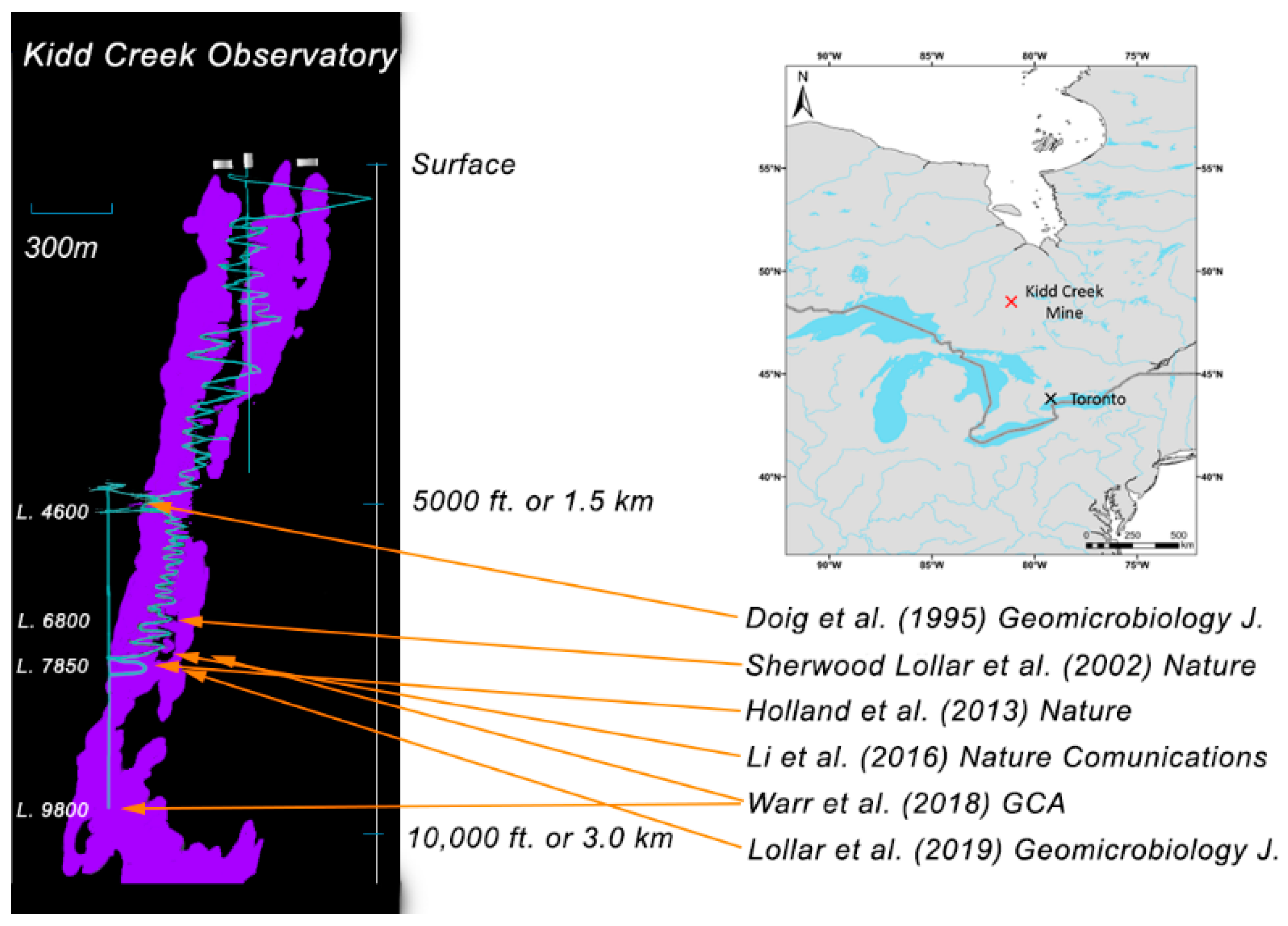
In Situ Growth of Halophilic Bacteria in Saline Fracture Fluids from 2.4 km below Surface in the Deep Canadian Shield
Abstract
1. Introduction
2. Materials and Methods
2.1. Filtration of Mine Water
2.2. In Situ Incubation and Fixation
2.3. SEM and EDS
2.4. DNA Extraction
2.5. DNA Amplification and Sequencing
2.6. Genetic Analyses
3. Results and Discussion
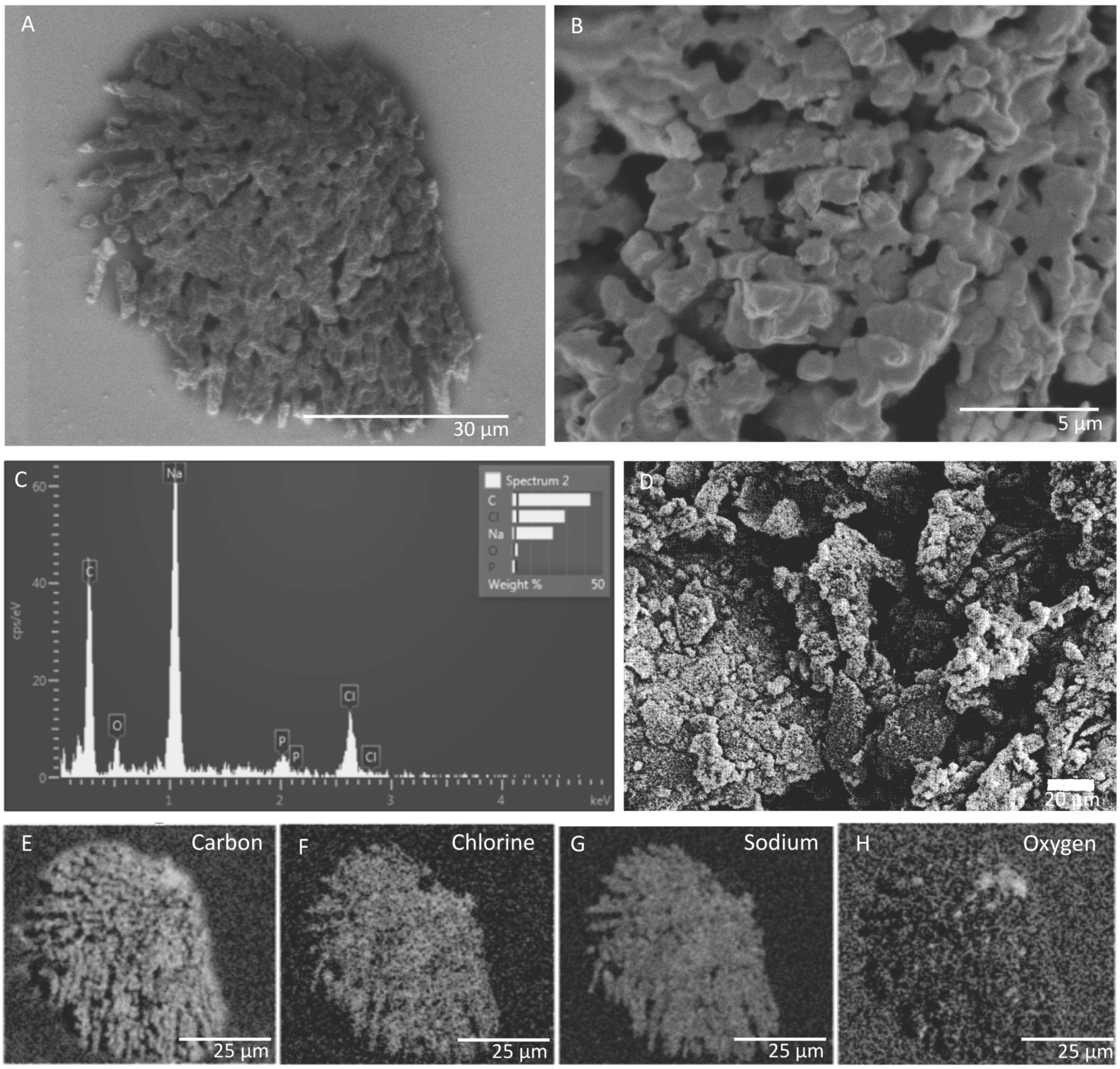
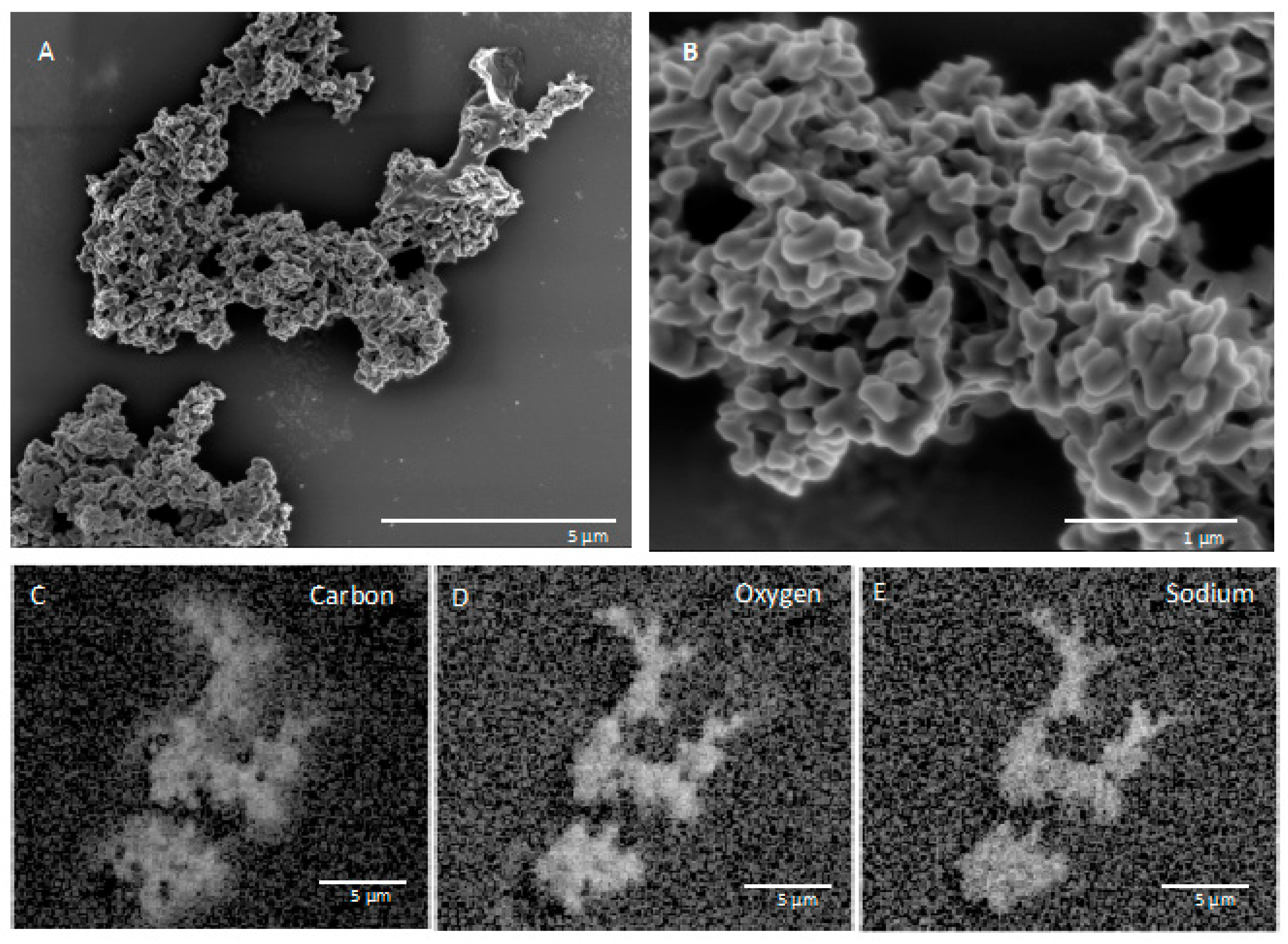
3.1. SEM and EDS Results of In Situ Incubations
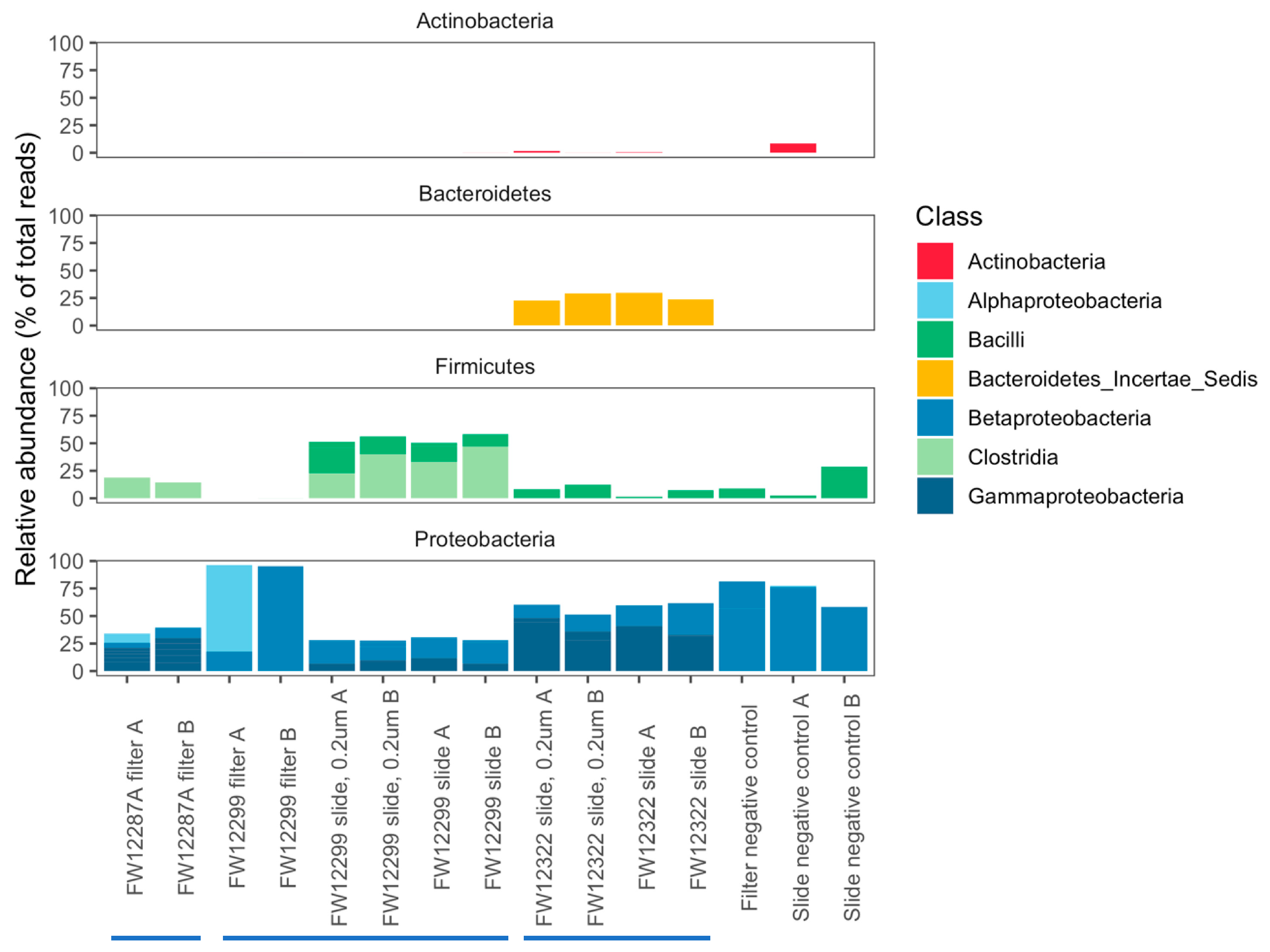
3.2. Fracture Water vs. Service Water
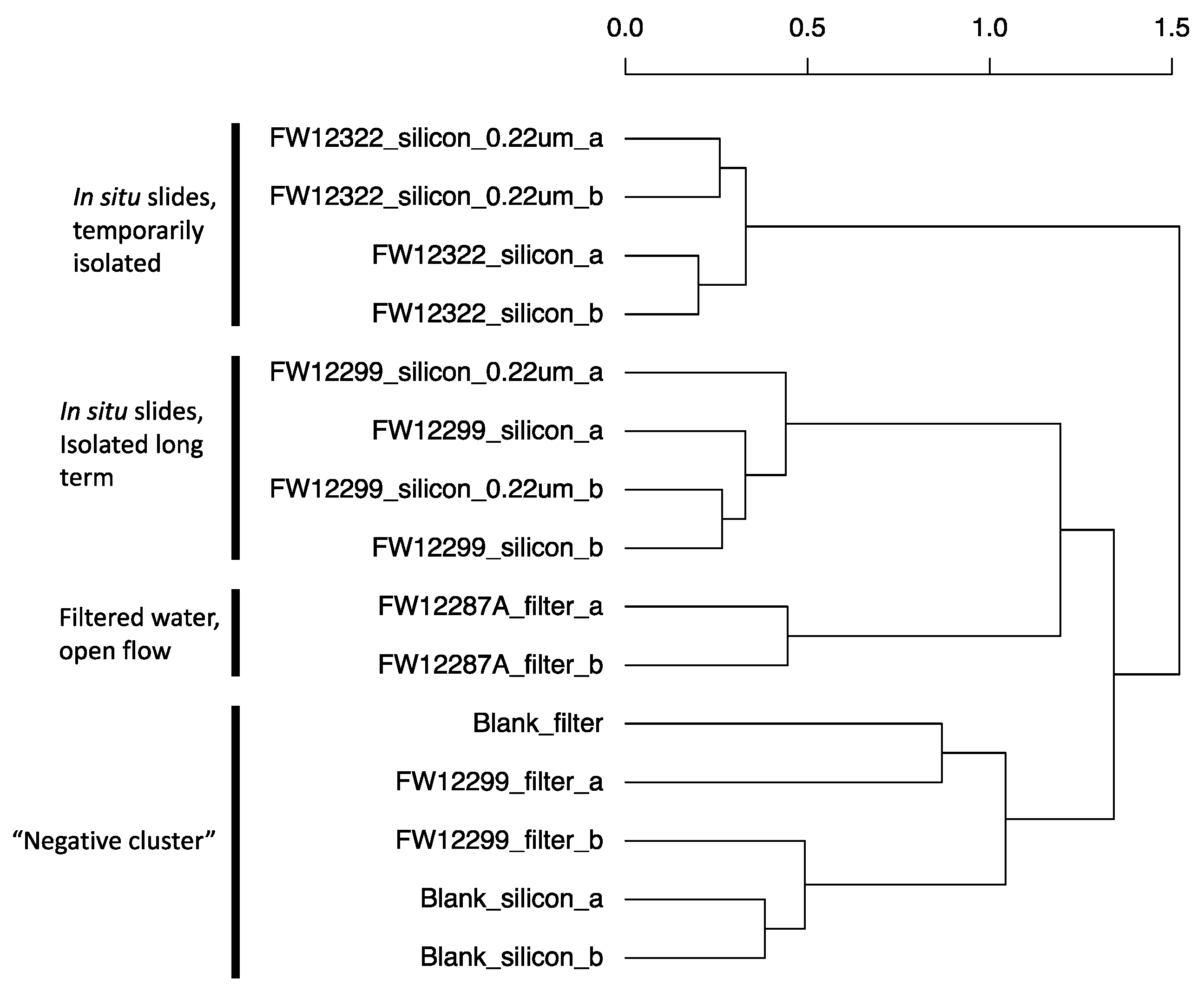
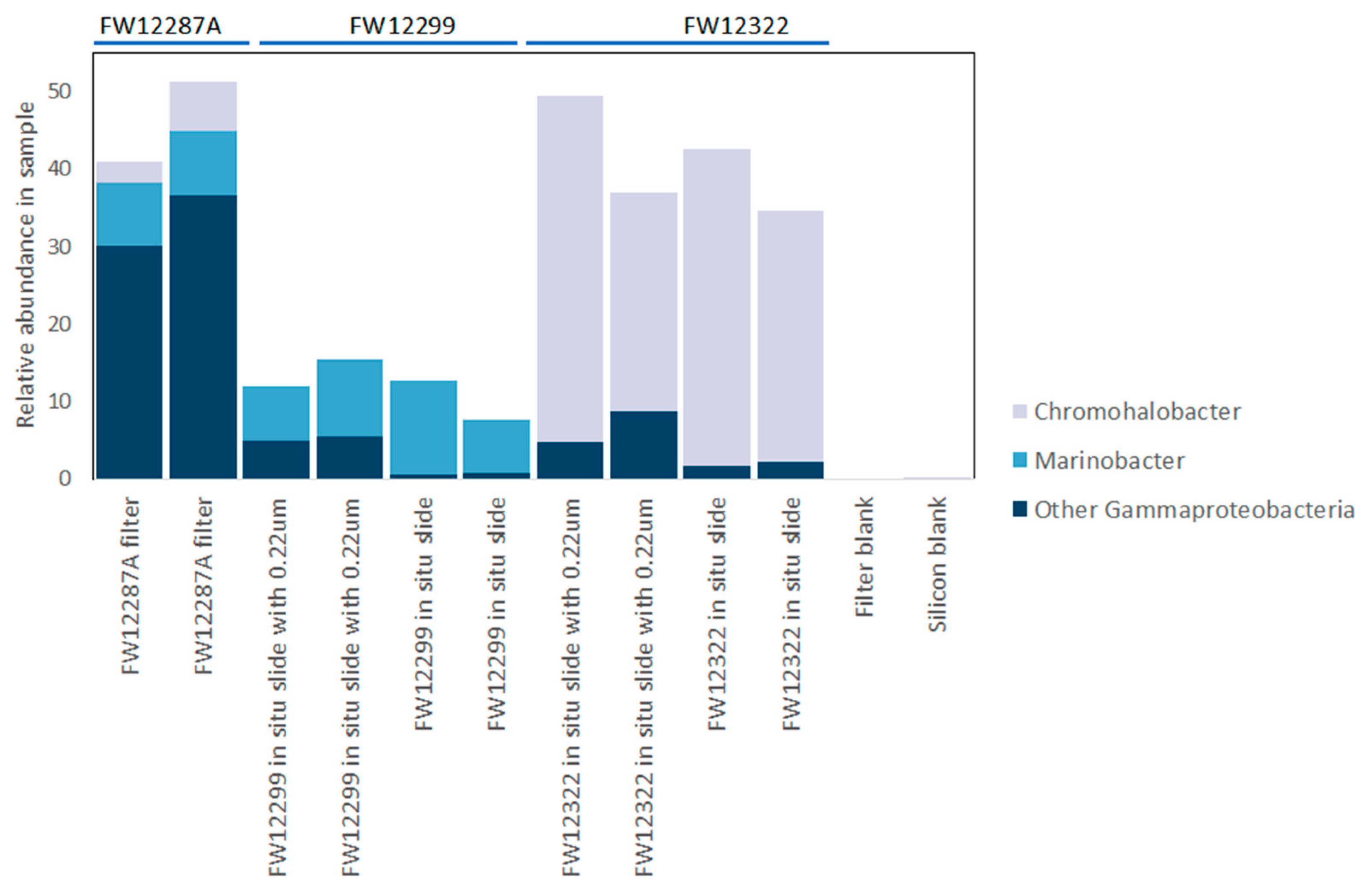
3.3. Results of 16S rRNA Sequencing
3.4. Comparison to Other Subsurface Studies
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Lin, L.-H.; Wang, P.-L.; Rumble, D.; Lippmann-Pipke, J.; Boice, E.; Pratt, L.M.; Sherwood Lollar, B.; Brodie, E.L.; Hazen, T.C.; Andersen, G.L.; et al. Long-term sustainability of a high-energy, low-diversity crustal biome. Science 2006, 314, 479–482. [Google Scholar] [CrossRef] [PubMed]
- Chivian, D.; Brodie, E.L.; Alm, E.J.; Culley, D.E.; Dehal, P.S.; DeSantis, T.Z.; Gihring, T.M.; Lapidus, A.; Lin, L.-H.; Lowry, S.R.; et al. Environmental Genomics Reveals a Single-Species Ecosystem Deep Within Earth. Science 2008, 322, 275–278. [Google Scholar] [CrossRef] [PubMed]
- Magnabosco, C.; Ryan, K.; Lau, M.C.Y.; Kuloyo, O.; Lollar, B.S.; Kieft, T.L.; Van Heerden, E.; Onstott, T.C. A metagenomic window into carbon metabolism at 3 km depth in Precambrian continental crust. ISME J. 2016, 10, 730–741. [Google Scholar] [CrossRef]
- Lollar, G.S.; Warr, O.; Telling, J.P.; Osburn, M.R.; Lollar, B.S. ‘Follow the Water’: Hydrogeochemical Constraints on Microbial Investigations 2.4 km Below Surface at the Kidd Creek Deep Fluid and Deep Life Observatory. Geomicrobiol. J. 2019, 36, 859–872. [Google Scholar] [CrossRef]
- Lau, M.C.Y.; Kieft, T.L.; Kuloyo, O.; Linage-Alvarez, B.; Van Heerden, E.; Lindsay, M.R.; Magnabosco, C.; Wang, W.; Wiggins, J.B.; Guo, L.; et al. An oligotrophic deep-subsurface community dependent on syntrophy is dominated by sulfur-driven autotrophic denitrifiers. Proc. Natl. Acad. Sci. USA 2016, 113, E7927–E7936. [Google Scholar] [CrossRef]
- Momper, L.; Jungbluth, S.P.; Lee, M.D.; Amend, J.P. Energy and carbon metabolisms in a deep terrestrial subsurface fluid microbial community. ISME J. 2017, 11, 2319–2333. [Google Scholar] [CrossRef] [PubMed]
- Purkamo, L.; Kietäväinen, R.; Nuppunen-Puputti, M.; Bomberg, M.; Cousins, C. Ultradeep Microbial Communities at 4.4 km within Crystalline Bedrock: Implications for Habitability in a Planetary Context. Life 2020, 10, 2. [Google Scholar] [CrossRef]
- Kotelnikova, S. Distribution and activity of methanogens and homoacetogens in deep granitic aquifers at Äspö Hard Rock Laboratory, Sweden. FEMS Microbiol. Ecol. 1998, 26, 121–134. [Google Scholar] [CrossRef]
- Takai, K.; Moser, D.P.; DeFlaun, M.; Onstott, T.C.; Fredrickson, J.K. Archaeal Diversity in Waters from Deep South African Gold Mines. Appl. Environ. Microbiol. 2001, 67, 5750–5760. [Google Scholar] [CrossRef]
