Identification and Validation of Aerobic Adaptation QTLs in Upland Rice
Abstract
1. Introduction
2. Materials and Methods
2.1. Introgression of Aerobic Adaptation from the Upland Rice to Lowland Rice
2.2. Performance of the ILs under Lowland and Aerobic Conditions at Sanya and Menglian
2.3. QTLs Mapping of Traits Based upon the ILs
2.4. Identification of QTLs in the BC4F5 Backcross Inbred Lines
2.5. Planting and Phenotypic Verification of QTLs Near-Isogenic Lines (NILs)
2.6. Fine-Mapping of Targeted QTLs
2.7. Pyramiding Aerobic Adaptation Molecular Modules for Breeding
3. Results
3.1. Performance of the Introgression Lines under Lowland and Rain-Fed Aerobic Condition
3.2. QTL Mapping of Traits Based upon the ILs
3.3. Identification of QTLs in the BC4F5 Backcross Inbred Lines
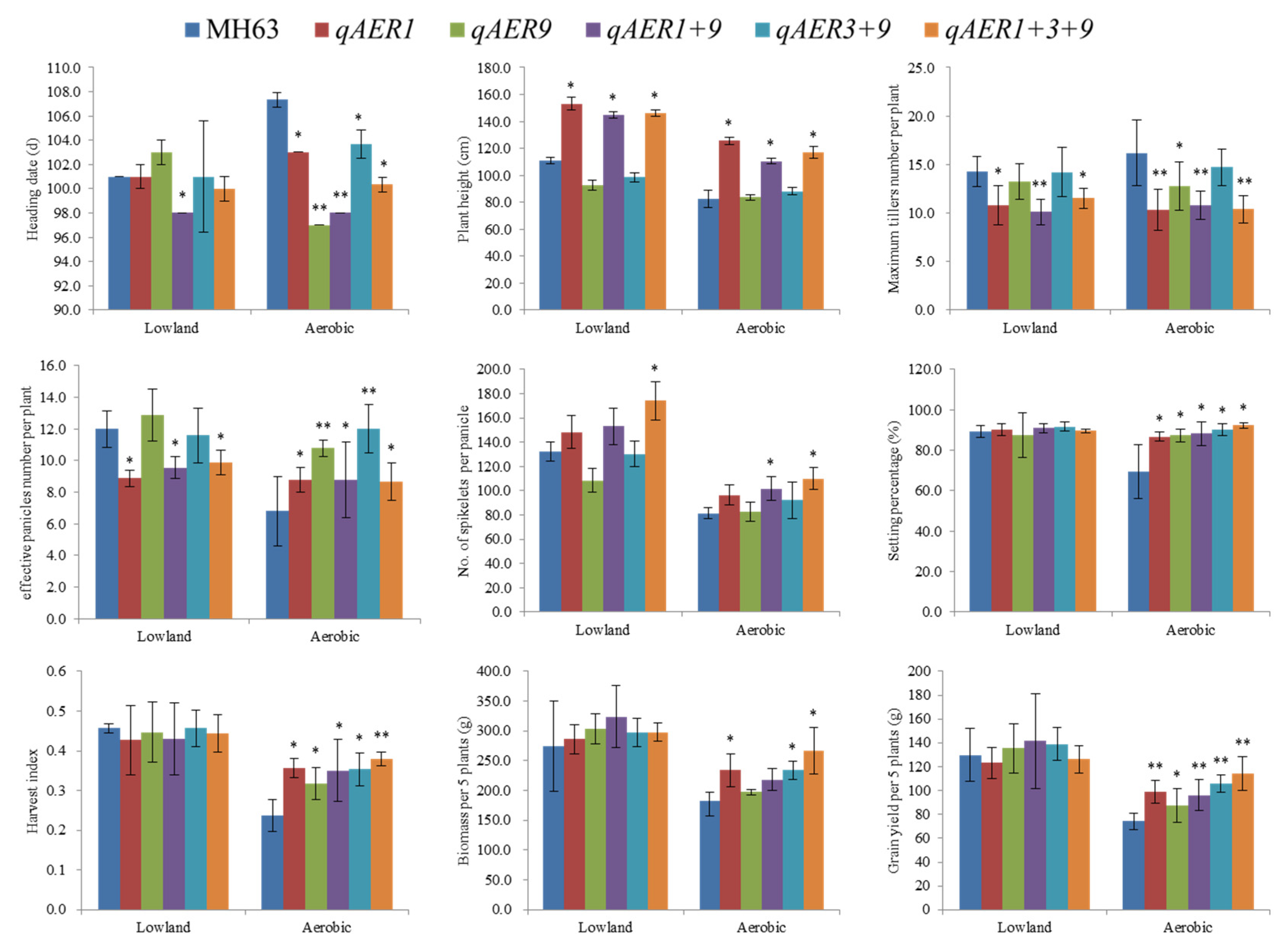
3.4. NILs Planting and Phenotype Evaluation
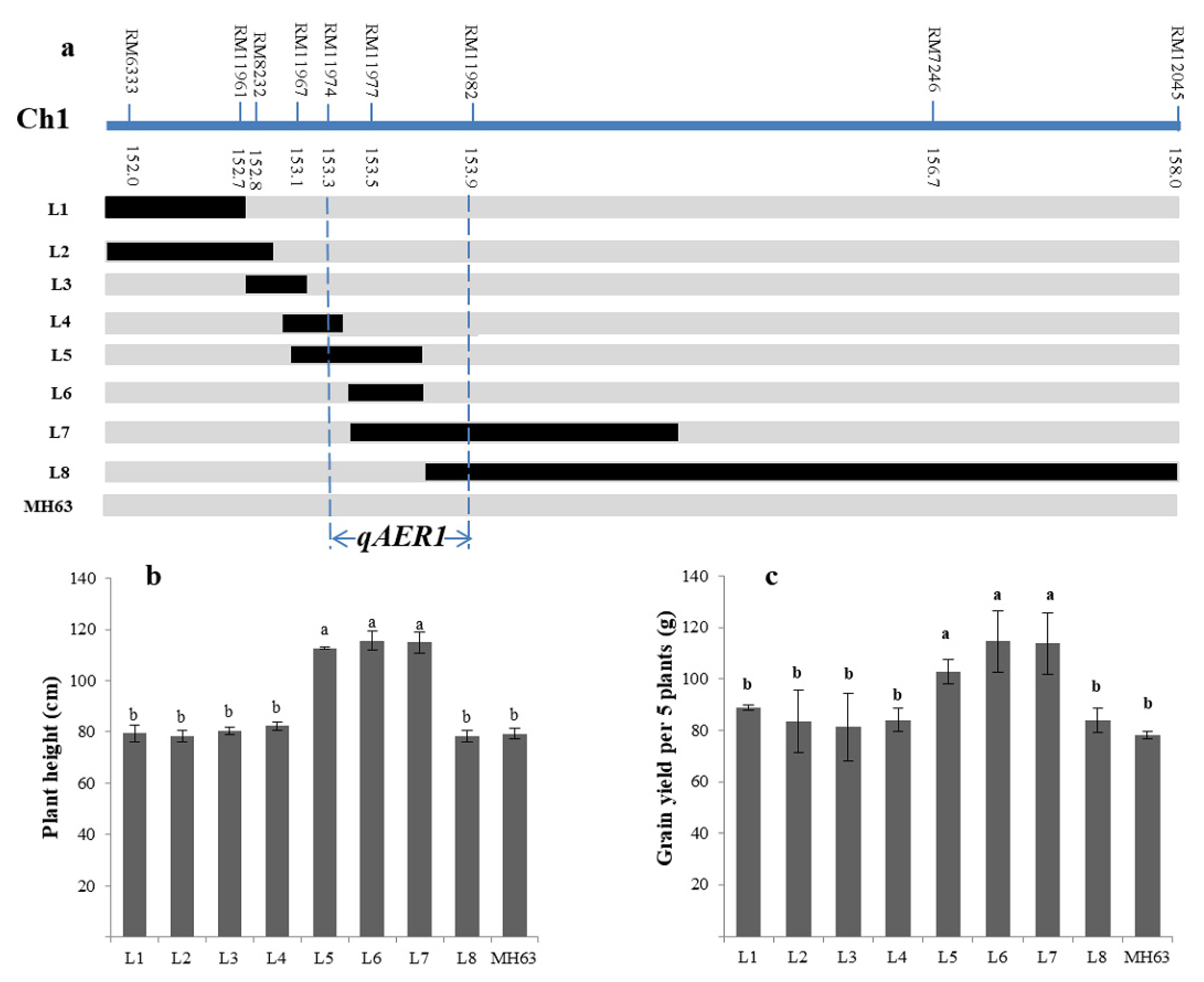
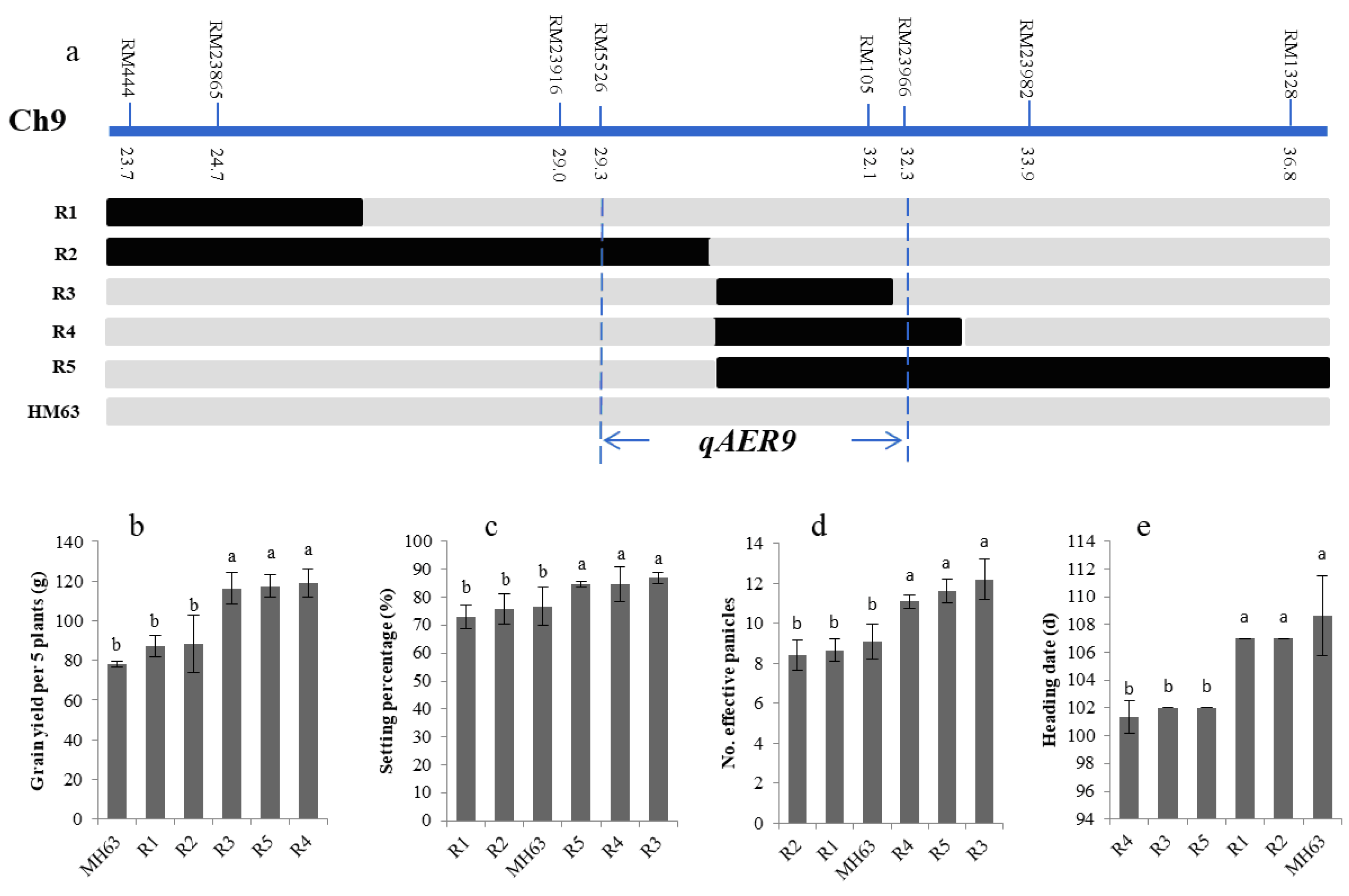
3.5. Fine-Mapping of the Targeted QTLs
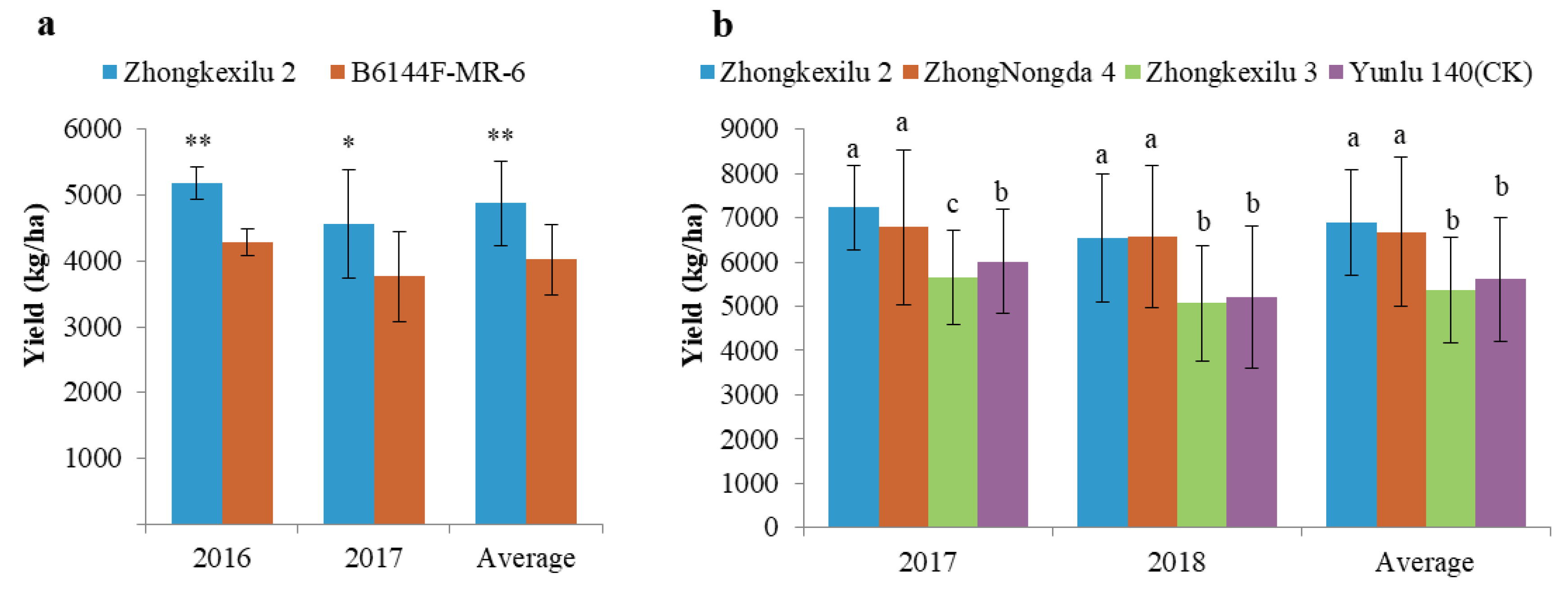
3.6. Pyramiding the Aerobic Adaptation Molecular Modules for Breeding
4. Discussion
4.1. Aerobic Adaptation of Upland Rice
4.2. Phenotype Differences between the Lowland Rice and Upland Rice in the Aerobic Environment
4.3. Does the Green Revolution Gene SD1 Play an Important Role in the Aerobic Adaptation of Upland Rice?
4.4. The Yield of Plants Largely Depends on the Successful Transition from Vegetative Growth to Reproductive Growth
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Hanson, A.D.; Peacock, W.J.; Evans, L.T.; Arntzen, C.J.; Khush, G.S. Drought resistance in rice. Nature 1990, 345, 26–27. [Google Scholar] [CrossRef]
- Eisenstein, M. Plant breeding: Discovery in a dry spell. Nature 2013, 501, S7. [Google Scholar] [CrossRef] [PubMed]
- Bridhikitti, A.; Overcamp, T.J. Estimation of Southeast Asian rice paddy areas with different ecosystems from moderate-resolution satellite imagery. Agric. Ecosyst. Environ. 2012, 146, 113–120. [Google Scholar] [CrossRef]
- Pandey, S.; Khiem, N.T.; Waibel, H.; Thien, T.C. Uplands and upland rice: An overview. In Upland Rice, Household Food Security and Commercialization of Upland Agriculture in Vietnam; Hardy, B., Ed.; International Rice Research Institute: Makati, Philippines, 2006; pp. 1–4. [Google Scholar]
- Sandhu, N.; Jain, S.; Kumar, A.; Mehla, B.S.; Jain, R. Genetic variation, linkage mapping of QTL and correlation studies for yield, root, and agronomic traits for aerobic adaptation. BMC Genet. 2013, 14, 104. [Google Scholar] [CrossRef] [PubMed]
- Paterson, A.H.; Li, Z.K. Paleo-Green Revolution for rice. Proc. Natl. Acad. Sci. USA 2011, 108, 10931–10932. [Google Scholar] [CrossRef] [PubMed]
