The Cramér–Von Mises Statistic for Continuous Distributions: A Monte Carlo Study for Calculating Its Associated Probability †
Abstract
1. Introduction
Literature Survey
- For :
- For :
- For :
2. Materials and Methods
- Extracting both and from : 25%; extracting both and from : 25%; extracting one (either or ) from and one (either or ) from : 50%.
- If four extractions are needed, both and should be from in one extraction, in another, both and are from , and in two extractions, one (either or ) from and one (either or ) from : 50% should be extracted.
- Going further, an extraction from is equivalent to an extraction from , at which 0.5 is added.
| Algorithm 1 MC experiment for CM (MC-CM) |
3. Results and Discussions
3.1. Raw Data
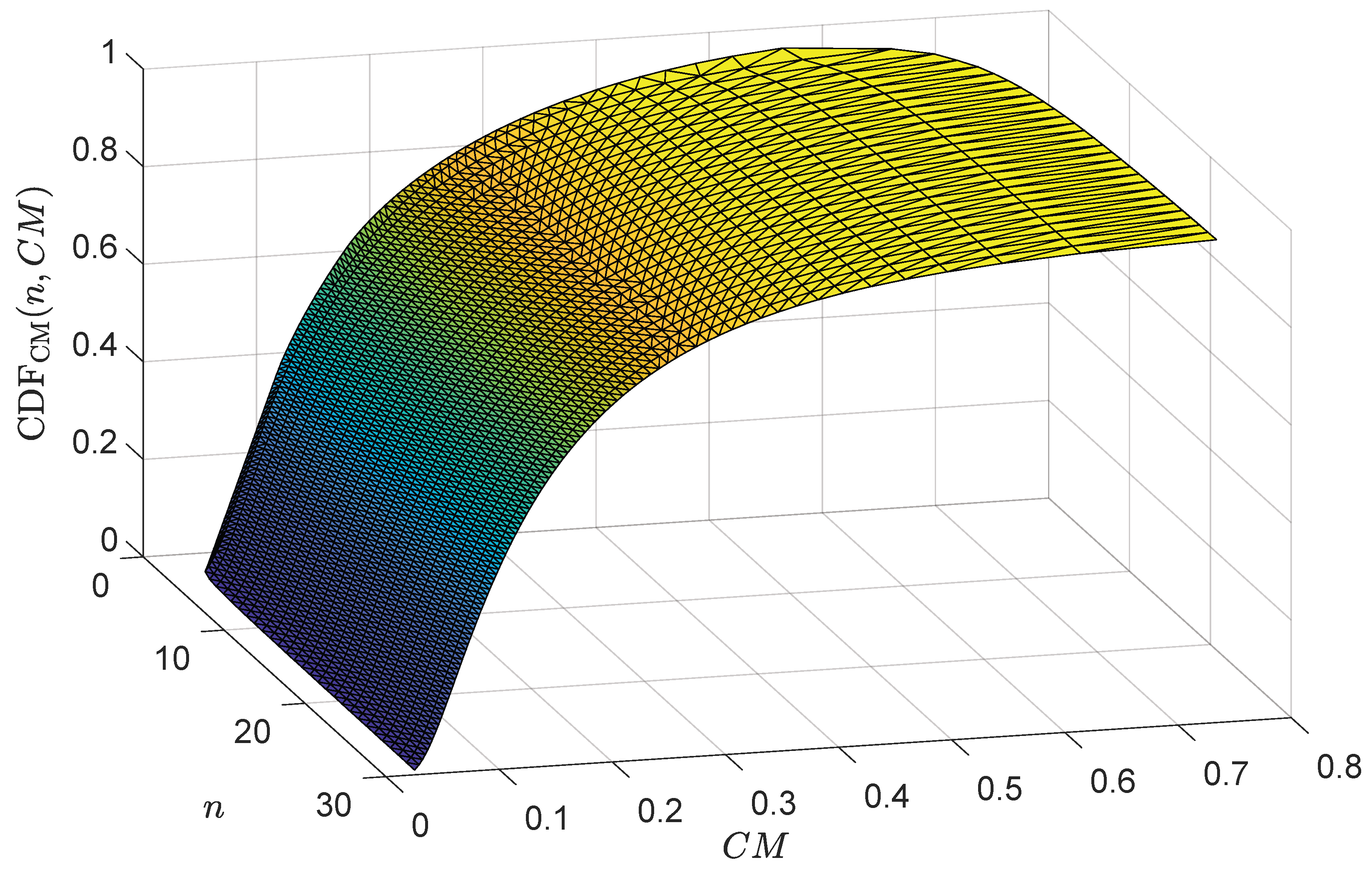
3.2. A Nonlinear Model for CM: Using Its Inverse for Calculating the Associated Probability
- The model giving the value of the CM population statistic () as a function of the cumulative probability (q) and sample size (n) uses Equation (13) (Model 1):
- the Model giving the value of the CM population statistic () as a function of the p-value () and sample size (n) uses Equation (14) (Model 2):
3.3. New Samples Analyzed with CM
3.3.1. Case Study on Beta Distribution
- Parameter estimation was conducted with MLE, so the number of degrees of freedom is equal to the sample size in each instance;
- The agreement between statistics is remarkable on average: and —this is the expected result, since one should expect both statistics to perform the same on random picked datasets. Student’s t-test does not discriminate between the two series of probabilities: there is a 98% likelihood to belong to the same population (one can test for paired probabilities, when the matching probability is somewhat lower, 92%);
- At a significance level of 5%, all samples passed the hypothesis that they are possibly drawn from the Beta distribution (it was not possible to reject the null hypothesis that the Beta distribution is not the population from which they were drawn).