- Baker, B.J.; Moser, D.P.; MacGregor, B.J.; Fishbain, S.; Wagner, M.; Fry, N.K.; Jackson, B.; Speolstra, N.; Loos, S.; Takai, K.; et al. Related assemblages of sulphate-reducing bacteria associated with ultradeep gold mines of South Africa and deep basalt aquifers of Washington State. Environ. Microbiol. 2003, 5, 267–277. [Google Scholar] [CrossRef]
- Sahl, J.W.; Schmidt, R.; Swanner, E.D.; Mandernack, K.W.; Templeton, A.S.; Kieft, T.L.; Smith, R.L.; Sanford, W.E.; Callaghan, R.L.; Mitton, J.B.; et al. Subsurface Microbial Diversity in Deep-Granitic-Fracture Water in Colorado. Appl. Environ. Microbiol. 2007, 74, 143–152. [Google Scholar] [CrossRef] [PubMed]
- Suko, T.; Kouduka, M.; Fukuda, A.; Nanba, K.; Takahashi, M.; Ito, K.; Suzuki, Y. Geomicrobiological properties of Tertiary sedimentary rocks from the deep terrestrial subsurface. Phys. Chem. Earth Parts A B C 2013, 58, 28–33. [Google Scholar] [CrossRef]
- Grotzinger, J.P.; Sumner, D.Y.; Kah, L.C.; Stack, K.; Gupta, S.; Edgar, L.; Rubin, D.; Lewis, K.; Schieber, J.; Mangold, N.; et al. MSL Science Team A habitable fluvio-lacustrine environment at Yellowknife Bay, Gale crater, Mars. Science 2014, 343, 1242777. [Google Scholar] [CrossRef] [PubMed]
- Ehlmann, B.; Anderson, F.S.; Andrews-Hanna, J.; Catling, D.C.; Christensen, P.R.; Cohen, B.A.; Dressing, C.D.; Edwards, C.S.; Elkins-Tanton, L.T.; Farley, K.A.; et al. The sustainability of habitability on terrestrial planets: Insights, questions, and needed measurements from Mars for understanding the evolution of Earth-like worlds. J. Geophys. Res. Planets 2016, 121, 1927–1961. [Google Scholar] [CrossRef]
- Michalski, J.R.; Cuadros, J.; Niles, P.B.; Parnell, J.; Rogers, A.D.; Wright, S.P. Groundwater activity on Mars and implications for a deep biosphere. Nat. Geosci. 2013, 6, 133–138. [Google Scholar] [CrossRef]
- Banerdt, W.B.; Smrekar, S.E.; Banfield, D.; Giardini, D.; Golombek, M.; Johnson, C.L.; Lognonné, P.; Spiga, A.; Spohn, T.; Perrin, C.; et al. Initial results from the InSight mission on Mars. Nat. Geosci. 2020, 13, 183–189. [Google Scholar] [CrossRef]
- Li, L.; Wing, B.A.; Bui, T.H.; McDermott, J.M.; Slater, G.F.; Wei, S.; Lacrampe-Couloume, G.; Sherwood Lollar, B. Sulfur mass-independent fractionation in subsurface fracture waters indicates a long-standingsulfur cycle in Precambrian rocks. Nat. Commun. 2016, 7, 1–9. [Google Scholar]
- Pedersen, K. Exploration of deep intraterrestrial microbial life: Current perspectives. FEMS Microbiol. Lett. 2000, 185, 9. [Google Scholar] [CrossRef]
- Fredrickson, J.K.; Balkwill, D.L. Geomicrobial Processes and Biodiversity in the Deep Terrestrial Subsurface. Geomicrobiol. J. 2006, 23, 345–356. [Google Scholar] [CrossRef]
- Nealson, K.H.; Inagaki, F.; Takai, K. Hydrogen-driven subsurface lithoautotrophic microbial ecosystems (SLiMEs): Do they exist and why should we care? Trends Microbiol. 2005, 13, 405–410. [Google Scholar] [CrossRef]
- Colwell, F.S.; D’Hondt, S. Nature and Extent of the Deep Biosphere. Rev. Mineral. Geochem. 2013, 75, 547. [Google Scholar] [CrossRef]
- Magnabosco, C.; Lin, L.-H.; Dong, H.; Bomberg, M.; Ghiorse, W.; Stan-Lotter, H.; Pedersen, K.; Kieft, T.L.; Van Heerden, E.; Onstott, T.C. The biomass and biodiversity of the continental subsurface. Nat. Geosci. 2018, 11, 707–717. [Google Scholar] [CrossRef]