- Kumar, A.; Verulkar, S.B.; Mandal, N.P.; Variar, M.; Shukla, V.D.; Dwivedi, J.L.; Singh, B.N.; Singh, O.N.; Swain, P.; Mall, A.K. High-yielding, drought-tolerant, stable rice genotypes for the shallow rainfed lowland drought-prone ecosystem. Field Crops Res. 2012, 133, 37–47. [Google Scholar] [CrossRef]
- Lafitte, H.R.; Courtois, B.; Arraudeau, M. Genetic improvement of rice in aerobic systems: Progress from yield to genes. Field Crops Res. 2002, 75, 171–190. [Google Scholar] [CrossRef]
- Vikram, P.; Swamy, B.P.M.; Dixit, S.; Singh, R.; Singh, B.P.; Miro, B.; Kohli, A.; Henry, A.; Singh, N.K.; Kumar, A. Drought susceptibility of modern rice varieties: An effect of linkage of drought tolerance with undesirable traits. Sci. Rep. 2015, 5, 14799. [Google Scholar] [CrossRef]
- Sandhu, N.; Yadaw, R.B.; Chaudhary, B.; Prasai, H.; Iftekharuddaula, K.; Venkateshwarlu, C.; Annamalai, A.; Xangsayasane, P.; Battan, K.R.; Ram, M.; et al. Evaluating the performance of rice genotypes for improving yield and adaptability under direct seeded aerobic cultivation conditions. Front. Plant Sci. 2019, 10, 159. [Google Scholar] [CrossRef]
- Phule, A.S.; Barbadikar, K.M.; Maganti, S.M.; Seguttuvel, P.; Subrahmanyam, D.; Babu, M.B.B.P.; Kumar, P.A. RNA-seq reveals the involvement of key genes for aerobic adaptation in rice. Sci. Rep. 2019, 9, 5235. [Google Scholar] [CrossRef]
- Chang, T.T. The origin, evolution, cultivation, dissemination, and diversification of Asian and African rices. Euphytica 1976, 25, 425–441. [Google Scholar] [CrossRef]
- Carroll, S.B. Evo-devo and an expanding evolutionary synthesis: A genetic theory of morphological evolution. Cell 2008, 134, 25–36. [Google Scholar] [CrossRef] [PubMed]
- Manickavelu, A.; Nadarajan, N.; Ganesh, S.K.; Gnanamalar, R.P.; Babu, R.C. Drought tolerance in rice: Morphological and molecular genetic consideration. Plant Growth Regul. 2006, 50, 121–138. [Google Scholar] [CrossRef]
- Zheng, H.G.; Babu, R.C.; Pathan, M.S.; Ali, L.; Huang, N.; Courtois, B.; Nguyen, H.T. Quantitative trait loci for root-penetration ability and root thickness in rice: Comparison of genetic backgrounds. Genome 2000, 43, 53. [Google Scholar] [CrossRef] [PubMed]
- Shen, L.; Courtois, B.; Mcnally, K.L.; Robin, S.; Li, Z. Evaluation of near-isogenic lines of rice introgressed with QTLs for rootdepth through marker-aided selection. Theor. Appl. Genet. 2001, 103, 75–83. [Google Scholar] [CrossRef]
- Uga, Y.; Okuno, K.; Yano, M. Dro1, a major QTL involved in deep rooting of rice under upland field conditions. J. Exp. Bot. 2011, 62, 2485. [Google Scholar] [CrossRef]
