Sensitivity Analysis of a 2D Stochastic Agent-Based and PDE Diffusion Model for Cancer-on-Chip Experiments
Abstract
1. Introduction
Motivation and Original Contribution of the Present Work
- Besides chemotactic migration, leukocytes also randomly explore the environment;
- Both leukocytes and tumor cells have an upper bound for their lifetimes (i.e., they have a limited number of iterations in the simulation in which they are considered to be active);
- The interaction between tumor cell agents and leukocyte agents leads to a reduction in the lifetime of the former (immune-mediated toxicity to the tumor cells);
- The width of the microchannels is marginally reduced to deny leukocyte movement along the y axis (in accordance with empirical observations);
- Even in the absence of annexin, leukocytes randomly explore the environment around them;
- The code is optimized, translated in Julia v1.8.3, and parallelized in order to vastly improve the performance of the algorithm and drastically reduce simulation time.
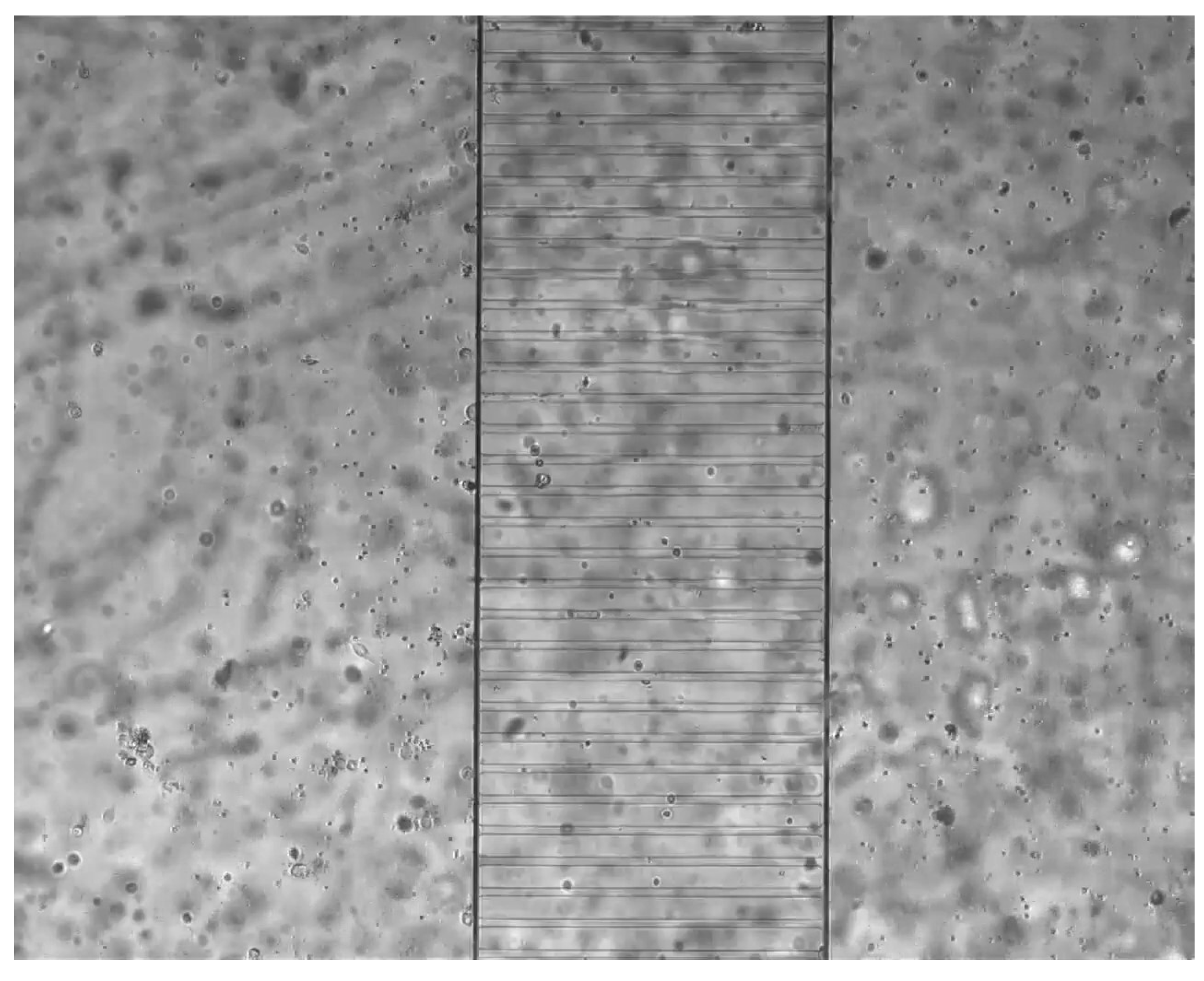
2. Biological Experiment
3. The Mathematical Model
3.1. Hybrid Agent-Based Cellular Automata Model
