Bioinformatics Analysis of Evolution and Human Disease Related Transposable Element-Derived microRNAs
Abstract
1. Introduction
2. Bioinformatics Analysis of Transposable Elements
3. Selection of Microrna Related Papers and Bioinformatic Analyses of Transposable Element-Derived microRNAs
4. Bioinformatic Analyses of Evolution Related Transposable Element-Derived microRNAs
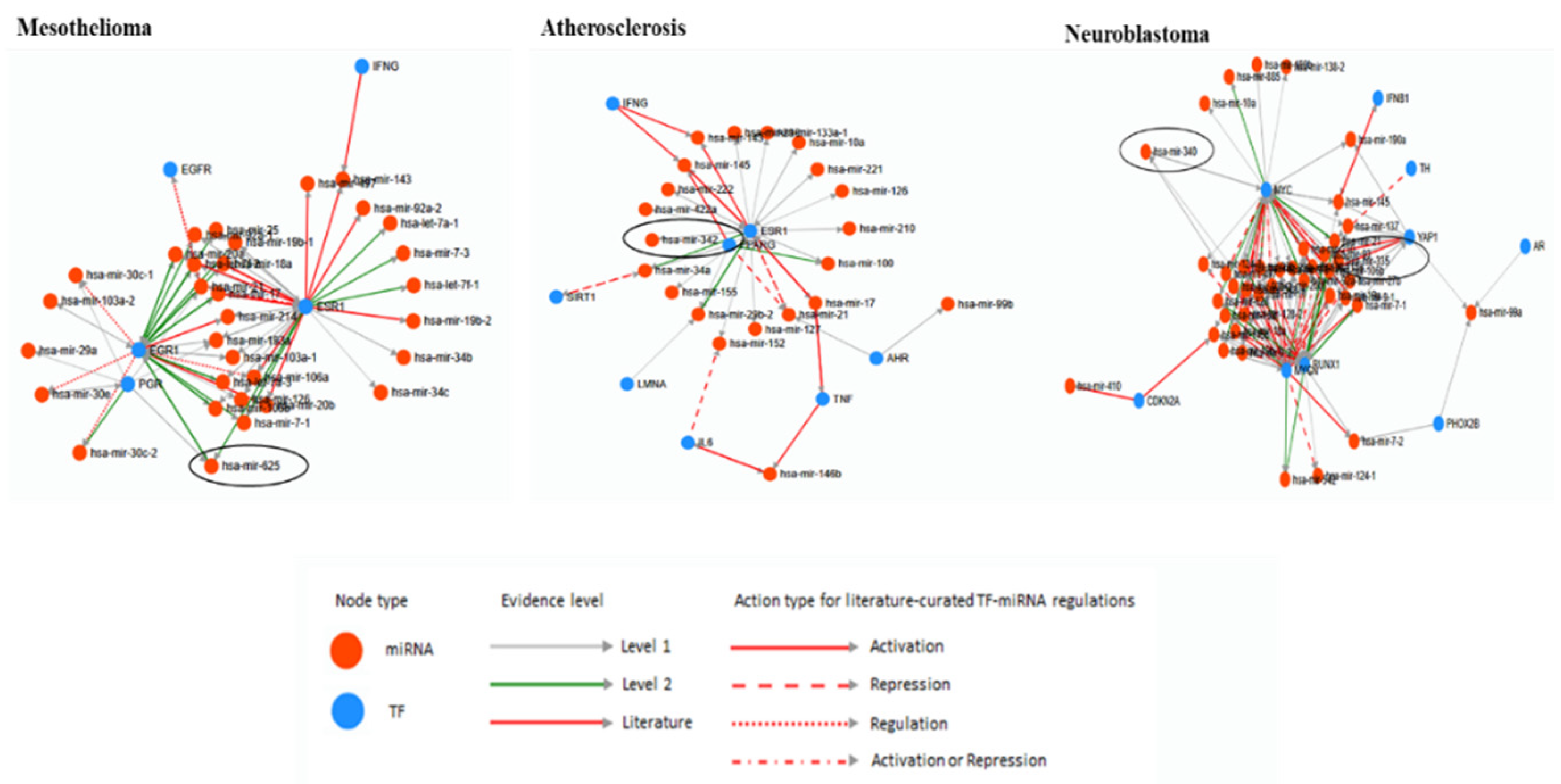
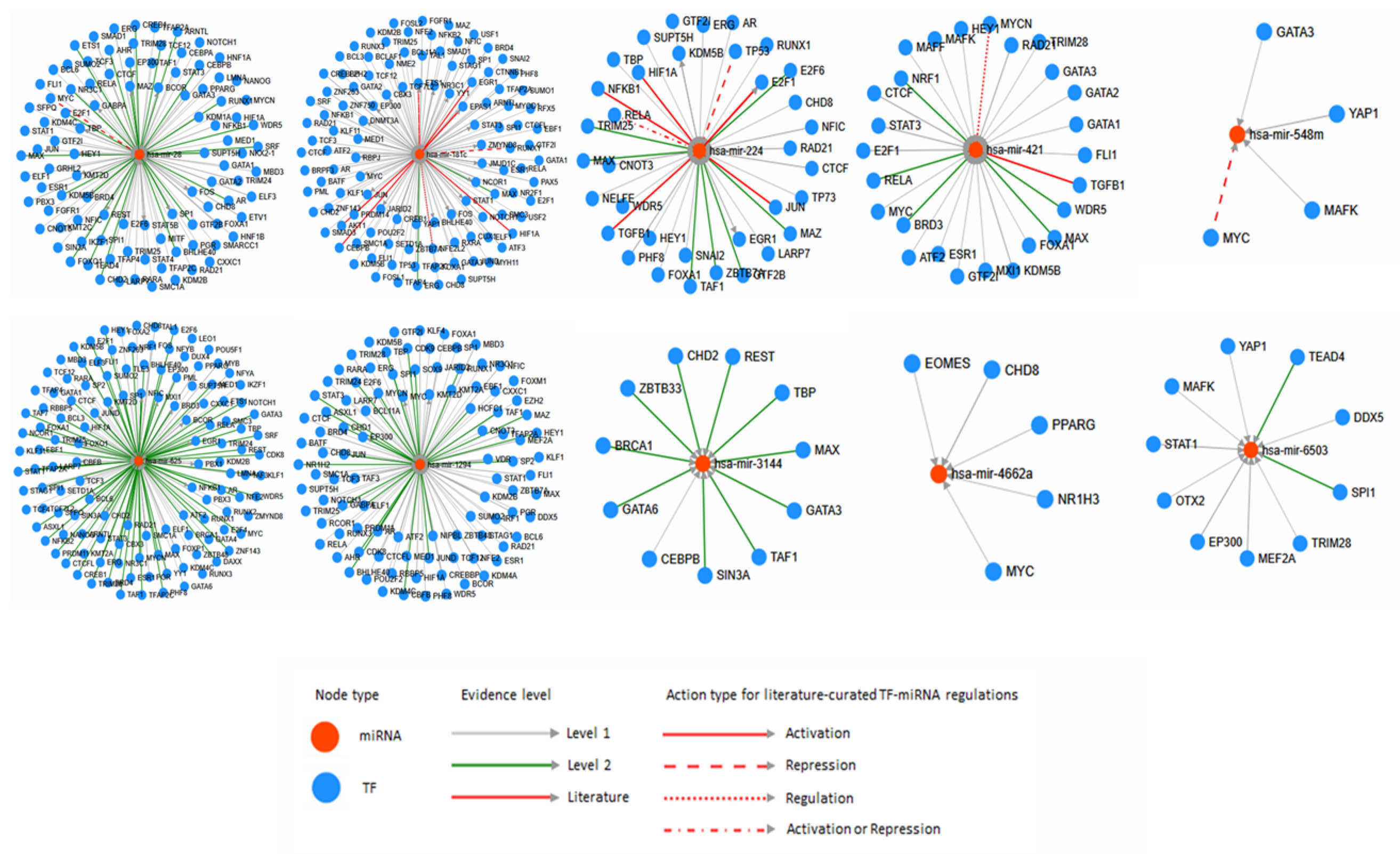
5. Bioinformatic Analyses of Human Diseases Related Transposable Element-Derived microRNAs
6. Discussion
Author Contributions
Funding
Conflicts of Interest
References
- Finnegan, D.J.; Fawcett, D.H. Transposable elements in Drosophila melanogaster. Oxf. Surv. Eukaryot. Genes 1986, 3, 1–62. [Google Scholar] [PubMed]
- Hollister, J.D.; Gaut, B.S. Epigenetic silencing of transposable elements: A trade-off between reduced transposition and deleterious effects on neighboring gene expression. Genome Res. 2009, 19, 1419–1428. [Google Scholar] [CrossRef] [PubMed]
- Mendizabal, I.; Keller, T.E.; Zeng, J.; Yi, S.V. Epigenetics and evolution. Integr. Comp. Biol. 2014, 54, 31–42. [Google Scholar] [CrossRef] [PubMed]
- Lisch, D.; Bennetzen, J.L. Transposable element origins of epigenetic gene regulation. Curr. Opin. Plant. Biol. 2011, 14, 156–161. [Google Scholar] [CrossRef]
- Lee, H.-E.; Eo, J.; Kim, H.-S. Composition and evolutionary importance of transposable elements in humans and primates. Genes Genom. 2014, 37, 135–140. [Google Scholar] [CrossRef]
- Feschotte, C.; Pritham, E.J. DNA transposons and the evolution of eukaryotic genomes. In Annual Review of Genetics; Annual Reviews: Palo Alto, CA, USA, 2007; Volume 41, pp. 331–368. [Google Scholar]
- Finnegan, D.J. Eukaryotic transposable elements and genome evolution. Trends Genet. 1989, 5, 103–107. [Google Scholar] [CrossRef]
- Biemont, C.; Vieira, C. Genetics: Junk DNA as an evolutionary force. Nature 2006, 443, 521–524. [Google Scholar] [CrossRef]
- Deininger, P.L.; Moran, J.V.; Batzer, M.A.; Kazazian, H.H., Jr. Mobile elements and mammalian genome evolution. Curr. Opin. Genet. Dev. 2003, 13, 651–658. [Google Scholar] [CrossRef]
- Callinan, P.A.; Batzer, M.A. Retrotransposable elements and human disease. Genome Dyn. 2006, 1, 104–115. [Google Scholar] [CrossRef]
- Bourque, G.; Burns, K.H.; Gehring, M.; Gorbunova, V.; Seluanov, A.; Hammell, M.; Imbeault, M.; Izsvak, Z.; Levin, H.L.; Macfarlan, T.S.; et al. Ten things you should know about transposable elements. Genome Biol. 2018, 19. [Google Scholar] [CrossRef]
- Ayarpadikannan, S.; Lee, H.E.; Han, K.; Kim, H.S. Transposable element-driven transcript diversification and its relevance to genetic disorders. Gene 2015, 558, 187–194. [Google Scholar] [CrossRef]
- Kazazian, H.H., Jr.; Moran, J.V. The impact of L1 retrotransposons on the human genome. Nat. Genet. 1998, 19, 19–24. [Google Scholar] [CrossRef] [PubMed]
- Cordaux, R.; Batzer, M.A. The impact of retrotransposons on human genome evolution. Nat. Rev. Genet. 2009, 10, 691–703. [Google Scholar] [CrossRef] [PubMed]
- Smit, A.; Hubley, R.; Green, P. RepeatMasker. Available online: http://repeatmasker.org (accessed on 1 October 2019).
- Wang, H.; Xing, J.; Grover, D.; Hedges, D.J.; Han, K.; Walker, J.A.; Batzer, M.A. SVA elements: A hominid-specific retroposon family. J. Mol. Biol. 2005, 354, 994–1007. [Google Scholar] [CrossRef] [PubMed]
- Mills, R.E.; Bennett, E.A.; Iskow, R.C.; Devine, S.E. Which transposable elements are active in the human genome? Trends Genet. 2007, 23, 183–191. [Google Scholar] [CrossRef] [PubMed]
- Jang, H.S.; Shah, N.M.; Du, A.Y.; Dailey, Z.Z.; Pehrsson, E.C.; Godoy, P.M.; Zhang, D.; Li, D.; Xing, X.; Kim, S.; et al. Transposable elements drive widespread expression of oncogenes in human cancers. Nat. Genet. 2019, 51, 611–617. [Google Scholar] [CrossRef]
- Burns, K.H. Transposable elements in cancer. Nat. Rev. Cancer 2017, 17, 415–424. [Google Scholar] [CrossRef]
- Anwar, S.L.; Wulaningsih, W.; Lehmann, U. Transposable Elements in Human Cancer: Causes and Consequences of Deregulation. Int. J. Mol. Sci 2017, 18, 974. [Google Scholar] [CrossRef]
- Transposable Elements Regulate Oncogene Expression in Human Cancers. Cancer Discov. 2019, 9, 689. [CrossRef]
- Babaian, A.; Mager, D.L. Endogenous retroviral promoter exaptation in human cancer. Mob. DNA 2016, 7, 24. [Google Scholar] [CrossRef]