- Zhao, Y.; Zhang, H.; Xu, J.; Jiang, C.; Yin, Z.; Xiong, H.; Xie, J.; Wang, X.; Zhu, X.; Li, Y.; et al. Loci and natural alleles underlying robust roots and adaptive domestication of upland ecotype rice in aerobic conditions. PLoS Genet. 2018, 14, e1007521. [Google Scholar] [CrossRef]
- Cairns, J.E.; Namuco, O.S.; Torres, R.; Simborio, F.A.; Courtois, B.; Aquino, G.A.; Johnson, D.E. Investigating early vigour in upland rice ( Oryza sativa L.): Part II. Identification of QTLs controlling early vigour under greenhouse and field conditions. Field Crops Res. 2009, 113, 207–217. [Google Scholar] [CrossRef]
- Abe, A.; Takagi, H.; Fujibe, T.; Aya, K.; Kojima, M.; Sakakibara, H.; Uemura, A.; Matsuoka, M.; Terauchi, R. OsGA20ox1, a candidate gene for a major QTL controlling seedling vigor in rice. Theor. Appl. Genet. 2012, 125, 647–657. [Google Scholar] [CrossRef]
- Lilley, J.M.; Ludlow, M.M.; Mccouch, S.R.; O’Toole, J.C. Locating QTL for osmotic adjustment and dehydration tolerance in rice. J. Exp. Bot. 1996, 47, 1427–1436. [Google Scholar] [CrossRef]
- Robin, S.; Pathan, M.S.; Courtois, B.; Lafitte, R.; Carandang, S.; Lanceras, S.; Amante, M.; Nguyen, H.T.; Li, Z. Mapping osmotic adjustment in an advanced back-cross inbred population of rice. Theor. Appl. Genet. 2003, 107, 1288–1296. [Google Scholar] [CrossRef] [PubMed]
- Li, J.J.; Li, Y.; Yin, Z.G.; Jiang, J.H.; Zhang, M.H.; Guo, X.; Ye, Z.J.; Zhao, Y.; Xiong, H.Y.; Zhang, Z.Y.; et al. OsASR5 enhances drought tolerance through a stomatal closure pathway associated with ABA and H2O2 signalling in rice. Plant Biotechnol. J. 2017, 15, 183–196. [Google Scholar] [CrossRef] [PubMed]
- Lyu, J.; Zhang, S.L.; Yang, D.; He, W.M.; Jing, Z.; Deng, X.N.; Zhang, Y.S.; Xin, L.; Li, B.Y.; Huang, W.Q.; et al. Analysis of elite variety tag SNPs reveals an important allele in upland rice. Nat. Commun. 2013, 4, 2138. [Google Scholar] [CrossRef]
- Li, J.Z.; Han, Y.C.; Liu, L.; Chen, Y.P.; Du, Y.X.; Zhang, J.; Sun, H.Z.; Zhao, Q.Z. qRT9, a quantitative trait locus controlling root thickness and root length in upland rice. J. Exp. Bot. 2015, 66, 2723. [Google Scholar] [CrossRef]
- Jeong, J.S.; Kim, Y.S.; Baek, K.H.; Jung, H.; Ha, S.H.; Do Choi, Y.; Kim, M.; Reuzeau, C.; Kim, J.K. Root-specific expression of OsNAC10 improves drought tolerance and grain yield in rice under field drought conditions. Plant Physiol. 2010, 153, 185–197. [Google Scholar] [CrossRef]
- Uga, Y.; Sugimoto, K.; Ogawa, S.; Rane, J.; Ishitani, M.; Hara, N.; Kitomi, Y.; Inukai, Y.; Ono, K.; Kanno, N.; et al. Control of root system architecture by DEEPER ROOTING 1 increases rice yield under drought conditions. Nat. Genet. 2013, 45, 1097–1102. [Google Scholar] [CrossRef]
- Hanzawa, E.; Sasaki, K.; Nagai, S.; Obara, M.; Fukuta, Y.; Uga, Y.; Miyao, A.; Hirochika, H.; Higashitani, A.; Maekawa, M. Isolation of a novel mutant gene for soil-surface rooting in rice (Oryza sativa L.). Rice 2013, 6, 30. [Google Scholar] [CrossRef]