- Warr, O.; Lollar, B.S.; Fellowes, J.; Sutcliffe, C.N.; McDermott, J.M.; Holland, G.; Mabry, J.C.; Ballentine, C.J. Tracing ancient hydrogeological fracture network age and compartmentalisation using noble gases. Geochim. Cosmochim. Acta 2018, 222, 340–362. [Google Scholar] [CrossRef]
- Walker, R.R.; Matulich, A.; Amos, A.C.; Watkins, J.J.; Mannard, G.W. The geology of the Kidd Creek Mine. Econ. Geol. 1975, 70, 80–89. [Google Scholar] [CrossRef]
- Doig, F.; Lollar, B.S.; Ferris, F.G. Microbial communities in deep Canadian shield groundwaters—An in situ biofilm experiment. Geomicrobiol. J. 1995, 13, 91–102. [Google Scholar] [CrossRef]
- Sherwood Lollar, B.; Westgate, T.D.; Ward, J.A.; Slater, G.F.; Lacrampe-Couloume, G. Abiogenic formation of alkanes in the Earth’s crust as a minor source for global hydrocarbon reservoirs. Nature 2002, 416, 522. [Google Scholar] [CrossRef]
- Holland, G.P.; Lollar, B.S.; Li, L.; Lacrampe-Couloume, G.; Slater, G.F.; Ballentine, C. Deep fracture fluids isolated in the crust since the Precambrian era. Nat. Cell Biol. 2013, 497, 357–360. [Google Scholar] [CrossRef]
- Martínez, G.M.; Renno, N. Water and Brines on Mars: Current Evidence and Implications for MSL. Space Sci. Rev. 2013, 175, 29–51. [Google Scholar] [CrossRef]
- Lollar, B.S.; Onstott, T.C.; Lacrampecouloume, G.; Ballentine, C.J. The contribution of the Precambrian continental lithosphere to global H2 production. Nat. Cell Biol. 2014, 516, 379–382. [Google Scholar] [CrossRef]
- Cowen, J.P.; Giovannoni, S.J.; Kenig, F.; Johnson, H.P.; Butterfield, D.; Rappé, M.S.; Hutnak, M.; Lam, P. Fluids from Aging Ocean Crust That Support Microbial Life. Science 2003, 299, 120–123. [Google Scholar] [CrossRef]
- Nakagawa, S.; Inagaki, F.; Suzuki, Y.; Steinsbu, B.O.; Lever, M.A.; Takai, K.; Engelen, B.; Sako, Y.; Wheat, C.G.; Horikoshi, K. Microbial Community in Black Rust Exposed to Hot Ridge Flank Crustal Fluids. Appl. Environ. Microbiol. 2006, 72, 6789–6799. [Google Scholar] [CrossRef] [PubMed]
- Lau, K.V.; Maher, K.; Altiner, D.; Kelley, B.M.; Kump, L.R.; Lehrmann, D.J.; Silva-Tamayo, J.C.; Weaver, K.L.; Yu, M.; Payne, J.L. Marine anoxia and delayed Earth system recovery after the end-Permian extinction. Proc. Natl. Acad. Sci. USA 2016, 113, 2360–2365. [Google Scholar] [CrossRef] [PubMed]
- House, C.H.; Orphan, V.J.; Turk, K.A.; Thomas, R.B.; Pernthaler, A.; Vrentas, J.M.; Joye, S.B. Extensive carbon isotopic heterogeneity among methane seep microbiota. Environ. Microbiol. 2009, 11, 2207–2215. [Google Scholar] [CrossRef] [PubMed]
- Caporaso, J.G.; Lauber, C.L.; Walters, W.A.; Berg-Lyons, D.; Lozupone, C.A.; Turnbaugh, P.J.; Fierer, N.; Knight, R. Global patterns of 16S rRNA diversity at a depth of millions of sequences per sample. Proc. Natl. Acad. Sci. USA 2011, 108, 4516–4522. [Google Scholar] [CrossRef] [PubMed]
- Rochelle, P.; Fry, J.C.; Parkes, R.J.; Weightman, A.J. DNA extraction for 16S rRNA gene analysis to determine genetic diversity in deep sediment communities. FEMS Microbiol. Lett. 1992, 100, 59. [Google Scholar] [CrossRef] [PubMed]