3.2. Innovation: Leukocyte and Tumor Cell Death
3.3. Innovation: Numerical Algorithm
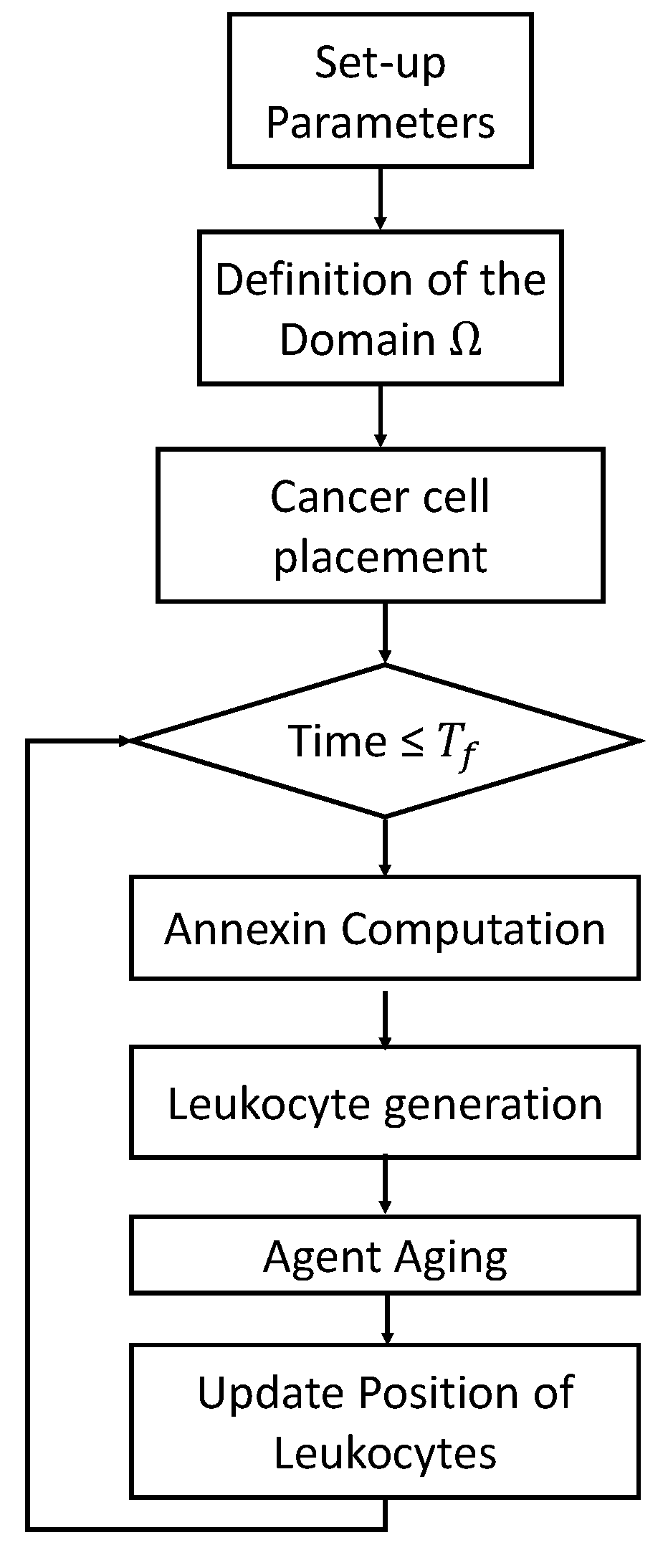
3.4. Computational Steps
- Solving Equation (3) in the discretized domain. If is the concentration of annexin at pixel p at each time step, we computewith being the concentration of annexin obtained after the sharing among the neighbors of pixel p, with being the Euclidean distance among the pixel p and its neighbors. Then, neighboring pixels receive the amount of annexin shared by pixel p: .Note that the production in the grid cells not occupied by the tumor is zero, while the elimination and diffusion of annexin in the environment occurs in each cell of the computational domain;
- Aging the agents to increase the age of both tumor cells and leukocytes;
- Randomly generating new leukocytes appearing in the right chamber;
- Determining the new leukocyte positions based on the calculated annexin concentration (as described in Section 3.1). In this step, it is also considered that if a leukocyte moves to the same location as a tumor cell, the age of that cancer cell increases, as shown in Equation (4).
