3.1. Evaluation Method
To evaluate the comprehensive performance of the model in terms of speed and accuracy, we developed a weighted evaluation method. In the experiments of this study, the final evaluation criteria for the model mainly included three aspects: MAE (Mean Absolute Error; this method quantifies the accuracy of model predictions by calculating the average of the absolute differences between actual values and predicted values),
(
commonly refers to the optimization of the
score), and the model’s training time. To make the final evaluation results more intuitive, this study used a comprehensive weighted evaluation as the final result of the model evaluation, weighting the training time and model accuracy in the same proportion, as shown in Formula (
1):
where
t represents the model training duration, indicating that the model training time significantly impacts the final model evaluation result. As time increases, the quality of the model gradually becomes influenced by the values of the training time. Formula (
2) was used for the MAE:
is a metric used to evaluate the balance between precision and recall in binary classification models. Formula (
3) was used for
s:
The higher the accuracy of the model training, the smaller the MAE value. Consequently, the value of × becomes larger, gradually approaching 0.25. The larger the value, the larger × becomes, gradually approaching 0.25. The longer the model training time, as training time increases, if there is no significant improvement in MAE and , the final result of f will primarily be influenced by training duration.
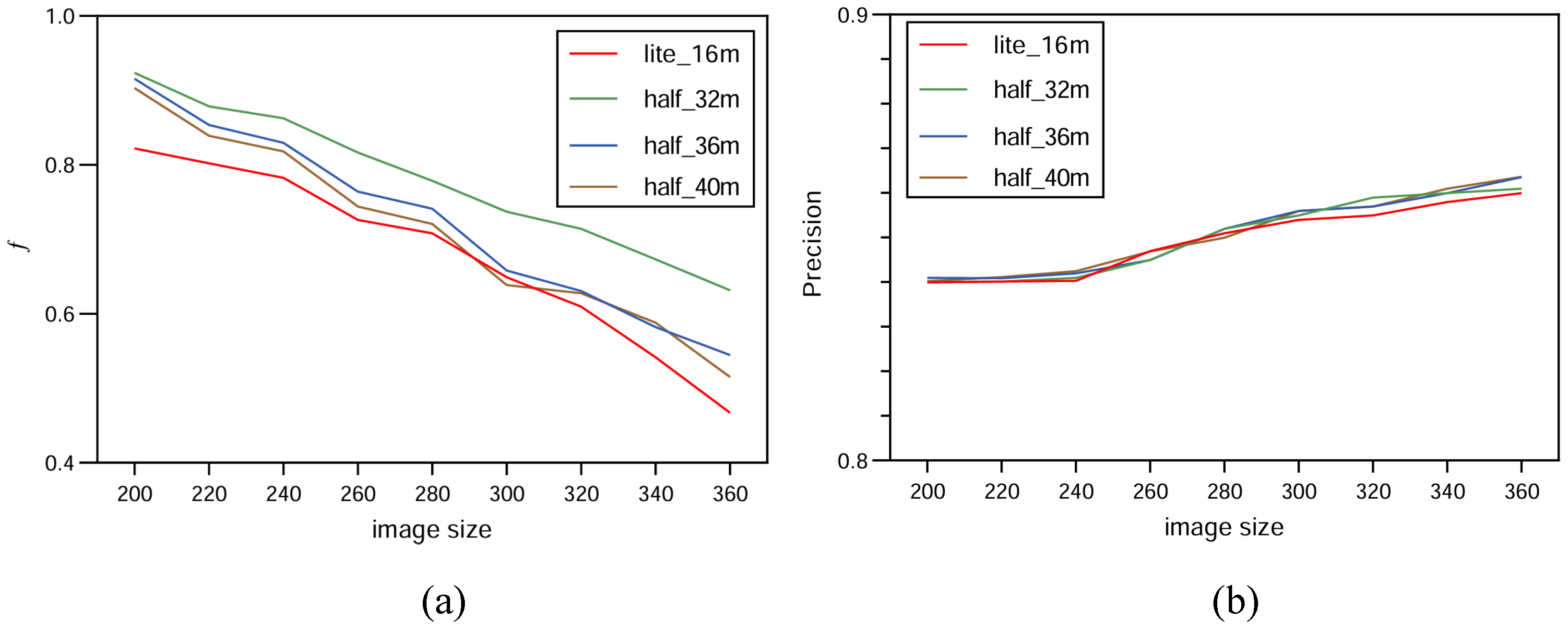
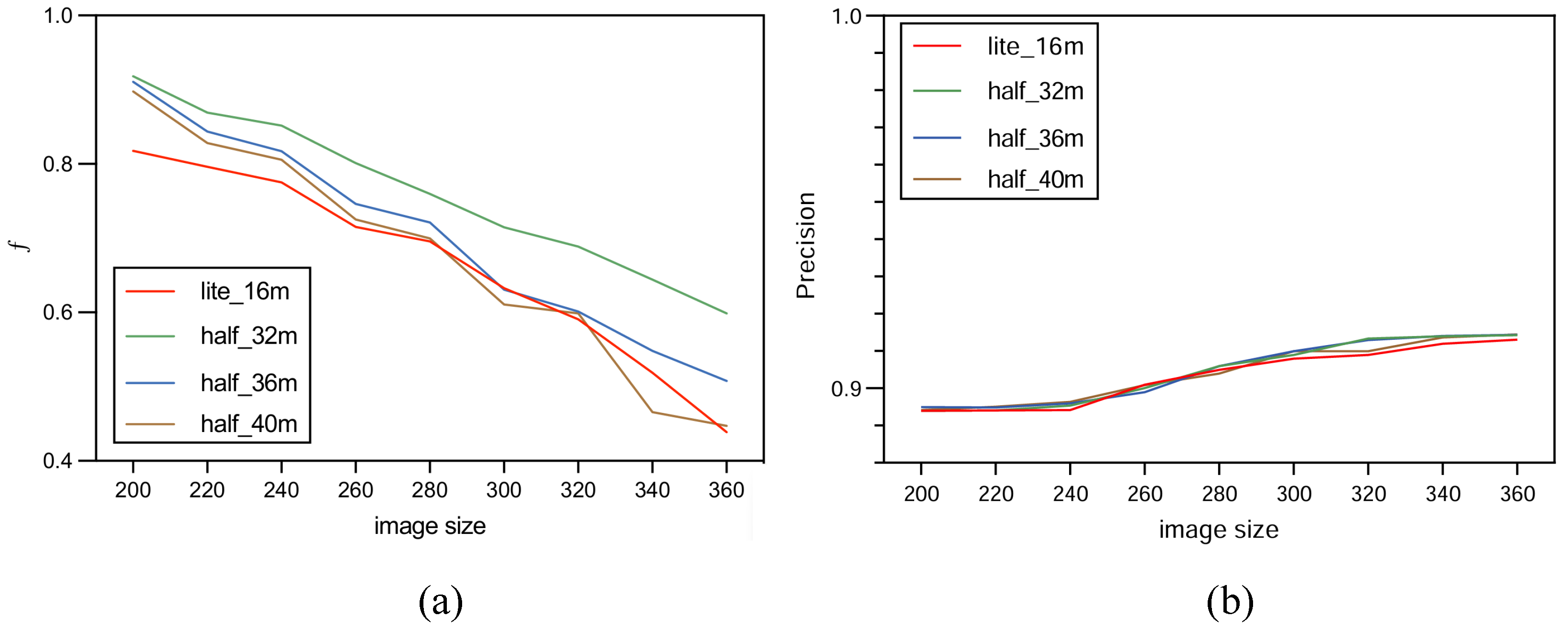
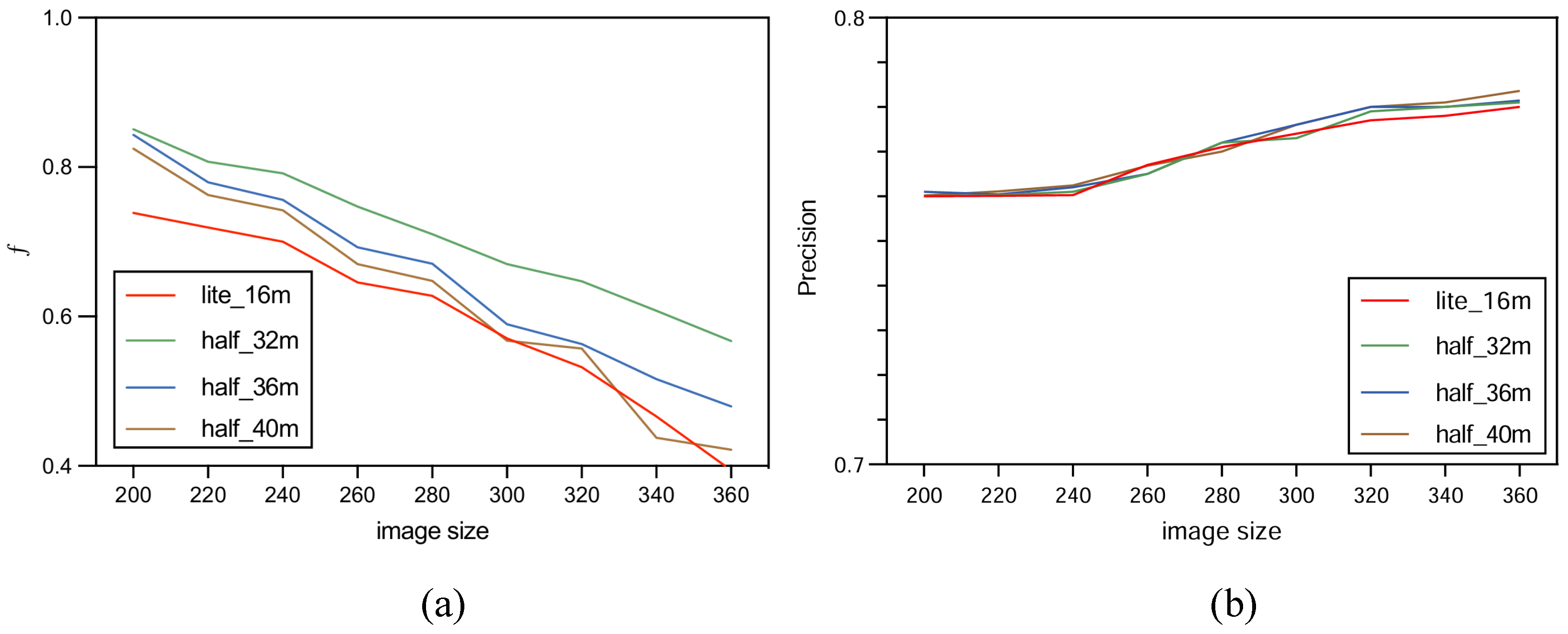
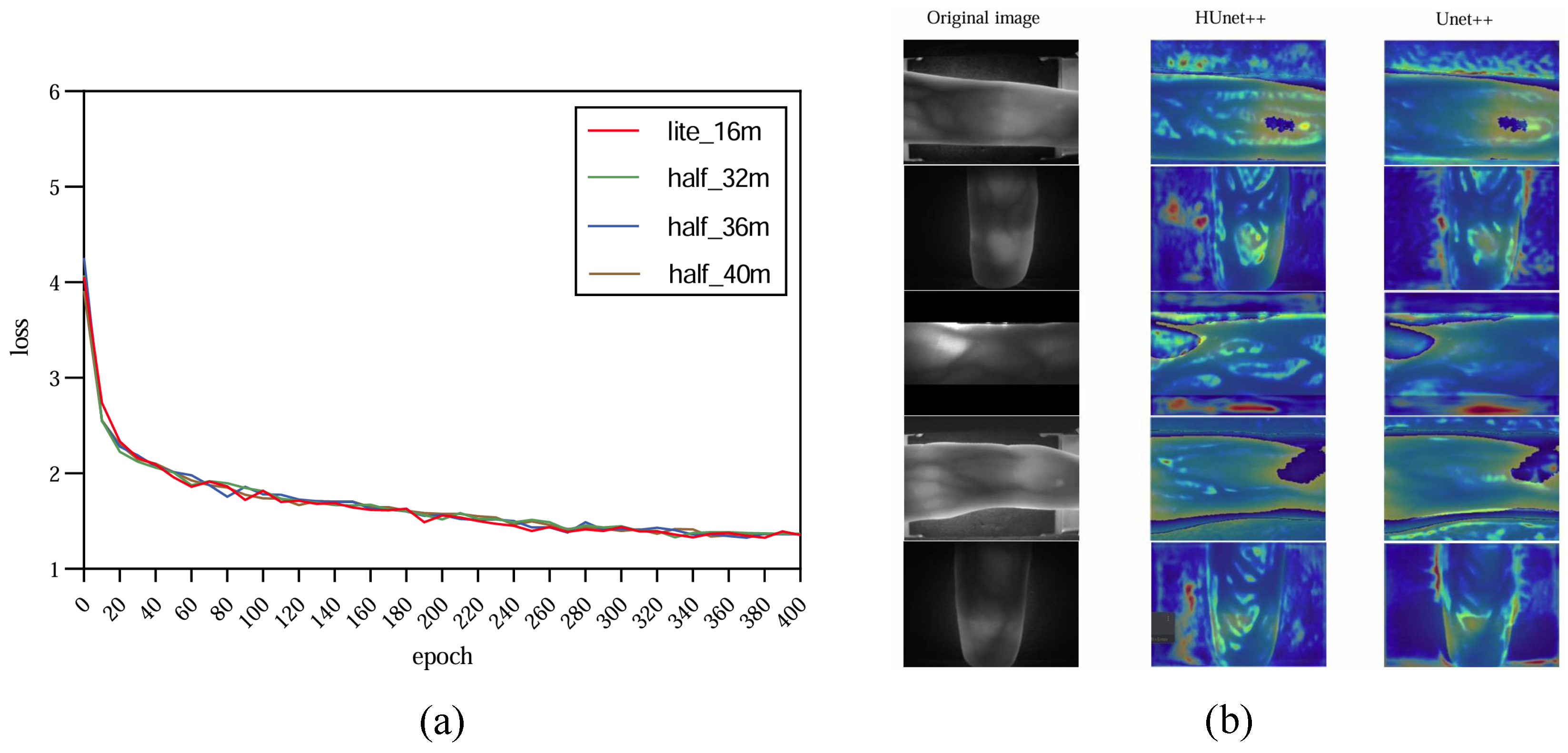
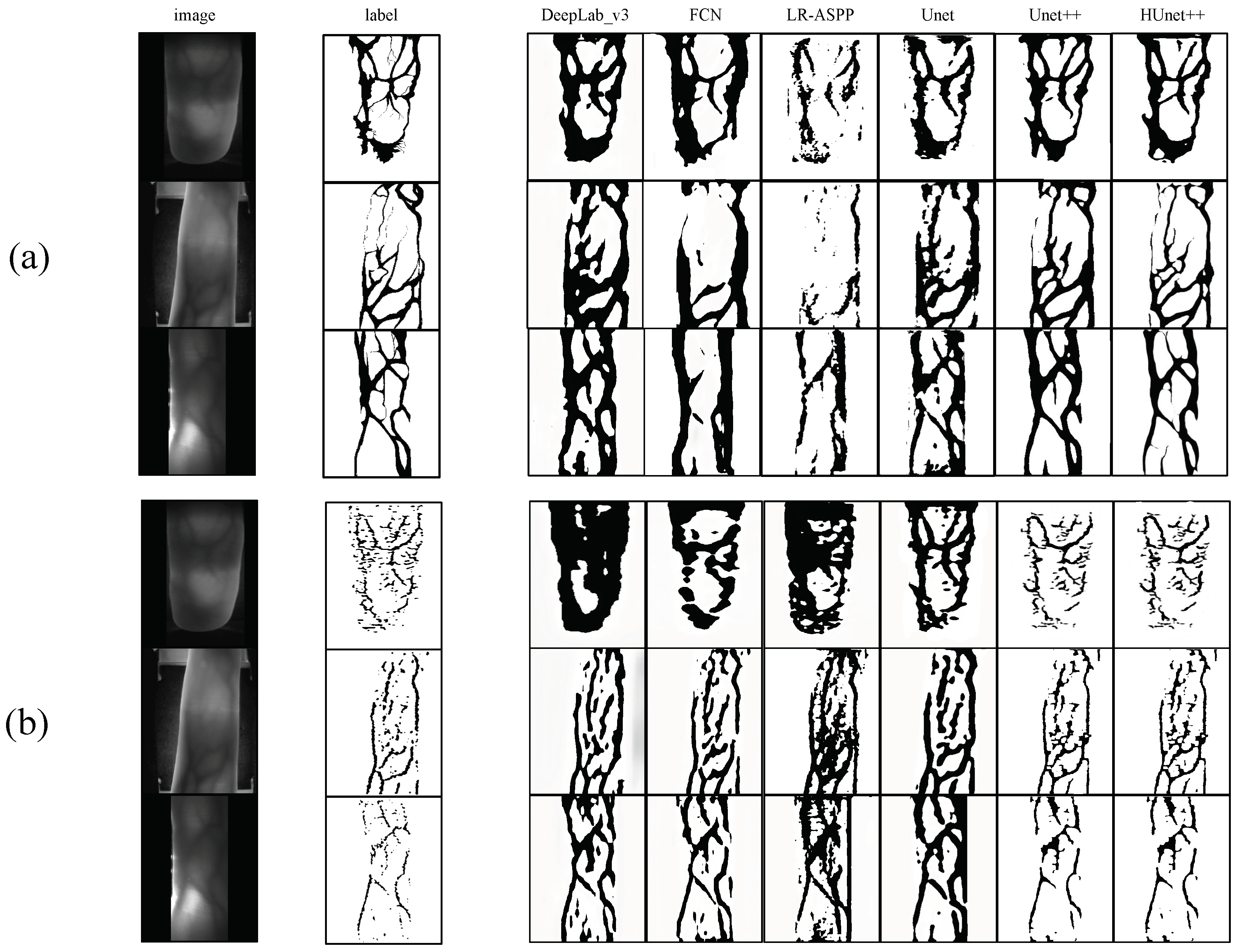
In order to increase confidence in the experiment, this study carried out multiple tests with HUnet++ and Unet++ on images of different sizes. Among these, HUnet++ was further divided into experiments with different hidden layers and compared with the lightweight version of Unet++. The final results are presented in the form of a line graph.
