M. Comparative dose-response analysis of inducible promoters in cyanobacteria. ACS Synth. Biol. 2020, 9, 843–855. [Google Scholar] [CrossRef]
- Englund, E.; Liang, F.; Lindberg, P. Evaluation of promoters and ribosome binding sites for biotechnological applications in the unicellular cyanobacterium Synechocystis sp. PCC 6803. Sci. Rep. 2016, 6, 36640. [Google Scholar] [CrossRef]
- Kelly, C.L.; Taylor, G.M.; Hitchcock, A.; Torres-Méndez, A.; Heap, J.T. A Rhamnose-Inducible System for Precise and Temporal Control of Gene Expression in Cyanobacteria. ACS Synth. Biol. 2018, 7, 1056–1066. [Google Scholar] [CrossRef]
- Gale, G.A.R.; Schiavon Osorio, A.A.; Mills, L.A.; Wang, B.; Lea-Smith, D.J.; McCormick, A.J. Emerging Species and Genome Editing Tools: Future Prospects in Cyanobacterial Synthetic Biology. Microorganisms 2019, 7, 409. [Google Scholar] [CrossRef] [PubMed]
- Kufryk, G.; Sachet, M.; Schmetterer, G.; Vermaas, W.F.J. Transformation of the cyanobacterium Synechocystis sp. PCC 6803 as a tool for genetic mapping: Optimization of efficiency. FEMS Microbiol. Lett. 2002, 206, 215–219. [Google Scholar] [CrossRef] [PubMed]
- Iwai, M.; Katoh, H.; Katayama, M.; Ikeuchi, M. Improved Genetic Transformation of the Thermophilic Cyanobacterium, Thermosynechococcus elongatus BP-1. Plant Cell Physiol. 2004, 45, 171–175. [Google Scholar] [CrossRef]
- Wang, B.; Yu, J.; Zhang, W.; Meldrum, D.R. Premethylation of Foreign DNA Improves Integrative Transformation Efficiency in Synechocystis sp. Strain PCC 6803. Appl. Environ. Microbiol. 2015, 81, 8500–8506. [Google Scholar] [CrossRef] [PubMed]
- Elhai, J.; Vepritskiy, A.; Muro-Pastor, A.M.; Flores, E.; Wolk, C.P. Reduction of conjugal transfer efficiency by three restriction activities of Anabaena sp. strain PCC 7120. J. Bacteriol. 1997, 179, 1998–2005. [Google Scholar] [CrossRef]
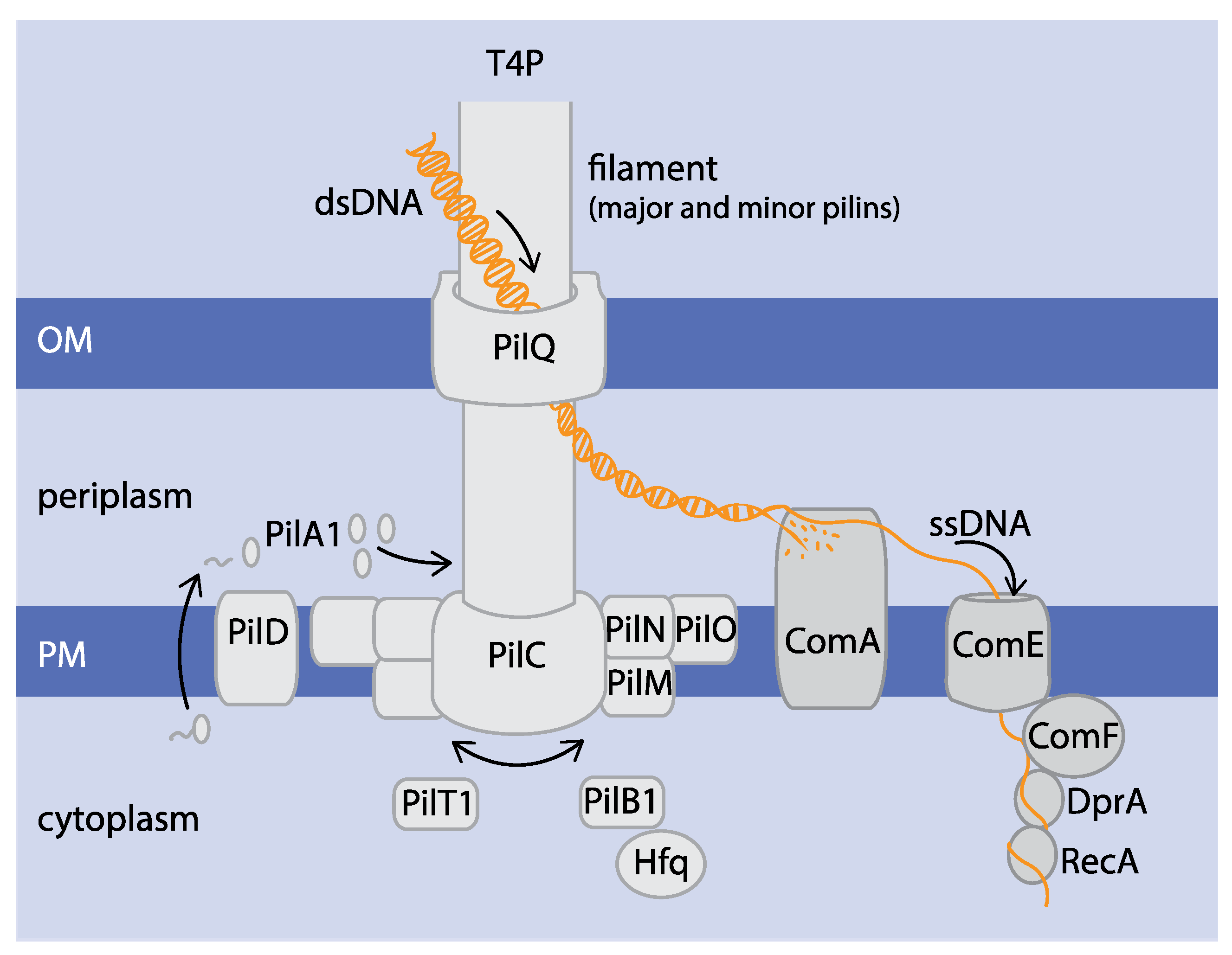
| Protein | Gene Assignment | Function | Reference |
|---|---|---|---|
| ComA | slr0197 | DNA translocation (DNA binding and nuclease activity?) | [17,18] |
| ComE | sll1929 | DNA translocation (translocase activity?) | [17] |
| ComF | slr0388 | DNA translocation (transitioning DNA uptake and homologous recombination?) | [19] |
| DprA | slr1197 | DNA processing protein | [20,21,22] |
| Hfq | ssr3341 | Pilus biogenesis | [23,24] |
| PilA1 | sll1694 | Filament formation (major pilin) | [5] |
| PilA2 | sll1695 | Filament formation (minor pilin) | [5,17] |