- Kozich, J.J.; Westcott, S.L.; Baxter, N.T.; Highlander, S.K.; Schloss, P.D. Development of a Dual-Index Sequencing Strategy and Curation Pipeline for Analyzing Amplicon Sequence Data on the MiSeq Illumina Sequencing Platform. Appl. Environ. Microbiol. 2013, 79, 5112–5120. [Google Scholar] [CrossRef] [PubMed]
- Schloss, P.D.; Westcott, S.L.; Ryabin, T.; Hall, J.R.; Hartmann, M.; Hollister, E.B.; Lesniewski, R.A.; Oakley, B.B.; Parks, D.H.; Robinson, C.J.; et al. Introducing mothur: Open-Source, Platform-Independent, Community-Supported Software for Describing and Comparing Microbial Communities. Appl. Environ. Microbiol. 2009, 75, 7537–7541. [Google Scholar] [CrossRef]
- Edgar, R.C.; Haas, B.J.; Clemente, J.C.; Quince, C.; Knight, R. UCHIME improves sensitivity and speed of chimera detection. Bioinformatics 2011, 27, 2194–2200. [Google Scholar] [CrossRef]
- O’Leary, N.A.; Wright, M.W.; Brister, J.R.; Ciufo, S.; Haddad, D.; McVeigh, R.; Rajput, B.; Robbertse, B.; Smith-White, B.; Ako-Adjei, D.; et al. Reference sequence (RefSeq) database at NCBI: Current status, taxonomic expansion, and functional annotation. Nucleic Acids Res. 2016, 44, D733–D745. [Google Scholar] [CrossRef]
- Camacho, C.; Coulouris, G.; Avagyan, V.; Ma, N.; Papadopoulos, J.S.; Bealer, K.; Madden, T.L. BLAST+: Architecture and applications. BMC Bioinform. 2009, 10, 421. [Google Scholar] [CrossRef]
- Huson, D.H.; Beier, S.; Flade, I.; Górska, A.; El-Hadidi, M.; Mitra, S.; Ruscheweyh, H.-J.; Tappu, R. MEGAN Community Edition—Interactive Exploration and Analysis of Large-Scale Microbiome Sequencing Data. PLoS Comput. Biol. 2016, 12, e1004957. [Google Scholar] [CrossRef] [PubMed]
- Quinlan, A.R.; Hall, I.M. BEDTools: A flexible suite of utilities for comparing genomic features. Bioinformatics 2010, 26, 841–842. [Google Scholar] [CrossRef] [PubMed]
- Rivadeneyra, M.A.; Martín-Algarra, A.; Sánchez-Román, M.; Sánchez-Navas, A.; Martín-Ramos, J.D. Amorphous Ca-phosphate precursors for Ca-carbonate biominerals mediated by Chromohalobacter marismortui. ISME J. 2010, 4, 922–932. [Google Scholar] [CrossRef] [PubMed]
- Sã¡nchez-Romã¡n, M.; Rivadeneyra, M.A.; Vasconcelos, C.; McKenzie, J.A. Biomineralization of carbonate and phosphate by moderately halophilic bacteria. FEMS Microbiol. Ecol. 2007, 61, 273–284. [Google Scholar] [CrossRef]
- Glassing, A.; Dowd, S.E.; Galandiuk, S.; Davis, B.; Chiodini, R.J. Inherent bacterial DNA contamination of extraction and sequencing reagents may affect interpretation of microbiota in low bacterial biomass samples. Gut Pathog. 2016, 8, 24. [Google Scholar] [CrossRef]
- Kelly, D.P.; Wood, A.P. Confirmation of Thiobacillus denitrificans as a species of the genus Thiobacillus, in the beta-subclass of the Proteobacteria, with strain NCIMB 9548 as the type strain. Int. J. Syst. Evol. Microbiol. 2000, 50, 547–550. [Google Scholar] [CrossRef]
- Zhilina, T.N.; Zavarzina, D.G.; Detkova, E.N.; Patutina, E.O.; Kuznetsov, B.B. Fuchsiella ferrireducens sp. nov., a novel haloalkaliphilic, lithoautotrophic homoacetogen capable of iron reduction, and emendation of the description of the genus Fuchsiella. Int. J. Syst. Evol. Microbiol. 2015, 65, 2432–2440. [Google Scholar] [CrossRef]