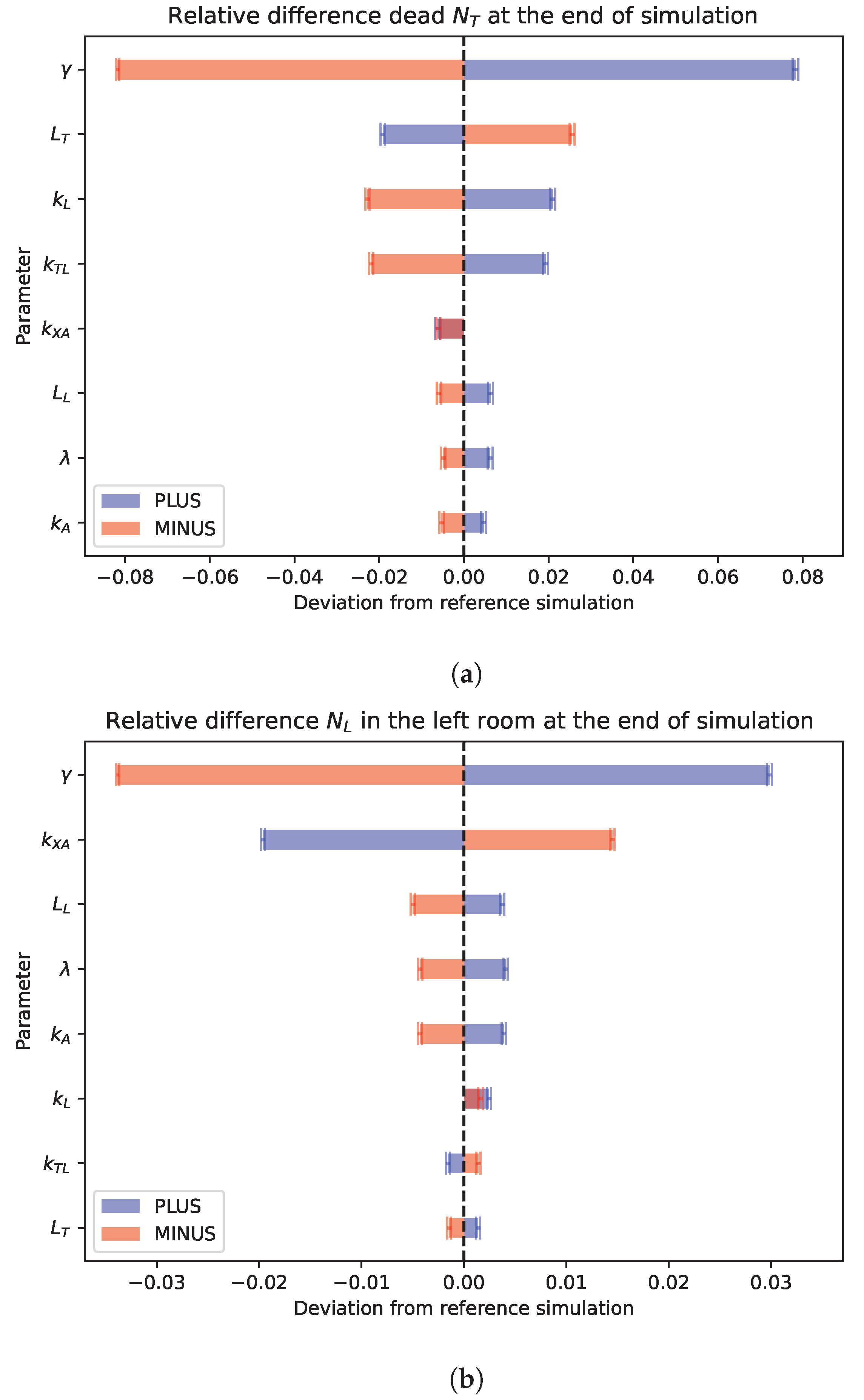
3.5. Sensitivity Analysis
- , the normalized rate of new leukocyte accrual;
- , the production rate of annexin;
- , the consumption rate of annexin;
- , the parameter enforcing migration towards high concentration;
- , the threshold value for migration;
- , the interaction coefficient between leukocytes and tumor cells;
- , the tumor cells lifetime;
- , the maximum leukocytes lifetime.