- Kim, Y.J.; Jung, Y.D.; Kim, T.O.; Kim, H.S. Alu-related transcript of TJP2 gene as a marker for colorectal cancer. Gene 2013, 524, 268–274. [Google Scholar] [CrossRef] [PubMed]
- Han, K.; Lee, J.; Km, H.S.; Yang, K.; Yi, J.M. DNA methylation of mobile genetic elements in human cancers. Genes Genom. 2013, 35, 265–271. [Google Scholar] [CrossRef]
- Lee, S.H.; Kang, Y.J.; Jo, J.O.; Ock, M.S.; Baek, K.W.; Eo, J.; Lee, W.J.; Choi, Y.H.; Kim, W.J.; Leem, S.H.; et al. Elevation of human ERV3-1 env protein expression in colorectal cancer. J. Clin. Pathol. 2014, 67, 840–844. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.R.; Jung, Y.D.; Kim, Y.J.; Lee, H.E.; Jeong, H.; Kim, H.S. Identification of L1ASP-derived chimeric transcripts in lung cancer. Genes Genom. 2014, 36, 853–859. [Google Scholar] [CrossRef]
- Yi, J.M.; Kim, H.M.; Kim, H.S. Human endogenous retrovirus HERV-H family in human tissues and cancer cells: Expression, identification, and phylogeny. Cancer Lett. 2006, 231, 228–239. [Google Scholar] [CrossRef]
- Sin, H.S.; Huh, J.W.; Kim, D.S.; Kang, D.W.; Min, D.S.; Kim, T.H.; Ha, H.S.; Kim, H.H.; Lee, S.Y.; Kim, H.S. Transcriptional control of the HERV-H LTR element of the GSDML gene in human tissues and cancer cells. Arch. Virol. 2006, 151, 1985–1994. [Google Scholar] [CrossRef]
- Yi, J.M.; Schuebel, K.; Kim, H.S. Molecular genetic analyses of human endogenous retroviral elements belonging to the HERV-P family in primates, human tissues, and cancer cells. Genomics 2007, 89, 1–9. [Google Scholar] [CrossRef]
- Roberts, J.T.; Cardin, S.E.; Borchert, G.M. Burgeoning evidence indicates that microRNAs were initially formed from transposable element sequences. Mob. Genet. Elem. 2014, 4, e29255. [Google Scholar] [CrossRef]
- Piriyapongsa, J.; Jordan, I.K. A family of human microRNA genes from miniature inverted-repeat transposable elements. PLoS ONE 2007, 2, e203. [Google Scholar] [CrossRef]
- Ugolini, I.; Halic, M. Fidelity in RNA-based recognition of transposable elements. Philos. Trans. R Soc. Lond B Biol. Sci. 2018, 373. [Google Scholar] [CrossRef]
- Smalheiser, N.R.; Torvik, V.I. Mammalian microRNAs derived from genomic repeats. Trends Genet. 2005, 21, 322–326. [Google Scholar] [CrossRef]
- Piriyapongsa, J.; Marino-Ramirez, L.; Jordan, I.K. Origin and evolution of human microRNAs from transposable elements. Genetics 2007, 176, 1323–1337. [Google Scholar] [CrossRef] [PubMed]
- Lippman, Z.; Gendrel, A.V.; Black, M.; Vaughn, M.W.; Dedhia, N.; McCombie, W.R.; Lavine, K.; Mittal, V.; May, B.; Kasschau, K.D.; et al. Role of transposable elements in heterochromatin and epigenetic control. Nature 2004, 430, 471–476. [Google Scholar] [CrossRef]
- Qin, S.; Jin, P.; Zhou, X.; Chen, L.; Ma, F. The Role of Transposable Elements in the Origin and Evolution of MicroRNAs in Human. PLoS ONE 2015, 10, e0131365. [Google Scholar] [CrossRef]
- McCue, A.D.; Slotkin, R.K. Transposable element small RNAs as regulators of gene expression. Trends Genet. 2012, 28, 616–623. [Google Scholar] [CrossRef] [PubMed]
- Feschotte, C. Transposable elements and the evolution of regulatory networks. Nat. Rev. Genet. 2008, 9, 397–405. [Google Scholar] [CrossRef]
- Lambert, S.A.; Jolma, A.; Campitelli, L.F.; Das, P.K.; Yin, Y.; Albu, M.; Chen, X.; Taipale, J.; Hughes, T.R.; Weirauch, M.T. The Human Transcription Factors. Cell 2018, 172, 650–665. [Google Scholar] [CrossRef] [PubMed]
- Zemojtel, T.; Vingron, M. P53 binding sites in transposons. Front. Genet. 2012, 3, 40. [Google Scholar] [CrossRef]
- Kunarso, G.; Chia, N.Y.; Jeyakani, J.; Hwang, C.; Lu, X.; Chan, Y.S.; Ng, H.H.; Bourque, G. Transposable elements have rewired the core regulatory network of human embryonic stem cells. Nat. Genet. 2010, 42, 631–634. [Google Scholar] [CrossRef]
- Ambros, V.; Bartel, B.; Bartel, D.P.; Burge, C.B.; Carrington, J.C.; Chen, X.; Dreyfuss, G.; Eddy, S.R.; Griffiths-Jones, S.; Marshall, M.; et al. A uniform system for microRNA annotation. RNA 2003, 9, 277–279. [Google Scholar] [CrossRef]
- Bartel, D.P. MicroRNAs: Genomics, biogenesis, mechanism, and function. Cell 2004, 116, 281–297. [Google Scholar] [CrossRef]
- Kim, V.N. MicroRNA biogenesis: Coordinated cropping and dicing. Nat. Rev. Mol. Cell Biol. 2005, 6, 376–385. [Google Scholar] [CrossRef] [PubMed]
- Yates, L.A.; Norbury, C.J.; Gilbert, R.J.C. The Long and Short of MicroRNA. Cell 2013, 153, 516–519. [Google Scholar] [CrossRef] [PubMed]