- Dharmappa, P.M.; Doddaraju, P.; Malagondanahalli, M.V.; Rangappa, R.B.; Mallikarjuna, N.M.; Rajendrareddy, S.H.; Ramanjinappa, R.; Mavinahalli, R.P.; Prasad, T.G.; Udayakumar, M.; et al. Introgression of root and water use efficiency traits enhances water productivity: An evidence for physiological breeding in rice (Oryza sativa L.). Rice 2019, 12, 14. [Google Scholar] [CrossRef]
- Kumar, R.; Venuprasad, R.; Atlin, G.N. Genetic analysis of rainfed lowland rice drought tolerance under naturally-occurring stress in eastern India: Heritability and QTL effects. Field Crops Res. 2007, 103, 42–52. [Google Scholar] [CrossRef]
- Vikram, P.; Swamy, B.M.; Dixit, S.; Ahmed, H.U.; Ma, T.S.C.; Singh, A.K.; Kumar, A. qDTY1.1, a major QTL for rice grain yield under reproductive-stage drought stress with a consistent effect in multiple elite genetic backgrounds. BMC Genet. 2011, 12, 89. [Google Scholar] [CrossRef]
- Venuprasad, R.; Dalid, C.O.; Valle, M.D.; Zhao, D.; Espiritu, M.; Cruz, M.T.S.; Amante, M.; Kumar, A.; Atlin, G.N. Identification and characterization of large-effect quantitative trait loci for grain yield under lowland drought stress in rice using bulk-segregant analysis. Theor Appl. Genet. 2009, 120, 177–190. [Google Scholar] [CrossRef] [PubMed]
- Mishra, K.K.; Vikram, P.; Yadaw, R.B.; Swamy, B.P.; Dixit, S.; Cruz, M.T.; Maturan, P.; Marker, S.; Kumar, A. qDTY12.1: A locus with a consistent effect on grain yield under drought in rice. BMC Genet. 2013, 14, 12. [Google Scholar] [CrossRef] [PubMed]
- Vinod, K.K.; Krishnan, S.G.; Thribhuvan, R.; Singh, A.K. Genetics of drought tolerance, mapping QTLs, candidate genes and their utilization in rice improvement. In Genomics assisted Breeding of Crops for Abiotic Stress Tolerance; Senthil-Kumar, M., Ed.; Springer: Cham, Switzerland, 2019; Volume Sustainable development and biodiversity; pp. 145–186. [Google Scholar]
- Bernardo, R. What proportion of declared QTL in plants are false? Theor. Appl. Genet. 2004, 109, 419–424. [Google Scholar] [CrossRef]
- Xu, P.; Zhou, J.W.; Li, J.; Yi, H.F.; Deng, X.N.; Feng, S.F.; Ren, G.Y.; Zhang, Z.; Deng, W.; Tao, D.Y. Mapping three new interspecific hybrid sterile loci between Oryza sativa and O. glaberrima. Breed Sci. 2014, 63, 476. [Google Scholar] [CrossRef]
- Khush, G.S. Origin, dispersal, cultivation and variation of rice. Plant Mol. Biol. 1997, 35, 25–34. [Google Scholar] [CrossRef]
- Torres, R.O.; Henry, A. Yield stability of selected rice breeding lines and donors across conditions of mild to moderately severe drought stress. Field Crops Res. 2018, 220, 37–45. [Google Scholar] [CrossRef]