| PilB1 | slr0063 | Motor protein (polymerisation) | [17] |
| PilB2 | slr0079 | Unknown | [17] |
| PilC | slr0162-slr0163 | Platform protein | [5] |
| PilD | slr1120 | Prepilin peptidase | [5,17,25] |
| PilH | slr1042 | Che-like response regulator pilus assembly | [26] |
| PilJ | slr1044 | Che-like response regulator pilus assembly | [26] |
| PilL-C | slr0322 | Che-like response regulator pilus assembly | [26] |
| PilM | slr1274 | Pilus alignment complex | [17] |
| PilN | slr1275 | Pilus alignment complex | [17] |
| PilO | slr1276 | Pilus alignment complex | [17] |
| PilQ | slr1277 | Secretin | [17] |
| PilT1 | slr0161 | Motor protein (depolymerisation) | [5,27] |
| RecA | sll0569 | Homologous recombination | [20,21,28] |
| Order | Number of Species | Number of pil and com Genes Identified in the Genome | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | 12 | ||
| Chroococcales | 10 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 2 | 8 |
| Chroococcidiopsidales | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 |
| Gloeobacterales | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 1 |
| Nostocales | 13 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 12 |
| Oscillatoriales | 8 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 7 |
| Pleurocapsales | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 2 |
| Synechococcales | 37 | 2 | 3 | 0 | 1 | 1 | 2 | 0 | 2 | 1 | 0 | 2 | 8 | 15 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Schirmacher, A.M.; Hanamghar, S.S.; Zedler, J.A.Z. Function and Benefits of Natural Competence in Cyanobacteria: From Ecology to Targeted Manipulation. Life 2020, 10, 249. https://doi.org/10.3390/life10110249
Schirmacher AM, Hanamghar SS, Zedler JAZ. Function and Benefits of Natural Competence in Cyanobacteria: From Ecology to Targeted Manipulation. Life. 2020; 10(11):249. https://doi.org/10.3390/life10110249
Chicago/Turabian StyleSchirmacher, Alexandra M., Sayali S. Hanamghar, and Julie A. Z. Zedler. 2020. "Function and Benefits of Natural Competence in Cyanobacteria: From Ecology to Targeted Manipulation" Life 10, no. 11: 249. https://doi.org/10.3390/life10110249
APA StyleSchirmacher, A. M., Hanamghar, S. S., & Zedler, J. A. Z. (2020). Function and Benefits of Natural Competence in Cyanobacteria: From Ecology to Targeted Manipulation. Life, 10(11), 249. https://doi.org/10.3390/life10110249