3.2. Data Preprocessing
The manual training set utilized in this study was formed by the fusion of three datasets, and manual annotation was conducted on the finger vein images within the combined dataset, comprising database of the University of Science, Malaysia (FV-USM) [
30], the Shandong University dataset [
31], and the self-built dataset (NCEPU-data). The composition of that dataset ensured the coverage of data with different sizes, lighting conditions, and resolutions, thereby further enhancing the model’s robustness. The training dataset included a total of 140 original images collected by finger vein devices and 140 annotated images corresponding to the original images, where the labels were binary images annotated manually. Out of these, 107 images were used for model training and the remaining 33 for testing the model. The specific processing workflow was as follows: Initially, all vein images were transformed into grayscale. Subsequently, based on the basis of the grayscale images, the vein regions were manually labeled as black (with a pixel value of 0), while the non-vein regions were designated as white (with a pixel value of 225). The aim of this preprocessing step was to minimize computational load during model execution by reducing the number of image channels. Before the model was trained, these images used for training and the labels used for model backpropagation underwent image normalization, data format conversion, and data augmentation operations. This then generated the data loader for subsequent model training.
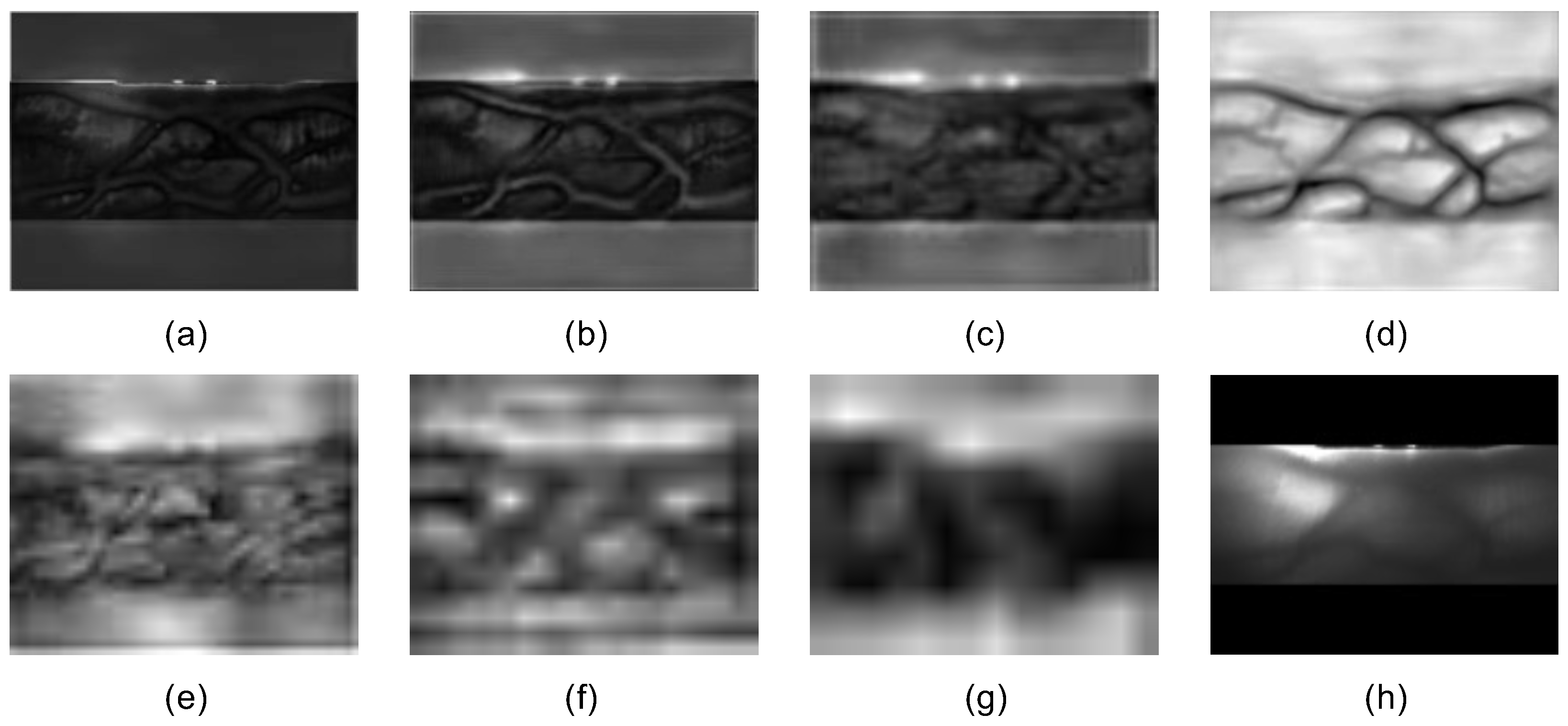
For labels annotated using traditional finger vein extraction methods, five finger vein images were first generated using Gabor, MC, PC, RLT, and WLD techniques, with vein regions set to pixel value 1 and background to 0. These images were added element-wise to produce a fusion image, where elements with values >3 were set to 255, and others to 0. Formula (
4) for fusing an image is shown below:
Median blur and morphological opening were then applied to connect sparse vein regions, removing residual noise and enhancing the model’s ability to learn vein features.