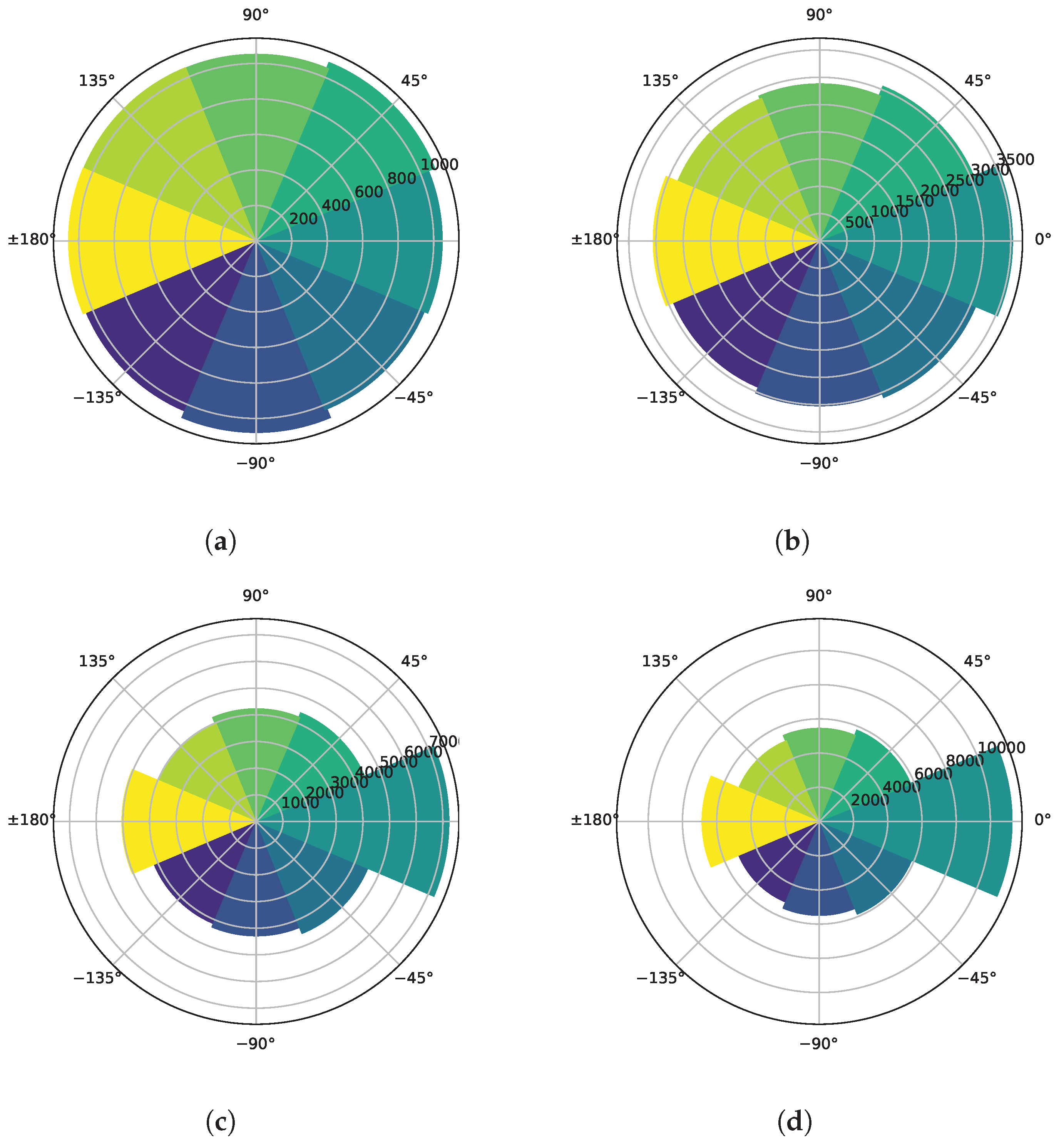
3.6. Preferred Direction
- The simulation of the migration of the leukocytes within the microfluidic chip environment.
- The identification of the leukocyte positions at different time intervals throughout the simulation.