3.3.2. Case Study on Cauchy Distribution
- Parameter estimation was conducted with MLE, so the number of degrees of freedom is equal to the sample size in each instance;
- The agreement between statistics is remarkable on average: and —this is the expected result since one should expect both statistics to perform the same on random picked datasets. Student’s t-test does not discriminate between the two series of probabilities: there is a 99% likelihood of belonging to the same population (one can test for paired probabilities when the matching probability is lower, 80%);
- At a significance level of 1%, there is some disagreement between the CM and KS statistics at Wheaton river sample if the hypothesis of being drawn from the Cauchy distribution must be rejected or not.
4. Conclusions
5. Future Work
Funding
Data Availability Statement
Conflicts of Interest
Abbreviations
| CDF | Cumulative distribution function |
| Statistical significance | |
| q | Cumulative probability |
| p-value | Complement of the cumulative probability () |
| CM | Cramér–von Mises test |
| Cramér–von Mises sample statistic (value, variable) | |
| CMP | Cramér–von Mises population statistic |
| CI | Confidence interval |
| MLE | Maximum likelihood estimation |
| MC | Monte Carlo |
| PHP | Recursive acronym for hypertext preprocessor, programming language |
| The coefficient of determination (the square of the sample correlation coefficient) | |
| Rand | A random number from standard uniform distribution |
| Sort | A function sorting ascending an numeric array |
| Beta | Beta distribution |
| Cauchy | Cauchy distribution |
Appendix A. Analitical Formulas for Probability Distribution Functions Used for the Analysis of the Data Samples from Literature
Appendix A.1. Beta Distribution
Appendix A.2. Cauchy Distribution
References
- Schloss, P.D.; Larget, B.R.; Handelsman, J. Integration of microbial ecology and statistics: A test to compare gene libraries. Appl. Environ. Microbiol. 2004, 70, 5485–5492. [Google Scholar] [CrossRef] [PubMed]
- Qiu, X.; Xiao, Y.; Gordon, A.; Yakovlev, A. Assessing stability of gene selection in microarray data analysis. BMC Bioinform. 2006, 7, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Merkle, E.C.; Zeileis, A. Tests of measurement invariance without subgroups: A generalization of classical methods. Psychometrika 2013, 78, 59–82. [Google Scholar] [CrossRef] [PubMed]
- Arshad, M.Z.; Iqbal, M.Z.; Were, F.; Aldallal, R.; Riad, F.H.; Bakr, M.E.; Tashkandy, Y.A.; Hussam, E.; Gemeay, A.M. An alternative statistical model to analysis pearl millet (Bajra) yield in province Punjab and Pakistan. Complexity 2023, 2023, 8713812. [Google Scholar] [CrossRef]
- Alqasem, O.A.; Nassar, M.; Abd Elwahab, M.E.; Elshahhat, A. A new inverted Pham distribution for data modeling of mechanical components and diamond in South-West Africa. Phys. Scr. 2024, 99, 115268. [Google Scholar] [CrossRef]
- Chaudhary, A.K.; Telee, L.B.S.; Karki, M.; Kumar, V. Statistical analysis of air quality dataset of Kathmandu, Nepal, with a New Extended Kumaraswamy Exponential Distribution. Environ. Sci. Pollut. Res. 2024, 31, 21073–21088. [Google Scholar] [CrossRef]
- Wang, C.; Zhu, H. Tests of fit for the power function lognormal distribution. PLoS ONE 2024, 19, e0298309. [Google Scholar] [CrossRef]