- Weiner, R.M.; Devine, R.A.; Powell, D.M.; Dagasan, L.; Moore, R.L. Hyphomonas oceanitis sp.nov., Hyphomonas hirschiana sp. nov., and Hyphomonas jannaschiana sp. nov. Int. J. Syst. Bacteriol. 1985, 35, 237–243. [Google Scholar] [CrossRef]
- Montes, M.J.; Bozal, N.; Mercade, E. Marinobacter guineae sp. nov., a novel moderately halophilic bacterium from an Antarctic environment. Int. J. Syst. Evol. Microbiol. 2008, 58, 1346–1349. [Google Scholar] [CrossRef]
- Ivanova, E.P.; Romanenko, L.A.; Chun, J.; Matte, M.H.; Matte, G.R.; Mikhailov, V.V.; Svetashev, V.I.; Huq, A.; Maugel, T.; Colwell, R.R. Idiomarina gen. nov., comprising novel indigenous deep-sea bacteria from the Pacific Ocean, including descriptions of two species, Idiomarina abyssalis sp. nov. and Idiomarina zobellii sp. nov. Int. J. Syst. Evol. Microbiol. 2000, 50, 901–907. [Google Scholar] [CrossRef]
- Yoon, J.-H.; Kang, S.-J.; Lee, S.-Y.; Oh, K.-H.; Oh, T.-K. Seohaeicola saemankumensis gen. nov., sp. nov., isolated from a tidal flat. Int. J. Syst. Evol. Microbiol. 2009, 59, 2675–2679. [Google Scholar] [CrossRef]
- García-Estepa, R.; Cánovas, D.; Iglesias-Guerra, F.; Ventosa, A.; Csonka, L.N.; Nieto, J.J.; Vargas, C.; Gutiérrez, J.J.N.; Macías, C.V. Osmoprotection of Salmonella enterica serovar Typhimurium by Nγ-acetyldiaminobutyrate, the precursor of the compatible solute ectoine. Syst. Appl. Microbiol. 2006, 29, 626–633. [Google Scholar] [CrossRef]
- Ventosa, A.; Gutierrez, M.C.; Garcia, M.T.; Ruiz-Berraquero, F. Classification of “Chromobacterium marismortui” in a New Genus, Chromohalobacter gen. nov., as Chromohalobacter marismortui comb. nov., nom. rev. Int. J. Syst. Bacteriol. 1989, 39, 382–386. [Google Scholar] [CrossRef]
- Quail, M.; Smith, M.E.; Coupland, P.; Otto, T.D.; Harris, S.R.; Connor, T.R.; Bertoni, A.; Swerdlow, H.P.; Gu, Y. A tale of three next generation sequencing platforms: Comparison of Ion torrent, pacific biosciences and illumina MiSeq sequencers. BMC Genom. 2012, 13, 341. [Google Scholar] [CrossRef] [PubMed]
- Suzuki, S.; Kuenen, J.G.; Schipper, K.; Van Der Velde, S.; Ishii, S.; Wu, A.; Sorokin, D.Y.; Tenney, A.; Meng, X.; Morrill, P.L.; et al. Physiological and genomic features of highly alkaliphilic hydrogen-utilizing Betaproteobacteria from a continental serpentinizing site. Nat. Commun. 2014, 5, 3900. [Google Scholar] [CrossRef] [PubMed]
- Cho, Y.; Chung, H.; Jang, G.I.; Choi, D.H.; Noh, J.H.; Cho, B.C. Gracilimonas rosea sp. nov., isolated from tropical seawater, and emended description of the genus Gracilimonas. Int. J. Syst. Evol. Microbiol. 2013, 63, 4006–4011. [Google Scholar] [CrossRef] [PubMed]
- Scholz, C.F.P.; Kilian, M. The natural history of cutaneous propionibacteria, and reclassification of selected species within the genus Propionibacterium to the proposed novel genera Acidipropionibacterium gen. nov., Cutibacterium gen. nov. and Pseudopropionibacterium gen. nov. Int. J. Syst. Evol. Microbiol. 2016, 66, 4422–4432. [Google Scholar] [CrossRef]
- Antunes, A.; Eder, W.; Fareleira, P.; Santos, H.; Huber, R. Salinisphaera shabanensis gen. nov., sp. nov., a novel, moderately halophilic bacterium from the brine–seawater interface of the Shaban Deep, Red Sea. Extremophiles 2003, 7, 29–34. [Google Scholar] [CrossRef]