- Kumar, A.; Bernier, J.; Verulkar, S.; Lafitte, H.R.; Atlin, G.N. Breeding for drought tolerance: Direct selection for yield, response to selection and use of drought-tolerant donors in upland and lowland-adapted populations. Field Crops Res. 2008, 107, 221–231. [Google Scholar] [CrossRef]
- Guan, Y.S.; Serraj, R.; Liu, S.H.; Xu, J.L.; Ali, J.; Wang, W.S.; Venus, E.; Zhu, L.H.; Li, Z.K. Simultaneously improving yield under drought stress and non-stress conditions: A case study of rice (Oryza sativa L.). J. Exp. Bot. 2010, 61, 4145–4156. [Google Scholar] [CrossRef]
- Shamsudin, N.A.A.; Swamy, B.P.M.; Ratnam, W.; Cruz, M.T.S.; Raman, A.; Kumar, A. Marker assisted pyramiding of drought yield QTLs into a popular Malaysian rice cultivar, MR219. BMC Genet. 2016, 17, 30. [Google Scholar] [CrossRef]
- Ouk, M.; Basnayake, J.; Tsubo, M.; Fukai, S.; Fischer, K.S.; Cooper, M.; Nesbitt, H. Use of drought response index for identification of drought tolerant genotypes in rainfed lowland rice. Field Crops Res. 2006, 99, 48–58. [Google Scholar] [CrossRef]
- Yadav, S.; Sandhu, N.; Majumder, R.R.; Dixit, S.; Kumar, S.; Singh, S.P.; Mandal, N.P.; Das, S.P.; Yadaw, R.B.; Singh, V.K.; et al. Epistatic interactions of major effect drought QTLs with genetic background loci determine grain yield of rice under drought stress. Sci. Rep. 2019, 9, 2616. [Google Scholar] [CrossRef] [PubMed]
- Trijatmiko, K.R.; Supriyanta; Prasetiyono, J.; Thomson, M.J.; Cruz, C.M.V.; Moeljopawiro, S.; Pereira, A. Meta-analysis of quantitative trait loci for grain yield and component traits under reproductive-stage drought stress in an upland rice population. Mol. Breed. 2014, 34, 283–295. [Google Scholar] [CrossRef] [PubMed]
- Courtois, B.; Ahmadi, N.; Khowaja, F.; Price, A.H.; Rami, J.F.; Frouin, J.; Hamelin, C.; Ruiz, M. Rice root genetic architecture: Meta-analysis from a drought QTL database. Rice 2009, 2, 115–128. [Google Scholar] [CrossRef]
- Khowaja, F.S.; Norton, G.J.; Courtois, B.; Price, A.H. Improved resolution in the position of drought-related QTLs in a single mapping population of rice by meta-analysis. BMC Genom. 2009, 10, 276. [Google Scholar] [CrossRef]
- Kanagaraj, P.; Prince, K.S.J.; Sheeba, J.A.; Biji, K.R.; Paul, S.B.; Senthil, A.; Babu, R.C. Microsatellite markers linked to drought resistance in rice (Oryza sativa L.). Curr. Sci. 2010, 98, 836–839. [Google Scholar]
- Zhang, F.; Jiang, Y.Z.; Yu, S.B.; Ali, J.; Paterson, A.H.; Khush, G.S.; Xu, J.L.; Gao, Y.M.; Fu, B.Y.; Lafitte, R.; et al. Three genetic systems controlling growth, development and productivity of rice (Oryza sativa L.): A reevaluation of the ‘Green Revolution’. Theor. Appl. Genet. 2013, 126, 1011–1024. [Google Scholar] [CrossRef]