- Jin, D.; Lee, H. Prioritizing cancer-related microRNAs by integrating microRNA and mRNA datasets. Sci. Rep. 2016, 6, 35350. [Google Scholar] [CrossRef] [PubMed]
- Schulte, J.H.; Horn, S.; Otto, T.; Samans, B.; Heukamp, L.C.; Eilers, U.C.; Krause, M.; Astrahantseff, K.; Klein-Hitpass, L.; Buettner, R.; et al. MYCN regulates oncogenic MicroRNAs in neuroblastoma. Int. J. Cancer 2008, 122, 699–704. [Google Scholar] [CrossRef] [PubMed]
- Lee, Y.S.; Dutta, A. MicroRNAs in cancer. Annu. Rev. Pathol. 2009, 4, 199–227. [Google Scholar] [CrossRef]
- Pinto, Y.; Buchumenski, I.; Levanon, E.Y.; Eisenberg, E. Human cancer tissues exhibit reduced A-to-I editing of miRNAs coupled with elevated editing of their targets. Nucleic Acids Res. 2018, 46, 71–82. [Google Scholar] [CrossRef]
- Zhong, Y.; Zhu, F.; Ding, Y. Differential microRNA expression profile in the plasma of preeclampsia and normal pregnancies. Exp. Ther. Med. 2019, 18, 826–832. [Google Scholar] [CrossRef]
- Murakami, Y.; Yasuda, T.; Saigo, K.; Urashima, T.; Toyoda, H.; Okanoue, T.; Shimotohno, K. Comprehensive analysis of microRNA expression patterns in hepatocellular carcinoma and non-tumorous tissues. Oncogene 2006, 25, 2537–2545. [Google Scholar] [CrossRef]
- Wang, H.; Peng, R.; Wang, J.; Qin, Z.; Xue, L. Circulating microRNAs as potential cancer biomarkers: The advantage and disadvantage. Clin. Epigenet. 2018, 10, 59. [Google Scholar] [CrossRef]
- McCreight, J.C.; Schneider, S.E.; Wilburn, D.B.; Swanson, W.J. Evolution of microRNA in primates. PLoS ONE 2017, 12, e0176596. [Google Scholar] [CrossRef] [PubMed]
- Guerra-Assuncao, J.A.; Enright, A.J. Large-scale analysis of microRNA evolution. BMC Genom. 2012, 13, 218. [Google Scholar] [CrossRef] [PubMed]
- Teferedegne, B.; Murata, H.; Quinones, M.; Peden, K.; Lewis, A.M. Patterns of microRNA expression in non-human primate cells correlate with neoplastic development in vitro. PLoS ONE 2010, 5, e14416. [Google Scholar] [CrossRef] [PubMed]
- Awan, H.M.; Shah, A.; Rashid, F.; Shan, G. Primate-specific Long Non-coding RNAs and MicroRNAs. Genom. Proteom. Bioinform. 2017, 15, 187–195. [Google Scholar] [CrossRef]
- Robles, A.I.; Harris, C.C. A primate-specific microRNA enters the lung cancer landscape. Proc. Natl. Acad. Sci. USA 2013, 110, 18748–18749. [Google Scholar] [CrossRef] [PubMed]
- Abrusan, G.; Grundmann, N.; DeMester, L.; Makalowski, W. TEclass-a tool for automated classification of unknown eukaryotic transposable elements. Bioinformatics 2009, 25, 1329–1330. [Google Scholar] [CrossRef]
- Wicker, T.; Matthews, D.E.; Keller, B. TREP: A database for Triticeae repetitive elements. Trends Plant Sci. 2002, 7, 561–562. [Google Scholar] [CrossRef]
- Casper, J.; Zweig, A.S.; Villarreal, C.; Tyner, C.; Speir, M.L.; Rosenbloom, K.R.; Raney, B.J.; Lee, C.M.; Lee, B.T.; Karolchik, D.; et al. The UCSC Genome Browser database: 2018 update. Nucleic Acids Res. 2018, 46, D762–D769. [Google Scholar] [CrossRef]
- Jurka, J.; Kapitonov, V.V.; Pavlicek, A.; Klonowski, P.; Kohany, O.; Walichiewicz, J. Repbase Update, a database of eukaryotic repetitive elements. Cytogenet. Genome Res. 2005, 110, 462–467. [Google Scholar] [CrossRef]
- An, J.; Lai, J.; Lehman, M.L.; Nelson, C.C. miRDeep*: An integrated application tool for miRNA identification from RNA sequencing data. Nucleic Acids Res. 2013, 41, 727–737. [Google Scholar] [CrossRef]
- Liu, B.; Li, J.; Cairns, M.J. Identifying miRNAs, targets and functions. Brief. Bioinform. 2014, 15, 1–19. [Google Scholar] [CrossRef] [PubMed]
- Agarwal, V.; Bell, G.W.; Nam, J.W.; Bartel, D.P. Predicting effective microRNA target sites in mammalian mRNAs. Elife 2015, 4. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.; Heikkinen, L.; Wang, C.; Yang, Y.; Sun, H.; Wong, G. Trends in the development of miRNA bioinformatics tools. Brief. Bioinform. 2018, 20, 1836–1852. [Google Scholar] [CrossRef] [PubMed]
- Kozomara, A.; Birgaoanu, M.; Griffiths-Jones, S. miRBase: From microRNA sequences to function. Nucleic Acids Res. 2019, 47, D155–D162. [Google Scholar] [CrossRef]
- Tong, Z.; Cui, Q.; Wang, J.; Zhou, Y. TransmiR v2.0: An updated transcription factor-microRNA regulation database. Nucleic Acids Res. 2019, 47, D253–D258. [Google Scholar] [CrossRef] [PubMed]
- National Center for Biotechnology Information-PubMed. Available online: https://www.ncbi.nlm.nih.gov/pubmed/ (accessed on 20 May 2020).