- Giovannoni, S.J.; Schabtach, E.; Castenholz, R.W. Isosphaera pallida, gen. and comb. nov., a gliding, budding eubacterium from hot springs. Arch. Microbiol. 1987, 147, 276–284. [Google Scholar] [CrossRef]
- Wierckx, N.; Koopman, F.; Bandounas, L.; De Winde, J.H.; Ruijssenaars, H.J. Isolation and characterization of Cupriavidus basilensis HMF14 for biological removal of inhibitors from lignocellulosic hydrolysate. Microb. Biotechnol. 2010, 3, 336–343. [Google Scholar] [CrossRef]
- Ten, L.N.; Jung, H.-M.; Im, W.-T.; Oh, H.W.; Yang, D.-C.; Yoo, S.-A.; Lee, S.-T. Dokdonella ginsengisoli sp. nov., isolated from soil from a ginseng field, and emended description of the genus Dokdonella. Int. J. Syst. Evol. Microbiol. 2009, 59, 1947–1952. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Vandieken, V.; Knoblauch, C.; Jørgensen, B.B. Desulfotomaculum arcticum sp. nov., a novel spore-forming, moderately thermophilic, sulfate-reducing bacterium isolated from a permanently cold fjord sediment of Svalbard. Int. J. Syst. Evol. Microbiol. 2006, 56, 687–690. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Ryan, M.P.; Pembroke, J.T.; Adley, C.C. Ralstonia pickettiiin environmental biotechnology: Potential and applications. J. Appl. Microbiol. 2007, 103, 754–764. [Google Scholar] [CrossRef] [PubMed]
- Ryu, S.H.; Lee, D.S.; Park, M.; Wang, Q.; Jang, H.H.; Park, W.; Jeon, C.O. Caenimonas koreensis gen. nov., sp. nov., isolated from activated sludge. Int. J. Syst. Evol. Microbiol. 2008, 58, 1064–1068. [Google Scholar] [CrossRef] [PubMed]
- Khan, S.T.; Hiraishi, A. Diaphorobacter nitroreducens gen. nov., sp. nov., a poly(3-hydroxybutyrate)-degrading denitrifying bacterium isolated from activated sludge. J. Gen. Appl. Microbiol. 2002, 48, 299–308. [Google Scholar] [CrossRef]
- Gu, J.; Cai, H.; Yu, S.-L.; Qu, R.; Yin, B.; Guo, Y.-F.; Zhao, J.-Y.; Wu, X.-L. Marinobacter gudaonensis sp. nov., isolated from an oil-polluted saline soil in a Chinese oilfield. Int. J. Syst. Evol. Microbiol. 2007, 57, 250–254. [Google Scholar] [CrossRef]
- Onstott, T.C.; McGown, D.J.; Bakermans, C.; Ruskeeniemi, T.; Ahonen, L.; Telling, J.; Soffientino, B.; Pfiffner, S.M.; Sherwood Lollar, B.; Frape, S.; et al. Microbial Communities in Subpermafrost Saline Fracture Water at the Lupin Au Mine, Nunavut, Canada. Microb. Ecol. 2009, 58, 786–807. [Google Scholar] [CrossRef]
- Mills, C.T.; Amano, Y.; Slater, G.F.; Dias, R.F.; Iwatsuki, T.; Mandernack, K.W. Microbial carbon cycling in oligotrophic regional aquifers near the Tono Uranium Mine, Japan as inferred from δ13C and Δ14C values of in situ phospholipid fatty acids and carbon sources. Geochim. Cosmochim. Acta 2010, 74, 3785–3805. [Google Scholar] [CrossRef]
- Onstott, T.C.; Magnabosco, C.; Aubrey, A.D.; Burton, A.S.; Dworkin, J.P.; Elsila, J.E.; Grunsfeld, S.; Cao, B.H.; Hein, J.E.; Glavin, D.P.; et al. Does aspartic acid racemization constrain the depth limit of the subsurface biosphere? Geobiology 2013, 12, 1–19. [Google Scholar] [CrossRef]
- Wanger, G.; Southam, G.; Onstott, T.C. Structural and Chemical Characterization of a Natural Fracture Surface from 2.8 Kilometers Below Land Surface: Biofilms in the Deep Subsurface. Geomicrobiol. J. 2006, 23, 443–452. [Google Scholar] [CrossRef]