- Tilman, D. The greening of the green revolution. Nature 1998, 396, 211–212. [Google Scholar] [CrossRef]
- Peng, J.; Richards, D.E.; Hartley, N.M.; Murphy, G.P.; Devos, K.M.; Flintham, J.E.; Beales, J.; Fish, L.J.; Worland, A.J.; Pelica, F.; et al. ‘Green revolution’ genes encode mutant gibberellin response modulators. Nature 1999, 400, 256–261. [Google Scholar] [CrossRef]
- Matson, P.A.; Parton, W.J.; Power, A.G.; Swift, M.J. Agricultural intensification and ecosystem properties. Science 1997, 277, 504–509. [Google Scholar] [CrossRef]
- Li, S.; Tian, Y.H.; Wu, K.; Ye, Y.F.; Yu, J.P.; Zhang, J.Q.; Liu, Q.; Hu, M.Y.; Li, H.; Tong, Y.P.; et al. Modulating plant growth-metabolism coordination for sustainable agriculture. Nature 2018, 560, 595–600. [Google Scholar] [CrossRef]
- Lafitte, H.R.; Yongsheng, G.; Yan, S.; Li, Z.K. Whole plant responses, key processes, and adaptation to drought stress: The case of rice. J. Exp. Bot. 2007, 58, 169–175. [Google Scholar] [CrossRef] [PubMed]
- Kuroha, T.; Nagai, K.; Gamuyao, R.; Wang, D.R.; Furuta, T.; Nakamori, M.; Kitaoka, T.; Adachi, K.; Minami, A.; Mori, Y.; et al. Ethylene-gibberellin signaling underlies adaptation of rice to periodic flooding. Science 2018, 361, 181–186. [Google Scholar] [CrossRef] [PubMed]
- Asano, K.; Yamasaki, M.; Takuno, S.; Miura, K.; Katagiri, S.; Ito, T.; Doi, K.; Wu, J.; Ebana, K.; Matsumoto, T.; et al. Artificial selection for a green revolution gene during japonica rice domestication. Proc. Natl. Acad. Sci. USA 2011, 108, 11034–13039. [Google Scholar] [CrossRef] [PubMed]
- Abbai, R.; Singh, V.K.; Nachimuthu, V.V.; Sinha, P.; Selvaraj, R.; Vipparla, A.K.; Singh, A.K.; Singh, U.M.; Varshney, R.K.; Kumar, A. Haplotype analysis of key genes governing grain yield and quality traits across 3K RG panel reveals scope for the development of tailor-made rice with enhanced genetic gains. Plant Biotechnol. J. 2019, 17, 1612–1622. [Google Scholar] [CrossRef]
- Hu, Z.Q.; Wang, W.S.; Wu, Z.C.; Sun, C.; Li, M.; Lu, J.Y.; Fu, B.Y.; Shi, J.X.; Xu, J.L.; Ruan, J.; et al. Novel sequences, structural variations and gene presence variations of Asian cultivated rice. Sci. Data 2018, 5, 180079. [Google Scholar] [CrossRef]
- Kazan, K.; Lyons, R. The link between flowering time and stress tolerance. J. Exp. Bot. 2016, 67, 47–60. [Google Scholar] [CrossRef]
- Minh-Thu, P.T.; Kim, J.S.; Chae, S.; Jun, K.M.; Lee, G.S.; Kim, D.E.; Cheong, J.J.; Song, S.I.; Nahm, B.H.; Kim, Y.K. A WUSCHEL homeobox transcription factor, OsWOX13, enhances drought tolerance and triggers early flowering in rice. Mol. Cells 2018, 41, 781–798. [Google Scholar]