- Google Scholar. Available online: https://scholar.google.co.kr/ (accessed on 20 May 2020).
- Zhang, R.; Wang, Y.Q.; Su, B. Molecular evolution of a primate-specific microRNA family. Mol. Biol. Evol. 2008, 25, 1493–1502. [Google Scholar] [CrossRef]
- Nguyen, P.N.; Huang, C.J.; Sugii, S.; Cheong, S.K.; Choo, K.B. Selective activation of miRNAs of the primate-specific chromosome 19 miRNA cluster (C19MC) in cancer and stem cells and possible contribution to regulation of apoptosis. J. Biomed. Sci. 2017, 24, 20. [Google Scholar] [CrossRef]
- Ovcharenko, I.; Nobrega, M.A.; Loots, G.G.; Stubbs, L. ECR Browser: A tool for visualizing and accessing data from comparisons of multiple vertebrate genomes. Nucleic Acids Res. 2004, 32, W280–W286. [Google Scholar] [CrossRef]
- Hofacker, I.L. Vienna RNA secondary structure server. Nucleic Acids Res. 2003, 31, 3429–3431. [Google Scholar] [CrossRef]
- Zhang, L.; Huang, J.; Yang, N.; Greshock, J.; Megraw, M.S.; Giannakakis, A.; Liang, S.; Naylor, T.L.; Barchetti, A.; Ward, M.R.; et al. microRNAs exhibit high frequency genomic alterations in human cancer. Proc. Natl. Acad. Sci. USA 2006, 103, 9136–9141. [Google Scholar] [CrossRef]
- Kutay, H.; Bai, S.; Datta, J.; Motiwala, T.; Pogribny, I.; Frankel, W.; Jacob, S.T.; Ghoshal, K. Downregulation of miR-122 in the rodent and human hepatocellular carcinomas. J. Cell Biochem. 2006, 99, 671–678. [Google Scholar] [CrossRef]
- Hayashita, Y.; Osada, H.; Tatematsu, Y.; Yamada, H.; Yanagisawa, K.; Tomida, S.; Yatabe, Y.; Kawahara, K.; Sekido, Y.; Takahashi, T. A polycistronic microRNA cluster, miR-17-92, is overexpressed in human lung cancers and enhances cell proliferation. Cancer Res. 2005, 65, 9628–9632. [Google Scholar] [CrossRef] [PubMed]
- Lal, A.; Kim, H.H.; Abdelmohsen, K.; Kuwano, Y.; Pullmann, R., Jr.; Srikantan, S.; Subrahmanyam, R.; Martindale, J.L.; Yang, X.; Ahmed, F.; et al. p16(INK4a) translation suppressed by miR-24. PLoS ONE 2008, 3, e1864. [Google Scholar] [CrossRef] [PubMed]
- Roldo, C.; Missiaglia, E.; Hagan, J.P.; Falconi, M.; Capelli, P.; Bersani, S.; Calin, G.A.; Volinia, S.; Liu, C.G.; Scarpa, A.; et al. MicroRNA expression abnormalities in pancreatic endocrine and acinar tumors are associated with distinctive pathologic features and clinical behavior. J. Clin. Oncol. 2006, 24, 4677–4684. [Google Scholar] [CrossRef]
- Du, B.; Wang, Z.; Zhang, X.; Feng, S.; Wang, G.; He, J.; Zhang, B. MicroRNA-545 suppresses cell proliferation by targeting cyclin D1 and CDK4 in lung cancer cells. PLoS ONE 2014, 9, e88022. [Google Scholar] [CrossRef]
- Rasmussen, M.H.; Jensen, N.F.; Tarpgaard, L.S.; Qvortrup, C.; Romer, M.U.; Stenvang, J.; Hansen, T.P.; Christensen, L.L.; Lindebjerg, J.; Hansen, F.; et al. High expression of microRNA-625-3p is associated with poor response to first-line oxaliplatin based treatment of metastatic colorectal cancer. Mol. Oncol. 2013, 7, 637–646. [Google Scholar] [CrossRef] [PubMed]
- Fang, W.; Fan, Y.; Fa, Z.; Xu, J.; Yu, H.; Li, P.; Gu, J. microRNA-625 inhibits tumorigenicity by suppressing proliferation, migration and invasion in malignant melanoma. Oncotarget 2017, 8, 13253–13263. [Google Scholar] [CrossRef]
- Lin, L.; Cai, Q.; Zhang, X.; Zhang, H.; Zhong, Y.; Xu, C.; Li, Y. Two less common human microRNAs miR-875 and miR-3144 target a conserved site of E6 oncogene in most high-risk human papillomavirus subtypes. Prot. Cell 2015, 6, 575–588. [Google Scholar] [CrossRef]