- The computation of the “difference angle”, the angle between the displacement vector of each leukocyte and the local annexin gradient (this last identified as the direction of the neighboring cell with the highest annexin concentration; in cases where multiple neighboring cells contained the same amount of annexin, one of these cells was selected at random).
- The grouping of difference angles into different time intervals.
- The plotting of polar histograms to visualize the distribution of the difference angles for each time interval.
4. Results
5. Discussion
6. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
Abbreviations
| COC | Cancer-On-Chip |
References
- An, G.; Mi, Q.; Dutta-Moscato, J.; Vodovotz, Y. Agent-based models in translational systems biology. Wiley Interdiscip. Rev. Syst. Biol. Med. 2009, 1, 159–171. [Google Scholar] [CrossRef] [PubMed]
- Politopoulos, I. Review and Analysis of Agent-Based Models in Biology; University of Liverpool: Liverpool, UK, 2007. [Google Scholar]
- Lee, S.W.L.; Seager, R.J.; Litvak, F.; Spill, F.; Sieow, J.L.; Leong, P.H.; Kumar, D.; Tan, A.S.M.; Wong, S.C.; Adriani, G.; et al. Integrated in silico and 3D in vitro model of macrophage migration in response to physical and chemical factors in the tumor microenvironment. Integr. Biol. 2020, 12, 90–108. [Google Scholar] [CrossRef]
- Wolfram, S. A New Kind of Science; Wolfram Media Champaign: Champaign, IL, USA, 2002; Volume 5. [Google Scholar]
- Reher, D.; Klink, B.; Deutsch, A.; Voss-Böhme, A. Cell adhesion heterogeneity reinforces tumour cell dissemination: Novel insights from a mathematical model. Biol. Direct 2017, 12, 1–17. [Google Scholar] [CrossRef] [PubMed]
- Keller, E.F.; Segel, L.A. Initiation of slime mold aggregation viewed as an instability. J. Theor. Biol. 1970, 26, 399–415. [Google Scholar] [CrossRef] [PubMed]
- Hillen, T.; Painter, K.J. A user’s guide to PDE models for chemotaxis. J. Math. Biol. 2009, 58, 183–217. [Google Scholar] [CrossRef]
- Mohammadi, V.; Dehghan, M. A POD-RBF-FD scheme for simulating chemotaxis models on surfaces. Eng. Anal. Bound. Elem. 2022, 143, 316–330. [Google Scholar] [CrossRef]
- Ciarletta, P.; Hillen, T.; Othmer, H.; Preziosi, L.; Trucu, D.; Othmer, H.G. Cell-based, continuum and hybrid models of tissue dynamics. In Mathematical Models and Methods for Living Systems. Lecture Notes in Mathematics; Springer: Cham, Switzerland, 2016; pp. 1–72. [Google Scholar]
- Zygourakis, K.; Bizios, R.; Markenscoff, P. Proliferation of anchorage-dependent contact-inhibited cells: I. Development of theoretical models based on cellular automata. Biotechnol. Bioeng. 1991, 38, 459–470. [Google Scholar] [CrossRef]
- Lee, Y.; Kouvroukoglou, S.; McIntire, L.V.; Zygourakis, K. A cellular automaton model for the proliferation of migrating contact-inhibited cells. Biophys. J. 1995, 69, 1284–1298. [Google Scholar] [CrossRef]
- Cheng, G.; Youssef, B.B.; Markenscoff, P.; Zygourakis, K. Cell population dynamics modulate the rates of tissue growth processes. Biophys. J. 2006, 90, 713–724. [Google Scholar] [CrossRef][Green Version]
- Picioreanu, C.; Van Loosdrecht, M.C.; Heijnen, J.J. Mathematical modeling of biofilm structure with a hybrid differential-discrete cellular automaton approach. Biotechnol. Bioeng. 1998, 58, 101–116. [Google Scholar] [CrossRef]
- Kapellos, G.E.; Alexiou, T.S.; Payatakes, A.C. Hierarchical simulator of biofilm growth and dynamics in granular porous materials. Adv. Water Resour. 2007, 30, 1648–1667. [Google Scholar] [CrossRef]