- Peleman, J.; Sorensen, A.; Voort, J. Breeding by design: Exploiting genetic maps and molecular markers through marker-assisted selection. In Handbook of Plant Genome Mapping Genetic & Physical Mapping; Meksem, K., Kahl, G., Eds.; John Wiley & Sons, Ltd.: Hoboken, NJ, USA, 2005; pp. 109–129. [Google Scholar]
- Xue, Y.B.; Duan, Z.Y.; Chong, K.; Yao, Y. A new generation of biological breeding technology for the future—Molecular breeding by modular design. Bull. Chin. Acad. Sci. 2013, 28, 308–314. [Google Scholar]
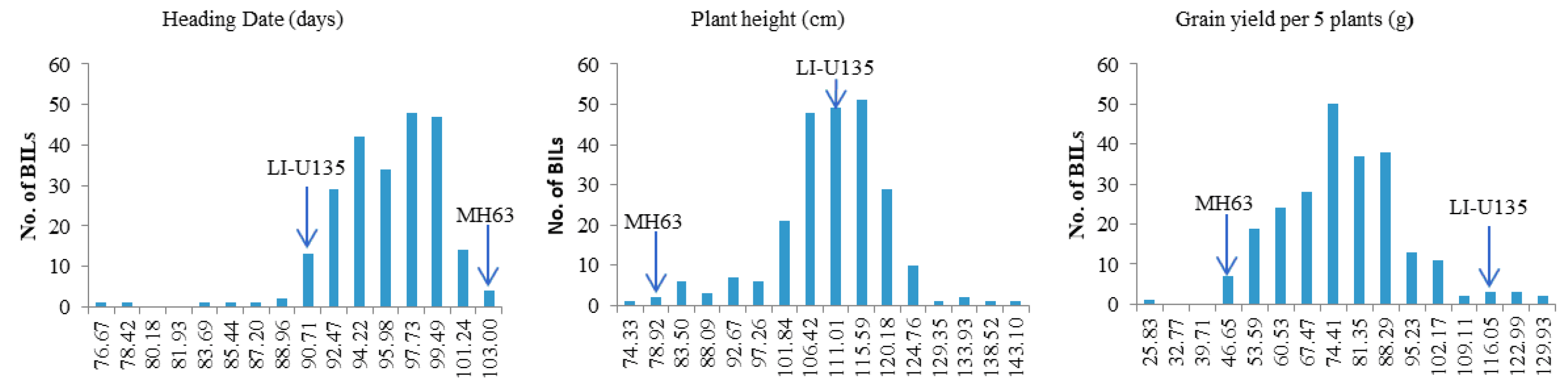
| Sites and condition | Menglian Breeding Station | Sanya Breeding Station | ||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| GY | BY | HI | HD | PH | GY | BY | HI | HD | PH | |||||||||||
| L | A | L | A | L | A | L | A | L | A | L | A | L | A | L | A | L | A | L | A | |
| MH 63 | 52.33 | 17.37 | 154.20 | 134.03 | 0.34 | 0.13 | 99.00 | 110.50 | 93.67 | 61.53 | 37.00 | 20.90 | 114.20 | 109.90 | 0.32 | 0.19 | 96.00 | 101.67 | 95.07 | 70.47 |
| BC1 ILs Mean (N = 33) | 60.51 | 80.97 | 156.48 | 230.26 | 0.39 | 0.34 | 97.39 | 102.58 | 98.07 | 97.09 | 55.19 | 38.19 | 138.96 | 118.25 | 0.39 | 0.32 | 86.77 | 94.03 | 119.53 | 87.15 |
| one-way ANOVA P | 0.25 | 0.02 | 0.65 | 0.65 | 0.00 | 0.00 | 0.14 | 0.14 | 0.00 | 0.00 | 0.61 | 0.05 | 0.92 | 0.92 | 0.00 | 0.00 | 0.00 | 0.00 | 0.02 | 0.01 |
| LSD 0.05 | 21.68 | 60.50 | 47.49 | 136.55 | 0.11 | 0.09 | 4.25 | 10.24 | 5.53 | 12.55 | 25.98 | 19.90 | 49.21 | 49.06 | 0.15 | 0.09 | 1.89 | 3.65 | 15.58 | 13.64 |
| Significant lines among 33 BC1 ILs | 22 | 2 | 32 | 13 | 33 | 11 | 30 | 31 | 31 | 28 | 27 | |||||||||
| MH 63 | 56.48 | 19.22 | 162.32 | 129.87 | 0.35 | 0.15 | 99.00 | 109.00 | 93.67 | 63.25 | 40.80 | 22.23 | 111.20 | 111.25 | 0.37 | 0.20 | 95.50 | 102.82 | 94.80 | 68.46 |
| BC2 ILs Mean (N = 23) | 57.90 | 71.27 | 161.64 | 221.74 | 0.36 | 0.30 | 95.14 | 103.76 | 106.10 | 97.02 | 46.54 | 34.50 | 138.79 | 112.15 | 0.33 | 0.31 | 90.40 | 93.37 | 126.66 | 94.62 |
| one-way ANOVA P | 0.11 | 0.00 | 0.01 | 0.01 | 0.00 | 0.00 | 0.64 | 0.64 | 0.00 | 0.00 | 0.46 | 0.05 | 1.00 | 0.99 | 0.04 | 0.04 | 0.00 | 0.00 | 0.00 | 0.00 |
| LSD 0.05 | 23.61 | 57.56 | 50.53 | 149.77 | 0.07 | 0.07 | 3.19 | 14.75 | 7.93 | 16.37 | 27.09 | 16.85 | 61.18 | 41.89 | 0.12 | 0.10 | 2.69 | 4.06 | 15.37 | 9.61 |
| Significant lines among 23 BC2 ILs | 9 | 6 | 23 | 9 | 23 | 6 | 16 | 16 | 23 | 16 | 22 | |||||||||
| MH 63 | 55.26 | 18.82 | 156.26 | 138.33 | 0.35 | 0.14 | 99.00 | 109.24 | 93.67 | 62.21 | 36.92 | 21.22 | 116.30 | 109.23 | 0.32 | 0.19 | 96.20 | 101.33 | 95.32 | 69.24 |
| BC3 ILs Mean (N = 6) | 61.38 | 51.33 | 173.45 | 227.30 | 0.35 | 0.22 | 96.47 | 109.69 | 101.05 | 98.28 | 43.10 | 28.56 | 136.13 | 119.53 | 0.31 | 0.24 | 88.83 | 96.17 | 123.47 | 92.84 |