- Wang, Y.; Xu, X.; Yu, S.; Jeong, K.J.; Zhou, Z.; Han, L.; Tsang, Y.H.; Li, J.; Chen, H.; Mangala, L.S.; et al. Systematic characterization of A-to-I RNA editing hotspots in microRNAs across human cancers. Genome Res. 2017, 27, 1112–1125. [Google Scholar] [CrossRef] [PubMed]
- Suh, A.; Churakov, G.; Ramakodi, M.P.; Platt, R.N., 2nd; Jurka, J.; Kojima, K.K.; Caballero, J.; Smit, A.F.; Vliet, K.A.; Hoffmann, F.G.; et al. Multiple lineages of ancient CR1 retroposons shaped the early genome evolution of amniotes. Genome Biol. Evol. 2014, 7, 205–217. [Google Scholar] [CrossRef]
- Sundaram, V.; Choudhary, M.N.; Pehrsson, E.; Xing, X.; Fiore, C.; Pandey, M.; Maricque, B.; Udawatta, M.; Ngo, D.; Chen, Y.; et al. Functional cis-regulatory modules encoded by mouse-specific endogenous retrovirus. Nat. Commun. 2017, 8, 14550. [Google Scholar] [CrossRef] [PubMed]
- Subramanian, R.P.; Wildschutte, J.H.; Russo, C.; Coffin, J.M. Identification, characterization, and comparative genomic distribution of the HERV-K (HML-2) group of human endogenous retroviruses. Retrovirology 2011, 8, 90. [Google Scholar] [CrossRef] [PubMed]
- Thompson, P.J.; Macfarlan, T.S.; Lorincz, M.C. Long Terminal Repeats: From Parasitic Elements to Building Blocks of the Transcriptional Regulatory Repertoire. Mol. Cell 2016, 62, 766–776. [Google Scholar] [CrossRef] [PubMed]
- Perdomo, C.; Campbell, J.D.; Gerrein, J.; Tellez, C.S.; Garrison, C.B.; Walser, T.C.; Drizik, E.; Si, H.Q.; Gower, A.C.; Vick, J.; et al. MicroRNA 4423 is a primate-specific regulator of airway epithelial cell differentiation and lung carcinogenesis. Proc. Natl. Acad. Sci. USA 2013, 110, 18946–18951. [Google Scholar] [CrossRef]
- Jo, A.; Im, J.; Lee, H.E.; Jang, D.; Nam, G.H.; Mishra, A.; Kim, W.J.; Kim, W.; Cha, H.J.; Kim, H.S. Evolutionary conservation and expression of miR-10a-3p in olive flounder and rock bream. Gene 2017, 628, 16–23. [Google Scholar] [CrossRef]
- Jo, A.; Lee, H.E.; Kim, H.S. Genomic Analysis of miR-21-3p and Expression Pattern with Target Gene in Olive Flounder. Genom. Inform. 2017, 15, 98–107. [Google Scholar] [CrossRef]
- Liu, S.; Zhang, F.; Shugart, Y.Y.; Yang, L.; Li, X.; Liu, Z.; Sun, N.; Yang, C.; Guo, X.; Shi, J.; et al. The early growth response protein 1-miR-30a-5p-neurogenic differentiation factor 1 axis as a novel biomarker for schizophrenia diagnosis and treatment monitoring. Transl. Psychiatry 2017, 7, e998. [Google Scholar] [CrossRef]
- Lin, C.C.; Jiang, W.; Mitra, R.; Cheng, F.; Yu, H.; Zhao, Z. Regulation rewiring analysis reveals mutual regulation between STAT1 and miR-155-5p in tumor immunosurveillance in seven major cancers. Sci. Rep. 2015, 5, 12063. [Google Scholar] [CrossRef]
- Lwin, T.; Zhao, X.; Cheng, F.; Zhang, X.; Huang, A.; Shah, B.; Zhang, Y.; Moscinski, L.C.; Choi, Y.S.; Kozikowski, A.P.; et al. A microenvironment-mediated c-Myc/miR-548m/HDAC6 amplification loop in non-Hodgkin B cell lymphomas. J. Clin. Investig. 2013, 123, 4612–4626. [Google Scholar] [CrossRef]
- Lee, H.E.; Jo, A.; Im, J.; Cha, H.J.; Kim, W.J.; Kim, H.H.; Kim, D.S.; Kim, W.; Yang, T.J.; Kim, H.S. Characterization of the Long Terminal Repeat of the Endogenous Retrovirus-derived microRNAs in the Olive Flounder. Sci. Rep. 2019, 9, 14007. [Google Scholar] [CrossRef]
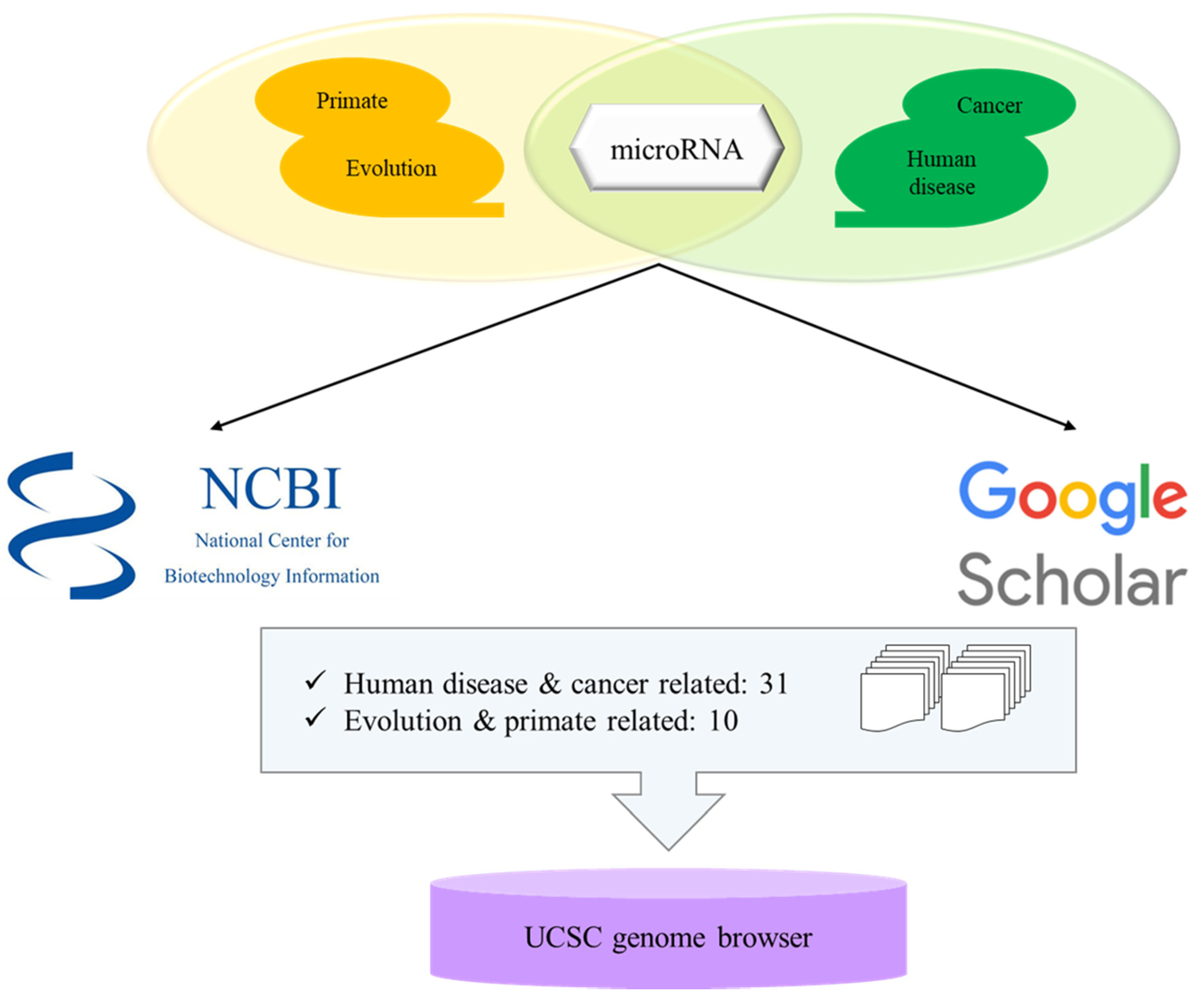