- Chau, T.T.; Nguyen, T.T.H.; Nguyen, L.; Do, T.D. Wind Speed Probability Distribution Based on Adaptive Bandwidth Kernel Density Estimation Model for Wind Farm Application. Wind Energy 2025, 28, e2970. [Google Scholar] [CrossRef]
- Chotsiri, P.; Yodsawat, P.; Hoglund, R.M.; Simpson, J.A.; Tarning, J. Pharmacometric and statistical considerations for dose optimization. CPT Pharmacometrics Syst. Pharmacol. 2025, 2025, 279–291. [Google Scholar] [CrossRef]
- Anderson, T.; Darling, D. Asymptotic theory of certain ’goodness-of-fit’ criteria based on stochastic processes. Ann. Math. Stat. 2006, 23, 193–212. [Google Scholar] [CrossRef]
- Anderson, T.; Darling, D. A test of goodness of fit. J. Am. Stat. Assoc. 1954, 49, 193–212. [Google Scholar] [CrossRef]
- Kolmogoroff, A. Confidence limits for an unknown distribution function. Ann. Math. Stat. 1941, 12, 461–463. [Google Scholar] [CrossRef]
- Smirnov, N. Table for estimating the goodness of fit of empirical distributions. Ann. Math. Stat. 1948, 19, 279–281. [Google Scholar] [CrossRef]
- Cramér, H. On the composition of elementary errors. Scand. Actuar. J. 1928, 1, 13–74. [Google Scholar] [CrossRef]
- VonMises, R. Wahrscheinlichkeit, Statistik und Wahrheit; Julius Springer: Berlin, Germany, 1928. [Google Scholar] [CrossRef]
- Traison, T.; Vaidyanathan, V. Goodness-of-Fit Tests for COM-Poisson Distribution Using Stein’s Characterization. Austrian J. Stat. 2025, 54, 85–100. [Google Scholar] [CrossRef]
- Muhammad, M.; Abba, B. A Bayesian inference with Hamiltonian Monte Carlo (HMC) framework for a three-parameter model with reliability applications. Kuwait J. Sci. 2025, 52, 100365. [Google Scholar] [CrossRef]
- Kumar, S.A.; Sridhar, A.; Rekha, S.; Nagarjuna, V.B.; Ramanaiah, M. Comparative Performance of Burr Type XII 3P, Dagum Type I 3P and Log-Logistic 3P Distributions in Modeling Ozone (O3), PM10 and PM2.5 Concentrations. Res. J. Chem. Environ. 2025, 29, 39–56. [Google Scholar] [CrossRef]
- Singh Nayal, A.; Ramos, P.L.; Tyagi, A.; Singh, B. Improving inference in exponential logarithmic distribution. Commun. -Stat.-Simul. Comput. 2024, 1–25, Online first. [Google Scholar] [CrossRef]
- Singh, B.; Tyagi, S.; Singh, R.P.; Tyagi, A. Modified Topp-Leone Distribution: Properties, Classical and Bayesian Estimation with Application to COVID-19 and Reliability Data. Thail. Stat. 2025, 23, 72–96. [Google Scholar]
- Ibrahim, M.; Shah, M.K.A.; Ahsan-ul Haq, M. New two-parameter XLindley distribution with statistical properties, simulation and applications on lifetime data. Int. J. Model. Simul. 2025, 45, 293–306. [Google Scholar] [CrossRef]
- Sunusi, N.; Auliana, N.H. Assessing SPI and SPEI for drought forecasting through the power law process: A case study in South Sulawesi, Indonesia. MethodsX 2025, 14, 103235. [Google Scholar] [CrossRef]
- Chen, Y.; Ding, T.; Wang, X.; Zhang, Y. A robust and powerful metric for distributional homogeneity. Stat. Neerl. 2025, 79, e12370. [Google Scholar] [CrossRef]
- Fisher, R.A. On the mathematical foundations of theoretical statistics. Philos. Trans. R. Soc. Lond. Ser. Contain. Pap. Math. Phys. Character 1922, 222, 309–368. [Google Scholar] [CrossRef]
- Fisher, R.A. Theory of statistical estimation. Math. Proc. Camb. Philos. Soc. 1925, 22, 700–725. [Google Scholar] [CrossRef]