| one-way ANOVA P | 0.05 | 0.05 | 0.17 | 0.17 | 0.28 | 0.28 | 0.09 | 0.09 | 0.00 | 0.00 | 0.08 | 0.08 | 0.91 | 0.91 | 0.01 | 0.01 | 0.00 | 0.00 | 0.00 | 0.00 |
| LSD 0.05 | 30.25 | 34.79 | 70.33 | 132.44 | 0.06 | 0.11 | 4.25 | 13.49 | 7.38 | 20.57 | 27.01 | 17.02 | 47.88 | 49.19 | 0.16 | 0.12 | 0.93 | 3.81 | 17.61 | 15.90 |
| Significant lines among 6 BC3 ILs | 5 | 2 | 6 | 1 | 6 | 4 | 5 | 5 | ||||||||||||
| QTL | Position (cM) | Flank Markers | p | Generation, Site &Condition |
|---|---|---|---|---|
| qPH1 | 132–147.2 | RM297–RM6333 | <0.0001 | BC1 ML A&L, SY A&L; BC2 ML A&L, SY A&L; BC3 ML L, SY A&L |
| qGY1 | BC1 ML A, SY A; BC2 ML A, SY A; BC3 ML A | |||
| qHI1 | BC1 ML A, SY A; BC2 ML A, SY A | |||
| qHD3 | 115.6–127.4 | RM2334–RM6329 | <0.0001 | BC1 SY A&L; BC2 SY A&L; BC3 SY L |
| qGY3 | BC1 ML A, SY A; BC2 ML A, SY A; BC3 ML A | |||
| qHI3 | BC1 ML A, SY A; BC2 ML A, SY A | |||
| qHD4 | 3.1–7.9 | RM7585–RM5414 | <0.0001 | BC1 SY L; BC1 SY L |
| qHD7 | 93.9–101.8 | RM234–RM429 | <0.0001 | BC1 SY A&L; BC2 SY A&L |
| qGY7 | BC1 ML A, SY A; BC2 ML A, SY A; BC3 ML A | |||
| qHI7 | BC1 ML A, SY A; BC2 ML A, SY A | |||
| qHD12 | 3.2–13.3 | RM20–RM6288 | <0.0001 | BC1 SY A&L; BC2 SY A&L; BC3 SY L |
| qGY12 | BC1 ML A, SY A; BC2 ML A, SY A; BC3 ML A | |||
| qHI12 | BC1 ML A, SY A; BC2 ML A, SY A |
| Traits | Chr | Marker Interval | Position (cM) | LOD | A | R2 (%) | QTL |
|---|---|---|---|---|---|---|---|
| Plant height | 1 | RM212-RM543 | 135.8–145.6 | 5.42 | 12.45 | 24.14 | qAER1 |
| Grains yield | 1 | RM212-RM543 | 135.8–145.6 | 4.19 | 13.87 | 18.70 | |
| Heading date | 3 | RM218-RM232 | 67.8–76.7 | 6.52 | −3.5 | 30.00 | qAER3 |
| Grains yield | 3 | RM218-RM232 | 67.8–76.7 | 8.97 | 16.51 | 19.19 | |
| Heading date | 9 | RM218-RM232 | 23.7–36.8 | 10.38 | −3.04 | 16.71 | qAER9 |
| Grains yield | 9 | RM218-RM232 | 23.7–36.8 | 3.18 | 13.87 | 16.71 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Xu, P.; Yang, J.; Ma, Z.; Yu, D.; Zhou, J.; Tao, D.; Li, Z. Identification and Validation of Aerobic Adaptation QTLs in Upland Rice. Life 2020, 10, 65. https://doi.org/10.3390/life10050065
Xu P, Yang J, Ma Z, Yu D, Zhou J, Tao D, Li Z. Identification and Validation of Aerobic Adaptation QTLs in Upland Rice. Life. 2020; 10(5):65. https://doi.org/10.3390/life10050065
Chicago/Turabian StyleXu, Peng, Jun Yang, Zhenbing Ma, Diqiu Yu, Jiawu Zhou, Dayun Tao, and Zichao Li. 2020. "Identification and Validation of Aerobic Adaptation QTLs in Upland Rice" Life 10, no. 5: 65. https://doi.org/10.3390/life10050065
APA StyleXu, P., Yang, J., Ma, Z., Yu, D., Zhou, J., Tao, D., & Li, Z. (2020). Identification and Validation of Aerobic Adaptation QTLs in Upland Rice. Life, 10(5), 65. https://doi.org/10.3390/life10050065