- Gihring, T.; Moser, D.P.; Lin, L.-H.; Davidson, M.; Onstott, T.C.; Morgan, L.; Milleson, M.; Kieft, T.L.; Trimarco, E.; Balkwill, D.L.; et al. The Distribution of Microbial Taxa in the Subsurface Water of the Kalahari Shield, South Africa. Geomicrobiol. J. 2006, 23, 415–430. [Google Scholar] [CrossRef]
- Morozova, D.; Zettlitzer, M.; Let, D.; Würdemann, H. Monitoring of the microbial community composition in deep subsurface saline aquifers during CO2 storage in Ketzin, Germany. Energy Procedia 2011, 4, 4362–4370. [Google Scholar] [CrossRef]
- Gales, G.; Tsesmetzis, N.; Neria, I.; Alazard, D.; Coulon, S.; Lomans, B.P.; Morin, D.; Ollivier, B.; Borgomano, J.; Joulian, C. Preservation of ancestral Cretaceous microflora recovered from a hypersaline oil reservoir. Sci. Rep. 2016, 6, 22960. [Google Scholar] [CrossRef] [PubMed]
- Arahal, D.R.; García, M.T.; Vargas, C.; Cánovas, D.; Nieto, J.J.; Ventosa, A. Chromohalobacter salexigens sp. nov., a moderately halophilic species that includes Halomonas elongata DSM 3043 and ATCC. Int. J. Syst. Evol. Microbiol. 2001, 51, 1457. [Google Scholar] [CrossRef]
- Wang, Y.-X.; Li, Y.-P.; Liu, J.-H.; Xiaoqian, L.; Lai, Y.-H.; Li, Z.-Y.; Ding, Z.-G.; Wen, M.-L.; Cui, X.-L. Gracilimonas mengyeensis sp. nov., a moderately halophilic bacterium isolated from a salt mine in Yunnan, south-western China. Int. J. Syst. Evol. Microbiol. 2013, 63, 3989–3993. [Google Scholar] [CrossRef]
- Adley, C.C.; Ryan, M.P.; Pembroke, J.T.; Saieb, F.M. Ralstonia pickettii: Biofilm formation in high purity water. In Biofilms: Persistence and Ubiquity; McBain, A.J., Allison, D.G., Pratten, J., Spratt, D., Upton, M., Verran, J., Eds.; Biofilm Club: Cardiff, UK, 2005; pp. 261–271. [Google Scholar]
| FW12299 Drilled 29-May-2007 | FW12287A Drilled 30-April-2007 | ||||||
|---|---|---|---|---|---|---|---|
| 27 August 2007 | 12 January 2010 | 21 October 2010 | 29 February 2012 | 14 June 2012 | 27 August 2007 | 21 October 2010 | |
| He (vol %) | 2.54 | 2.62 | 2.39 | 2.47 | |||
| H2 (vol %) | 4.62 | 3.97 | 3.19 | 10.67 | |||
| O2 (vol %) | 0.12 | <0.05 | <0.05 | <0.05 | |||
| N2 (vol %) | 14.9 | 15.3 | 14.6 | 12.7 | |||
| CH4 (vol %) | 71.5 | 70.3 | 71.9 | 72.2 | |||
| C2H6 (vol %) | ND | 6.47 | 6.62 | 6.81 | ND | 6.56 | ND |
| C3H8 (vol %) | 0.85 | 0.84 | 0.83 | 0.61 | |||
| i-C4H10 (vol %) | 0.07 | 0.07 | 0.07 | 0.04 | |||
| n-C4H10 (vol %) | 0.17 | 0.15 | 0.14 | 0.1 | |||
| i-C5H12 (vol %) | 0.05 | 0.06 | 0.05 | 0.03 | |||
| n-C5H12 (vol %) | 0.03 | 0.04 | 0.03 | 0.02 | |||
| Water flow rate (mL/min) | 770 | 172 | 258 | 170 | 150 | 79 | ≈5 |
| pH | 5.7 | 6.2 | 6 | 5.6 | 5.2 | 6.1 | 5.8 |
| T (°C) | 27.4 | 25.8 | 25.1 | 25.6 | 26.3 | 27.4 | 22.8 |
| Conductivity (mS/cm) | 161.1 | 141.3 | 149.6 | 146.8 | 150 | 109 | 139.9 |
| Sulfide (μM) | <2 | <2 | <2 | <2 | <2 | <2 | <2 |
| Sulfate (μM) | 123 | 97 | 126 | 194 | 198 | 109 | 184 |
| Genus | FW12322 (% of Total Sequences) | Closest Related Species in RefSeq 16S Microbial Database | Identity over Full 16S Gene (%) | Negative Control, DNA Extraction Blank (%) | Negative Control, Template Free PCR (%) | Notes |