- Votaw, D.F., Jr. Testing compound symmetry in a normal multivariate distribution. Ann. Math. Stat. 1948, 19, 447–473. [Google Scholar] [CrossRef]
- Srinivasan, R.; Godio, L. A Cramér-von Mises type statistic for testing symmetry. Biometrika 1974, 61, 196–198. [Google Scholar] [CrossRef]
- Gregory, G.G. Cramer-von Mises type tests for symmetry. South Afr. Stat. J. 1977, 11, 49–61. [Google Scholar]
- Koziol, J.A. On a Cramér-von Mises-type statistic for testing symmetry. J. Am. Stat. Assoc. 1980, 75, 161–167. [Google Scholar] [CrossRef]
- Aki, S. Asymptotic distribution of a Cramér-von Mises type statistic for testing symmetry when the center is estimated. Ann. Inst. Stat. Math. 1981, 33, 1–14. [Google Scholar] [CrossRef]
- Csörgo, S.; Faraway, J.J. The exact and asymptotic distributions of Cramér-von Mises statistics. J. R. Stat. Soc. Ser. B Stat. Methodol. 1996, 58, 221–234. [Google Scholar] [CrossRef]
- Abramowitz, M.; Stegun, I.A.; Miller, D. Handbook of Mathematical Functions with Formulas, Graphs and Mathematical Tables (National Bureau of Standards Applied Mathematics Series No. 55); U. S. Gov. Printing Office: Washington, DC, USA, 1965. [Google Scholar]
- Álvarez-Liébana, J.; López-Pérez, A.; González-Manteiga, W.; Febrero-Bande, M. A goodness-of-fit test for functional time series with applications to Ornstein-Uhlenbeck processes. Comput. Stat. Data Anal. 2025, 203, 108092. [Google Scholar] [CrossRef]
- Matsumoto, M.; Nishimura, T. Mersenne twister: A 623-dimensionally equidistributed uniform pseudo-random number generator. ACM Trans. Model. Comput. Simul. (TOMACS) 1998, 8, 3–30. [Google Scholar] [CrossRef]
- Variance Reduction. In Stochastic Simulation; Ripley, B.D., Ed.; John Wiley & Sons, Ltd.: Hoboken, NJ, USA, 1987; Chapter 5; pp. 118–141. [Google Scholar] [CrossRef]
- Walker, A.J. New fast method for generating discrete random numbers with arbitrary frequency distributions. Electron. Lett. 1974, 10, 127–128. [Google Scholar] [CrossRef]
- Azaïs, R.; Ferrigno, S.; Martinez, M.J. cvmgof: An R package for Cramér–von Mises goodness-of-fit tests in regression models. J. Stat. Comput. Simul. 2022, 92, 1246–1266. [Google Scholar] [CrossRef]
- Goldberg, D. What every computer scientist should know about floating-point arithmetic. ACM Comput. Surv. (CSUR) 1991, 23, 5–48. [Google Scholar] [CrossRef]
- Jäntschi, L.; Bolboacă, S.D. Performances of Shannon’s entropy statistic in assessment of distribution of data. Ovidius Univ. Ann. Chem. 2017, 28, 30–42. [Google Scholar] [CrossRef]
- Ungureanu, E.M.; Ștefaniu, A.; Isopescu, R.; Mușina, C.E.; Bujduveanu, M.R.; Jäntschi, L. Extended characteristic polynomial estimating the electrochemical behaviour of some 4-(azulen-1-yl)-2,6-divinylpyridine derivatives. J. Electrochem. Sci. Eng. 2025, 15, 2374. [Google Scholar] [CrossRef]
- AbouRizk, S.M.; Halpin, D.W.; Wilson, J.R. Fitting beta distributions based on sample data. J. Constr. Eng. Manag. 1994, 120, 288–305. [Google Scholar] [CrossRef]
- Raschke, M. Empirical behaviour of tests for the beta distribution and their application in environmental research. Stoch. Environ. Res. Risk Assess. 2011, 25, 79–89. [Google Scholar] [CrossRef]
- Jayakumar, K.; Fasna, K. On a new generalization of Cauchy distribution. Asian J. Stat. Sci. 2022, 2, 61–81. [Google Scholar]