- Shirinifard, A.; Gens, J.S.; Zaitlen, B.L.; Popławski, N.J.; Swat, M.; Glazier, J.A. 3D multi-cell simulation of tumor growth and angiogenesis. PLoS ONE 2009, 4, e7190. [Google Scholar] [CrossRef]
- Businaro, L.; De Ninno, A.; Schiavoni, G.; Lucarini, V.; Ciasca, G.; Gerardino, A.; Belardelli, F.; Gabrieleb, L.; Mattei, F. Cross talk between cancer and immune cells: Exploring complex dynamics in a microfluidic environment. Lab Chip 2013, 13, 229–239. [Google Scholar] [CrossRef] [PubMed]
- Vacchelli, E.; Ma, Y.; Baracco, E.E.; Sistigu, A.; Enot, D.P.; Pietrocola, F.; Yang, H.; Adjemian, S.; Chaba, K.; Semeraro, M.; et al. Chemotherapy-induced antitumor immunity requires formyl peptide receptor 1. Science 2015, 350, 972–978. [Google Scholar] [CrossRef]
- Biselli, E.; Agliari, E.; Barra, A.; Bertani, F.R.; Gerardino, A.; De Ninno, A.; Mencattini, A.; Di Giuseppe, D.; Mattei, F.; Schiavoni, G.; et al. Organs on chip approach: A tool to evaluate cancer-immune cells interactions. Sci. Rep. 2017, 7, 12737. [Google Scholar] [CrossRef]
- Comes, M.; Casti, P.; Mencattini, A.; DiGiuseppe, D.; Mermet-Meillon, F.; De Ninno, A.; Parrini, M.C.; Businaro, L.; Di Natale, C.; Martinelli, E.; et al. The influence of spatial and temporal resolutions on the analysis of cell-cell interaction: A systematic study for time-lapse microscopy applications. Sci. Rep. 2019, 9, 6789. [Google Scholar] [CrossRef]
- Gori, M.; Simonelli, M.C.; Giannitelli, S.M.; Businaro, L.; Trombetta, M.; Rainer, A. Investigating nonalcoholic fatty liver disease in a liver-on-a-chip microfluidic device. PLoS ONE 2016, 11, e0159729. [Google Scholar] [CrossRef]
- Paul, C.D.; Mistriotis, P.; Konstantopoulos, K. Cancer cell motility: Lessons from migration in confined spaces. Nat. Rev. Cancer 2017, 17, 131–140. [Google Scholar] [CrossRef]
- Braun, E.C.; Bretti, G.; Natalini, R. Mass-preserving approximation of a chemotaxis multi-domain transmission model for microfluidic chips. Mathematics 2021, 9, 688. [Google Scholar] [CrossRef]
- Braun, E.C.; Bretti, G.; Natalini, R. Parameter estimation techniques for a chemotaxis model inspired by Cancer-on-Chip (COC) experiments. Int. J.-Non-Linear Mech. 2022, 140, 103895. [Google Scholar] [CrossRef]
- Bretti, G.; DeNinno, A.; Natalini, R.; Peri, D.; Roselli, N. Estimation Algorithm for a Hybrid PDE–ODE Model Inspired by Immunocompetent Cancer-on-Chip Experiment. Axioms 2021, 10, 243. [Google Scholar] [CrossRef]
- Bretti, G.; DeGaetano, A. An Agent-Based Interpretation of Leukocyte Chemotaxis in Cancer-on-Chip Experiments. Mathematics 2022, 10, 1338. [Google Scholar] [CrossRef]
- Mattei, F.; Andreone, S.; Mencattini, A.; DeNinno, A.; Businaro, L.; Martinelli, E.; Schiavoni, G. Oncoimmunology meets organs-on-chip. Front. Mol. Biosci. 2021, 8, 627454. [Google Scholar] [CrossRef] [PubMed]
- Parlato, S.; DeNinno, A.; Molfetta, R.; Toschi, E.; Salerno, D.; Mencattini, A.; Romagnoli, G.; Fragale, A.; Roccazzello, L.; Buoncervello, M.; et al. 3D Microfluidic model for evaluating immunotherapy efficacy by tracking dendritic cell behaviour toward tumor cells. Sci. Rep. 2017, 7, 1093. [Google Scholar] [CrossRef]
- Murray, J.D. Mathematical Biology II: Spatial Models and Biomedical Applications; Springer: New York, NY, USA, 2001; Volume 3. [Google Scholar]
- Wiśniewski, J.R.; Hein, M.Y.; Cox, J.; Mann, M. A “proteomic ruler” for protein copy number and concentration estimation without spike-in standards. Mol. Cell. Proteom. 2014, 13, 3497–3506. [Google Scholar] [CrossRef] [PubMed]
- Boulter, E.; Grall, D.; Cagnol, S.; Van Obberghen-Schilling, E. Regulation of cell-matrix adhesion dynamics and Rac-1 by integrin linked kinase. FASEB J. 2006, 20, 1489–1491. [Google Scholar] [CrossRef]