|---|---|---|---|---|---|---|
| Chromohalobacter | 40.46 | Chromohalobacter salexigens | 96–99 | 0.01 | 0.23 | Strictly aerobic, halophilic heterotroph [74] |
| Aliifodinibius | 17.16 | Gracilimonas rosea | 88–89 | 0.02 | 0.14 | Non-motile tropical marine bacterium [56,75] |
| Propionibacterium | 3.20 | Propionibacterium acnes | 95–99 | 0.01 | 0.04 | Common skin bacterium (most likely introduced) [57] |
| Salinisphaera | 3.12 | Salinisphaera shabanensis | 94–96 | 0.01 | 0.00 | halophile from Red Sea [58] |
| Isosphaera | 2.96 | Isosphaera pallida | 87–89 | 0.15 | 0.01 | Filamentous budding bacterium from hot springs [59] |
| Cupriavidus | 2.31 | Cupriavidus basilensis | 94–98 | 0.00 | 0.00 | Heterotrophic soil bacterium [60] |
| Dokdonella | 1.76 | Dokdonella ginsengisoli | 97–99 | 0.00 | 0.00 | Heterotrophic soil bacterium [61] |
| Genus | FW12299 (% of Total Sequences) | Closest Related Species in RefSeq 16S Microbial Database | Identity over Full 16S Gene (%) | Negative Control, DNA Extraction Blank (%) | Negative Control, Template Free PCR (%) | Notes |
|---|---|---|---|---|---|---|
| Desulfotomaculum | 6.53 | Desulfotomaculum arcticum | 91–93 | 0.00 | 0.00 | Endospore-forming sulfate-reducing bacterium [62] |
| Propionibacterium | 5.45 | Propionibacterium acnes | 95–99 | 0.01 | 0.04 | Common skin bacterium (likely introduced) [57] |
| Marinobacter | 5.22 | Marinobacter gudaonensis | 95–97 | 0.01 | 0.03 | Halophile from oil-polluted saline soil [66] |
| Diaphorobacter | 3.73 | Diaphorobacter nitroreducens | 97–98 | 0.00 | 0.00 | Denitrifying bacterium that degrades simple hydrocarbons [65] |
| Fuchsiella | 3.66 | Fuchsiella ferrireducens | 91–92 | 0.01 | 0.01 | Homoacetogen capable of iron reduction [47] |
| Caenimonas | 3.32 | Caenimonas koreensis | 95–96 | 0.00 | 0.03 | Non-motile heterotroph from activated sludge [64] |
| Ralstonia | 3.26 | Ralstonia insidiosa | 96–99 | 0.00 | 0.01 | Strong biofilm producer (possible kit contaminant) [76] |
| Dokdonella | 1.56 | Dokdonella ginsengisoli | 97–99 | 0.00 | 0.00 | Heterotrophic soil bacterium [61] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wilpiszeski, R.L.; Sherwood Lollar, B.; Warr, O.; House, C.H. In Situ Growth of Halophilic Bacteria in Saline Fracture Fluids from 2.4 km below Surface in the Deep Canadian Shield. Life 2020, 10, 307. https://doi.org/10.3390/life10120307
Wilpiszeski RL, Sherwood Lollar B, Warr O, House CH. In Situ Growth of Halophilic Bacteria in Saline Fracture Fluids from 2.4 km below Surface in the Deep Canadian Shield. Life. 2020; 10(12):307. https://doi.org/10.3390/life10120307
Chicago/Turabian StyleWilpiszeski, Regina L., Barbara Sherwood Lollar, Oliver Warr, and Christopher H. House. 2020. "In Situ Growth of Halophilic Bacteria in Saline Fracture Fluids from 2.4 km below Surface in the Deep Canadian Shield" Life 10, no. 12: 307. https://doi.org/10.3390/life10120307
APA StyleWilpiszeski, R. L., Sherwood Lollar, B., Warr, O., & House, C. H. (2020). In Situ Growth of Halophilic Bacteria in Saline Fracture Fluids from 2.4 km below Surface in the Deep Canadian Shield. Life, 10(12), 307. https://doi.org/10.3390/life10120307