- Rublik, F. A quantile goodness-of-fit test for Cauchy distribution, based on extreme order statistics. Appl. Math. 2001, 46, 339–351. [Google Scholar] [CrossRef][Green Version]
- Akinsete, A.; Famoye, F.; Lee, C. The beta-Pareto distribution. Statistics 2008, 42, 547–563. [Google Scholar] [CrossRef]
- Alzaatreh, A.; Lee, C.; Famoye, F.; Ghosh, I. The generalized Cauchy family of distributions with applications. J. Stat. Distrib. Appl. 2016, 3, 12. [Google Scholar] [CrossRef]
- Hassan, A.S.; Alsadat, N.; Elgarhy, M.; Chesneau, C.; Mohamed, R.E. Different classical estimation methods using ranked set sampling and data analysis for the inverse power Cauchy distribution. J. Radiat. Res. Appl. Sci. 2023, 16, 100685. [Google Scholar] [CrossRef]
- Fisher, R.A. Combining independent tests of significance. Am. Stat. 1948, 2, 30. [Google Scholar] [CrossRef]

| Is ( Otherwise) | Occurrence | ||||||
|---|---|---|---|---|---|---|---|
| i | 1 | … | j | … | n | ||
| 0 | … | 0 | 0 | … | 0 | ||
| … | … | … | … | … | … | … | … |
| 0 | … | 0 | 1 | … | 1 | ||
| … | … | … | … | … | … | … | … |
| 1 | … | 1 | 1 | … | 1 | ||
| Coeff. Number | Model 1 (Equation (13)) | Model 2 (Equation (14)) | ||
|---|---|---|---|---|
| Value | ±95% CI | Value | ±95% CI | |
| Sample Data | a | b | c | d | ||||
|---|---|---|---|---|---|---|---|---|
| Dozer cycle in [41] | 2.3798 | 3.2848 | 0.0882 | 2.1199 | 0.036765 | 0.947 | 0.50799 | 0.944 |
| Truck cycle in [41] | 1.8312 | 3.6563 | 6.7852 | 16.746 | 0.080000 | 0.692 | 0.72844 | 0.632 |
| May 2007 in [42] | 1.5356 | 0.8614 | 0.3200 | 0.9700 | 0.207980 | 0.253 | 0.90067 | 0.354 |
| May 2008 in [42] | 0.4855 | 0.6385 | 0.3900 | 0.9800 | 0.043692 | 0.915 | 0.63770 | 0.770 |
| First set in [43] | 3.2524 | 8.3550 | 0.4614 | 8.3534 | 0.054735 | 0.850 | 0.55456 | 0.899 |
| Second set in [43] | 3.5028 | −0.5478 | 0.369010 | 0.084 | 1.09460 | 0.169 |
| Sample Data | a | b | ||||
|---|---|---|---|---|---|---|
| Venus observations in [44] | 0.0267 | 0.2613 | 0.024332 | 0.9962 | 0.3710 | 0.9968 |
| Wheaton river in [45] | 7.0718 | 6.4622 | 0.705420 | 0.0125 | 2.0189 | 0.0005 |
| Floyd river in [45] | 2878.9 | 1928.8 | 0.448170 | 0.0509 | 1.2921 | 0.0609 |
| Guinea pigs survival in [46] | 139.32 | 48.138 | 0.451190 | 0.0504 | 1.2017 | 0.1004 |
| Nile river in [46] | 879.34 | 103.89 | 0.351690 | 0.0945 | 1.3117 | 0.0579 |
| Accelerated life tests in [47] | 6.8426 | 0.8896 | 0.069199 | 0.7591 | 0.6465 | 0.7657 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the author. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Jäntschi, L. The Cramér–Von Mises Statistic for Continuous Distributions: A Monte Carlo Study for Calculating Its Associated Probability. Symmetry 2025, 17, 1542. https://doi.org/10.3390/sym17091542
Jäntschi L. The Cramér–Von Mises Statistic for Continuous Distributions: A Monte Carlo Study for Calculating Its Associated Probability. Symmetry. 2025; 17(9):1542. https://doi.org/10.3390/sym17091542
Chicago/Turabian StyleJäntschi, Lorentz. 2025. "The Cramér–Von Mises Statistic for Continuous Distributions: A Monte Carlo Study for Calculating Its Associated Probability" Symmetry 17, no. 9: 1542. https://doi.org/10.3390/sym17091542
APA StyleJäntschi, L. (2025). The Cramér–Von Mises Statistic for Continuous Distributions: A Monte Carlo Study for Calculating Its Associated Probability. Symmetry, 17(9), 1542. https://doi.org/10.3390/sym17091542