- Bagge, U.; Gustav, V.; Gaehtgens, P. White Blood Cells: Morphology and Rheology as Related to Function; Springer Science & Business Media: Berlin/Heidelberg, Germany, 2013; Volume 1. [Google Scholar]
- Kapellos, G.E.; Alexiou, T.S. Modeling momentum and mass transport in cellular biological media: From the molecular to the tissue scale. In Transport in Biological Media; Elsevier: Amsterdam, The Netherlands, 2013; pp. 1–40. [Google Scholar]
- McCabe, C.; Paulden, M.; Awotwe, I.; Sutton, A.; Hall, P. One-Way Sensitivity Analysis for Probabilistic Cost-Effectiveness Analysis: Conditional Expected Incremental Net Benefit. Pharmacoeconomics 2020, 38, 135–141. [Google Scholar] [CrossRef]
- Reilly, T. Sensitivity Analysis for Dependent Variables. Decis. Sci. 2000, 31, 551–572. [Google Scholar] [CrossRef]
- Byrne, H.M.; Cave, G.; Mcelwain, D.L.S. The effect of chemotaxis and chemokinesis on leukocyte locomotion: A new interpretation of experimental results. Math. Med. Biol. J. IMA 1998, 15, 235–256. [Google Scholar] [CrossRef]
- Byrne, H.M.; Owen, M.R. A new interpretation of the Keller-Segel model based on multiphase modelling. J. Math. Biol. 2004, 49, 604–626. [Google Scholar] [CrossRef]

| Parameter | Description | Units | Value | Ref. |
|---|---|---|---|---|
| discretization time step | 4 | design | ||
| discretization along the x axis | m | 10 | design | |
| discretization along the y axis | m | 10 | design | |
| D | diffusivity of chemoattractant | m | [28] | |
| production rate of chemicals | 40 | calibration | ||
| consumption rate of chemicals | calibration | |||
| threshold value for migration | - | 2 | calibration | |
| parameter enforcing migration towards high concentration | - | 2 | calibration | |
| length of the channels | m | 500 | datum | |
| horizontal size of the box | m | 1704 | datum | |
| vertical size of the box | m | 1560 | datum | |
| number of microchannels in the video footage | - | 31 | datum | |
| width of each microchannel | m | 8 | datum | |
| width of obstacles | m | 32 | datum | |
| observation time | datum | |||
| radius of leukocytes | m | 4 | [29] | |
| radius of tumor cells | m | 10 | [30] | |
| number of tumor cells | m | 31 | datum | |
| normalized rate of new leukocyte accrual | calibration | |||
| number of frames in the laboratory experiment | - | 1440 | datum | |
| tumor cells life reducing by leukocytes | 1600 | calibration | ||
| tumor cells lifetime | calibration | |||
| maximum leukocytes lifetime | [31] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Pompa, M.; Torre, D.; Bretti, G.; De Gaetano, A. Sensitivity Analysis of a 2D Stochastic Agent-Based and PDE Diffusion Model for Cancer-on-Chip Experiments. Axioms 2023, 12, 930. https://doi.org/10.3390/axioms12100930
Pompa M, Torre D, Bretti G, De Gaetano A. Sensitivity Analysis of a 2D Stochastic Agent-Based and PDE Diffusion Model for Cancer-on-Chip Experiments. Axioms. 2023; 12(10):930. https://doi.org/10.3390/axioms12100930
Chicago/Turabian StylePompa, Marcello, Davide Torre, Gabriella Bretti, and Andrea De Gaetano. 2023. "Sensitivity Analysis of a 2D Stochastic Agent-Based and PDE Diffusion Model for Cancer-on-Chip Experiments" Axioms 12, no. 10: 930. https://doi.org/10.3390/axioms12100930
APA StylePompa, M., Torre, D., Bretti, G., & De Gaetano, A. (2023). Sensitivity Analysis of a 2D Stochastic Agent-Based and PDE Diffusion Model for Cancer-on-Chip Experiments. Axioms, 12(10), 930. https://doi.org/10.3390/axioms12100930