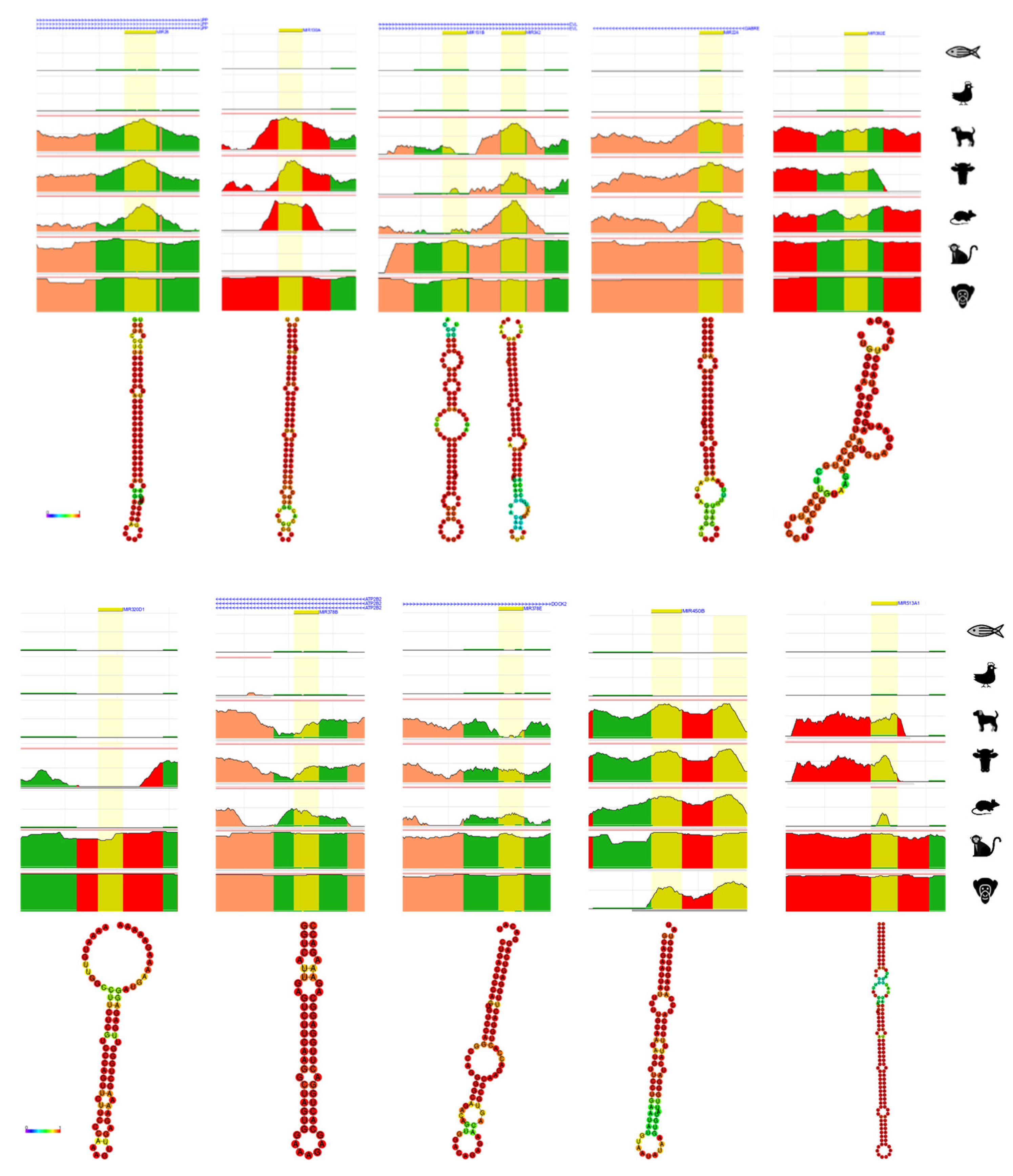
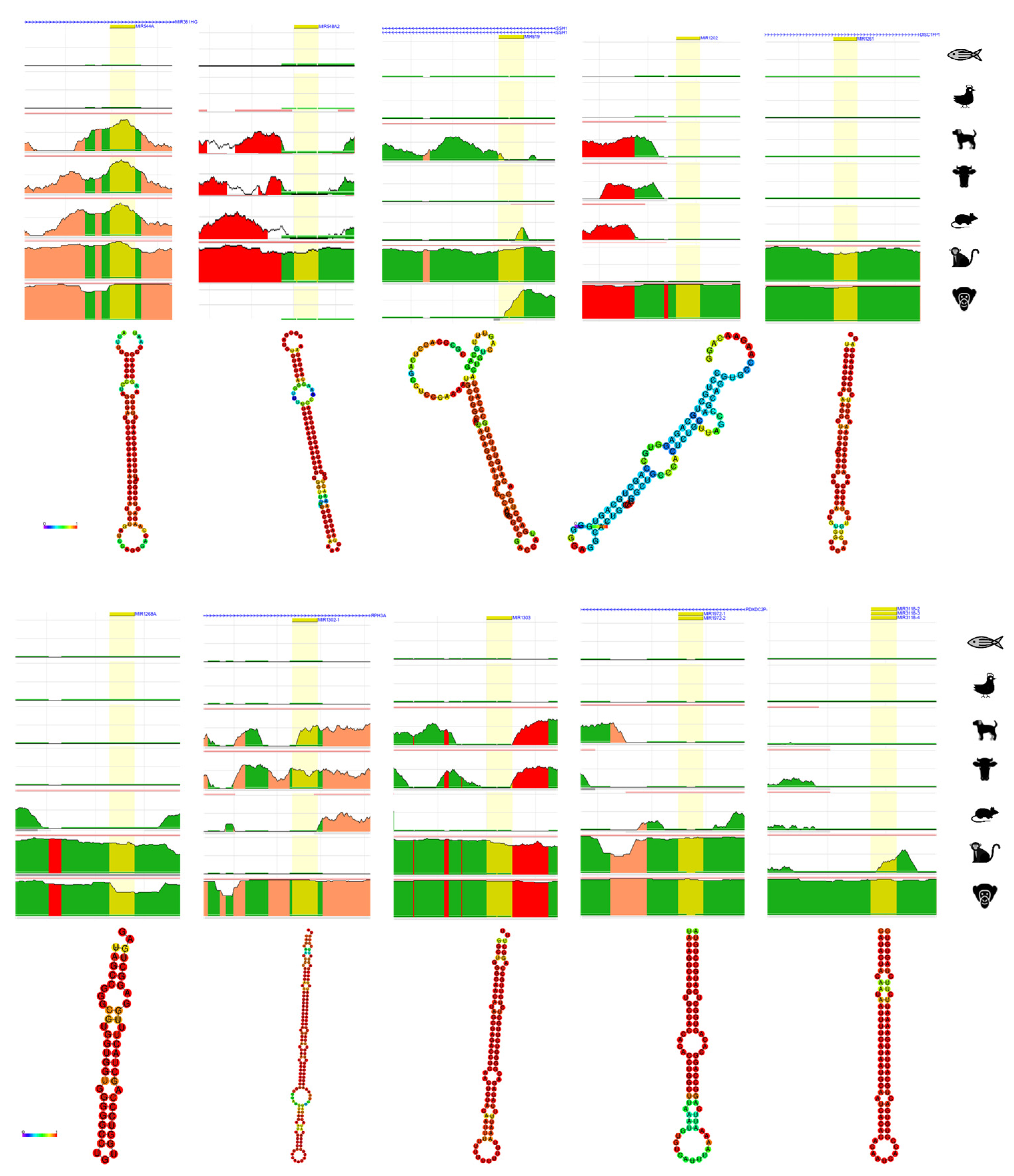
| Human | Chimpanzee | Gorilla | Orangutan | Gibbon | Macaque | Rhesus | Marmoset | Mouse | Horse | Cow | Cat | Dog | Chicken | Zebrafish | C. elegans | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Non-TEs | 47.5% | 49.2% | 50.6% | 47.9% | 48.1% | 48.5% | 50.7% | 51.0% | 55.0% | 54.8% | 50.6% | 56.3% | 56.8% | 88.8% | 47.4% | 87.4% |
| SINE/MIR | 2.9% | 2.9% | 3.0% | 2.9% | 2.9% | 2.9% | 3.1% | 2.7% | 0.6% | 3.8% | 2.3% | 3.1% | 2.9% | 0.0% | 0.0% | 0.0% |
| SINE/Alu | 10.5% | 10.3% | 8.8% | 10.0% | 10.4% | 11.0% | 10.5% | 11.0% | 4.8% | 0.0% | 0.0% | 0.0% | 0.0% | 0.0% | 0.0% | 0.0% |
| SINE/other | 0.0% | 0.0% | 0.0% | 0.0% | 0.0% | 0.0% | 0.0% | 0.0% | 2.4% | 3.7% | 9.4% | 8.3% | 7.6% | 0.0% | 2.4% | 0.0% |
| SVA | 0.1% | 0.0% | 0.0% | 0.0% | 0.0% | 0.0% | 0.0% | 0.0% | 0.0% | 0.0% | 0.0% | 0.0% | 0.0% | 0.0% | 0.0% | 0.0% |
| LINE/L2 | 3.7% | 3.8% | 3.8% | 3.7% | 3.8% | 3.8% | 4.0% | 3.3% | 0.4% | 5.2% | 2.7% | 4.0% | 3.7% | 0.0% | 1.7% | 0.0% |
| LINE/CR1 | 0.4% | 0.4% | 0.4% | 0.4% | 0.4% | 0.4% | 0.4% | 0.4% | 0.0% | 0.6% | 0.3% | 0.4% | 0.4% | 6.8% | 0.0% | 0.3% |
| LINE/RTE | 0.2% | 0.2% | 0.2% | 0.2% | 0.2% | 0.2% | 0.2% | 0.1% | 0.0% | 0.3% | 12.9% | 0.2% | 0.2% | 0.0% | 0.2% | 0.0% |
| LINE/L1 | 17.5% | 17.7% | 16.8% | 18.3% | 17.4% | 17.3% | 15.5% | 18.8% | 19.9% | 18.0% | 13.3% | 17.0% | 16.8% | 0.0% | 0.4% | 0.0% |
| LINE/other | 0.0% | 0.0% | 0.0% | 0.0% | 0.0% | 0.0% | 0.0% | 0.0% | 0.0% | 0.0% | 0.0% | 0.0% | 0.0% | 0.0% | 0.5% | 0.0% |
| LTR/ERVK | 0.3% | 0.3% | 0.3% | 0.3% | 0.2% | 0.4% | 0.3% | 0.0% | 4.9% | 0.0% | 0.5% | 0.0% | 0.0% | 0.0% | 0.0% | 0.0% |
| LTR/ERV1 | 2.9% | 2.8% | 2.7% | 2.9% | 2.6% | 2.7% | 2.8% | 2.2% | 1.2% | 1.9% | 1.4% | 1.0% | 1.0% | 0.2% | 0.4% | 0.0% |
| LTR/ERVL | 5.8% | 5.9% | 5.8% | 5.9% | 5.7% | 5.8% | 6.1% | 5.2% | 5.9% | 5.0% | 2.7% | 4.0% | 3.8% | 1.3% | 0.0% | 0.0% |
| LTR/Gypsy | 0.2% | 0.2% | 0.2% | 0.2% | 0.2% | 0.2% | 0.2% | 0.1% | 0.0% | 0.3% | 0.0% | 0.2% | 0.2% | 0.0% | 1.7% | 0.0% |
| LTR/other | 0.0% | 0.0% | 0.0% | 0.0% | 0.0% | 0.0% | 0.0% | 0.0% | 0.0% | 0.2% | 0.0% | 0.0% | 0.0% | 0.0% | 3.1% | 0.3% |
| DNA/TcMar | 1.5% | 1.5% | 1.5% | 1.5% | 1.5% | 1.5% | 1.6% | 1.3% | 0.2% | 0.8% | 0.6% | 0.8% | 0.7% | 0.3% | 6.1% | 1.8% |
| DNA/hAT | 2.2% | 2.2% | 2.2% | 2.2% | 2.2% | 2.2% | 2.3% | 2.0% | 0.9% | 2.7% | 1.6% | 2.2% | 2.1% | 0.5% | 9.7% | 0.5% |
| DNA/other | 0.0% | 0.0% | 0.0% | 0.0% | 0.0% | 0.0% | 0.0% | 0.0% | 0.0% | 0.0% | 0.0% | 0.0% | 0.0% | 0.2% | 19.6% | 6.1% |
| Other/Unknown | 4.3% | 2.6% | 3.7% | 3.6% | 4.4% | 3.1% | 2.3% | 1.9% | 3.8% | 2.7% | 1.7% | 2.5% | 3.8% | 1.9% | 6.8% | 3.6% |
| TOTAL: 100% | ||||||||||||||||
| microRNA | Coordinates | Type of TE | References | |
|---|---|---|---|---|
| 1 | miR-28 | chr3:188,688,781-188,688,866 | LINE_L2c, L2c | [53,55] |
| 2 | miR-130a | chr11:57,641,198-57,641,286 | LINE_MamRTE1 | [55] |
| 3 | miR-151a | chr8:140,732,564-140,732,653 | LINE_L2c, L2c | [55] |
| 4 | miR-151b | chr14:100,109,419-100,109,514 | LINE_L2b | [53,55] |
| 5 | miR-224 | chrX:151,958,578-151,958,658 | DNA_MER135 | [53] |
| 6 | miR-302e | chr11:7,234,766-7,234,837 | SINE_MIR | [70] |
| 7 | miR-320d-1 | chr13:40,727,816-40,727,887 | LINE_L1MEd | [53,55] |
| 8 | miR-342 | chr14:100,109,655-100,109,753 | SINE_MamSINE1 | [55] |
| 9 | miR-374a | chrX:74,287,286-74,287,357 | LINE_L2c | [53] |
| 10 | miR-378a | chr5:149,732,825-149,732,890 | SINE_MIRc, MIRc | [55] |
| 11 | miR-378b | chr3:10,330,229-10,330,285 | SINE_MIR3, MIRc | [54,55] |
| 12 | miR-378d-1 | chr4:5,923,275-5,923,328 | SINE_MIRb | [54,55] |
| 13 | miR-378d-2 | chr8:93,916,022-93,916,119 | SINE_MIRc | [54,55] |
| 14 | miR-378e | chr5:170,028,488-170,028,566 | SINE_MIRb, MIRc | [55] |
| 15 | miR-378f | chr1:23,929,070-23,929,147 | SINE_MIRc | [54,55] |
| 16 | miR-378h | chr5:154,829,458-154,829,540 | SINE_MIRc | [55] |
| 17 | miR-378i | chr22:41,923,222-41,923,297 | SINE_MIRc | [55] |
| 18 | miR-450b | chrX:134,540,185-134,540,262 | LINE_L1ME4a | [53] |
| 19 | miR-466 | chr3:31,161,704-31,161,787 | LINE_L1ME3 | [54] |
| 20 | miR-513a-1 | chrX:147,213,463-147,213,591 | DNA_MER91C | [54] |
| 21 | miR-513a-2 | chrX:147,225,826-147,225,952 | DNA_MER91C | [54] |
| 22 | miR-513b | chrX:147,199,044-147,199,127 | DNA_MER91C | [54] |
| 23 | miR-513c | chrX:147,189,704-147,189,787 | DNA_MER91C | [54] |
| 24 | miR-518d | chr19:53,734,877-53,734,963 | LINE_MamRTE1 | [54,71] |
| 25 | miR-544a | chr14:101,048,658-101,048,748 | DNA_MER5A1 | [54] |
| 26 | miR-544b | chr3:124,732,439-124,732,516 | DNA_MER5A1 | [54] |
| 27 | miR-548a-1 | chr6:18,571,784-18,571,880 | DNA_MADE1 | [54] |
| 28 | miR-548a-2 | chr6:135,239,160-135,239,256 | LTR_LTR16A2, DNA_MADE1, LTR_LTR16A2 | [54] |
| 29 | miR-570 | chr3:195,699,401-195,699,497 | DNA_MADE1 | [54] |
| 30 | miR-603 | chr10:24,275,685-24,275,781 | DNA_MADE1 | [56] |
| 31 | miR-619 | chr12:108,836,908-108,837,006 | LINE_L1MC4, SINE_AluSz6 | [54] |
| 32 | miR-637 | chr19:3,961,414-3,961,512 | LINE_L1MC4a | [57] |
| 33 | miR-652 | chrX:110,055,329-110,055,426 | DNA_MER91C | [54] |
| 34 | miR-1202 | chr6:155,946,797-155,946,879 | LTR_MER52A | [56] |
| 35 | miR-1261 | chr11:90,869,121-90,869,202 | DNA_Tigger1 | [54] |
| 36 | miR-1268a | chr15:22,225,278-22,225,329 | SINE_AluJo | [54] |
| 37 | miR-1268b | chr17:80,098,828-80,098,877 | SINE_AluSx1 | [54] |
| 38 | miR-1273c | chr6:154,853,360-154,853,436 | SINE_AluJo | [54] |
| 39 | miR-1273h | chr16:24,203,116-24,203,231 | SINE_AluJb | [54] |
| 40 | miR-1302-1 | chr12:112,695,034-112,695,176 | DNA_MER54 | [54] |
| 41 | miR-1303 | chr5:154,685,776-154,685,861 | SINE_AluJr, FLAM_A | [54] |
| 42 | miR-1304 | chr11:93,733,674-93,733,764 | SINE_AluJo | [54] |
| 43 | miR-1587 | chrX:39,837,561-39,837,613 | LTR_MLT1H2 | [54] |
| 44 | miR-1972-1 | chr16:15,010,321-15,010,397 | SINE_AluSx, FLAM_A | [54] |
| 45 | miR-2355 | chr2:207,109,987-207,110,073 | LINE_MamRTE1, MamRTE1 | [53] |
| 46 | miR-3118-1 | chr21:13,644,775-13,644,850 | LINE_L1PA12 | [54] |
| 47 | miR-3118-2 | chr15:20,832,795-20,832,869 | LINE_L1PA14 | [54] |
| 48 | miR-3118-3 | chr15:21,406,385-21,406,459 | LINE_L1PA13 | [54] |
| 49 | miR-3118-4 | chr15:21,843,750-21,843,824 | LINE_L1PA13 | [54] |
| 50 | miR-4452 | chr4:86,542,482-86,542,552 | SINE_AluJo | [54] |
| 51 | miR-6303 | chr10:24,275,685-24,275,781 | DNA_MADE1 | [54] |
| microRNA | Coordinates | Type of TE | Related Disorders | References | |
|---|---|---|---|---|---|
| 1 | miRNA-28 | chr3:188,688,781-188,688,866 | LINE_L2c, L2c | OC, M | [74] |
| 2 | miRNA-95 | chr4:8,005,301-8,005,381 | LINE_L2b, L2c | OC, BrC | [74] |
| 3 | miRNA-130a | chr11:57,641,198-57,641,286 | LINE_MamRTE1 | LuC, OC, LiC | [46,48,75,76] |
| 4 | miRNA-151a | chr8:140,732,564-140,732,653 | LINE_L2c | G, OC, BrC, M | [49,74] |
| 5 | miRNA-151b | chr14:100,109,419-100,109,514 | LINE_L2b | OC, BrC, M | [74] |
| 6 | miRNA-181c | chr19:13,874,699-13,874,808 | LINE_MamRTE1 | LiC, A, OC, BrC | [74,77] |
| 7 | miRNA-224 | chrX: 151,958,578-151,958,658 | DNA_MER135 | LiC | [48,51] |
| 8 | miRNA-320d-1 | chr13:40,727,816-40,727,887 | LINE_L1MEd | OC, BrC, M, N | [47,48,74] |
| 9 | miRNA-335 | chr7:130,496,111-130,496,204 | SINE_MIRb | OC, M | [74] |
| 10 | miRNA-340 | chr5:180,015,303-180,015,397 | DNA_MARNA | OC | [74] |
| 11 | miRNA-342 | chr14:100,109,655-100,109,753 | SINE_MamSINE1 | EC, ACC, PaC | [48,78] |
| 12 | miRNA-361 | chrX: 85,903,636-85,903,707 | DNA_MER5A | LuC | [52] |
| 13 | miRNA-378a | chr5:149,732,825-149,732,890 | SINE_MIRc, MIRc | BlC | [49] |
| 14 | miRNA-378b | chr3:10,330,229-10,330,285 | SINE_MIR3, MIRc | L | [49] |
| 15 | miRNA-421 | chrX:74,218,377-74,218,461 | LINE_L2c, L2c | Ph, Pa | [49] |
| 16 | miRNA-513a-1 | chrX:147,213,463-147,213,591 | DNA_MER91C | LuC | [79] |
| 17 | miRNA-513a-2 | chrX:147,225,826-147,225,952 | DNA_MER91C | LuC | [79] |
| 18 | miRNA-518d | chr19:53,734,877-53,734,963 | LINE_MamRTE1 | LuC | [79] |
| 19 | miRNA-545 | chrX:74,287,104-74,287,209 | LINE_L2c | LuC | [79] |
| 20 | miRNA-546b | chr6:119,069,022-119,069,167 | DNA_MADE1 | LuC | [79] |
| 21 | miRNA-548l | chr11:94,466,495-94,466,580 | DNA_MADE1 | LuC | [79] |
| 22 | miRNA-548m | chrX:95,063,141-95,063,226 | DNA_MADE1 | LuC | [79] |
| 23 | miRNA-625 | chr14:65,471,102-65,471,186 | LINE_L1MCa | M, GC, CoC, LuC, KC | [49,80,81] |
| 24 | miRNA-626 | chr15:41,691,585-41,691,678 | LINE_L1MB8, L1MB8 | HPV | [82] |
| 25 | miRNA-646 | chr20:60,308,474-60,308,567 | LTR_LTR67B | LuC | [79] |
| 26 | miRNA-659 | chr22:37,847,678-37,847,774 | DNA_Arthur1B | D | [77] |
| 27 | miRNA-1290 | chr1:18,897,071-18,897,148 | DNA_Tigger4a | LuC | [79] |
| 28 | miRNA-1294 | chr5:154,347,071-154,347,283 | SINE_MIRb | LuC | [79] |
| 29 | miRNA-2355 | chr2:207,109,987-207,110,073 | LINE_MamRTE1, MamRTE1 | HPV | [82] |
| 30 | miRNA-1304 | chr11:93,733,674-93,733,764 | SINE_AluJo | LuC, Pr, UCEC | [50,83] |
| 31 | miRNA-3144 | chr6:120,015,179-120,015,257 | LINE_L1MA8 | LiC PrC, CeC, HPV | [82,83] |
| 32 | miRNA-3681 | chr2:12,199,130-12,199,201 | LTR_LTR16D1 | CeC, LiC | [49] |
| 33 | miRNA-4662a | chr8:124,821,985-124,822,051 | LINE_L1ME4b | LuC | [49] |
| 34 | miRNA-6503 | chr11:60,209,071-60,209,156 | LINE_MLT1D | LiC, L | [49,83] |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lee, H.-E.; Huh, J.-W.; Kim, H.-S. Bioinformatics Analysis of Evolution and Human Disease Related Transposable Element-Derived microRNAs. Life 2020, 10, 95. https://doi.org/10.3390/life10060095
Lee H-E, Huh J-W, Kim H-S. Bioinformatics Analysis of Evolution and Human Disease Related Transposable Element-Derived microRNAs. Life. 2020; 10(6):95. https://doi.org/10.3390/life10060095
Chicago/Turabian StyleLee, Hee-Eun, Jae-Won Huh, and Heui-Soo Kim. 2020. "Bioinformatics Analysis of Evolution and Human Disease Related Transposable Element-Derived microRNAs" Life 10, no. 6: 95. https://doi.org/10.3390/life10060095
APA StyleLee, H.-E., Huh, J.-W., & Kim, H.-S. (2020). Bioinformatics Analysis of Evolution and Human Disease Related Transposable Element-Derived microRNAs. Life, 10(6), 95. https://doi.org/10.3390/life10060095