Figure 5 is the end result. During actual training, the vein region pixel values of all images were set to 0, while the background region was set to 255. This was carried out partly to maintain consistency with the manually annotated vein images and partly to ensure that the labels more closely resembled the original vein images. Based on the conclusions of Jalilian et al. [
16] and the scale of the manually annotated dataset, we set the scale of all traditional-method-annotated finger vein datasets used for model training to be consistent with that of the manually annotated dataset.
The finger vein masks extracted using traditional methods contained both explicit geometric features and implicit non-geometric features (i.e., the non-geometrization and discontinuity of vein patterns). To optimize the model’s performance, we applied a weighting strategy during the training process on datasets annotated by traditional methods. This approach involved increasing the weight of the foreground region and decreasing the weight of the background region. For each dataset, the final weights for the foreground and background regions were determined through experiments, with the specific weights varying according to the characteristics of each dataset.
3.3. Model Parameter Configuration
The experiment was performed on the Google Colab platform with the following configuration: GPU: Tesla K80; RAM: 16 GB; operating system: Ubuntu 22.04.2 LTS; system memory: 12.7 GB; system kernel: 5.15.120+; disk space: 78.2 GB. The environment was set up with CUDA 12.0 and driver version 525.105.17. During the model training phase, experiments were conducted using three variations of the HUnet++ model, each with different numbers of hidden layers. In each experiment, the number of hidden layers in the convolutional layer of the encoding and decoding blocks of HUnet++ were 32, 36, and 40, respectively. Each experiment was conducted on nine different image sizes. Lastly, one experiment was conducted with the hidden layers of the Unet++ (lightweight) model set to 16. In that experiment, training comparisons were also made on nine different image sizes.
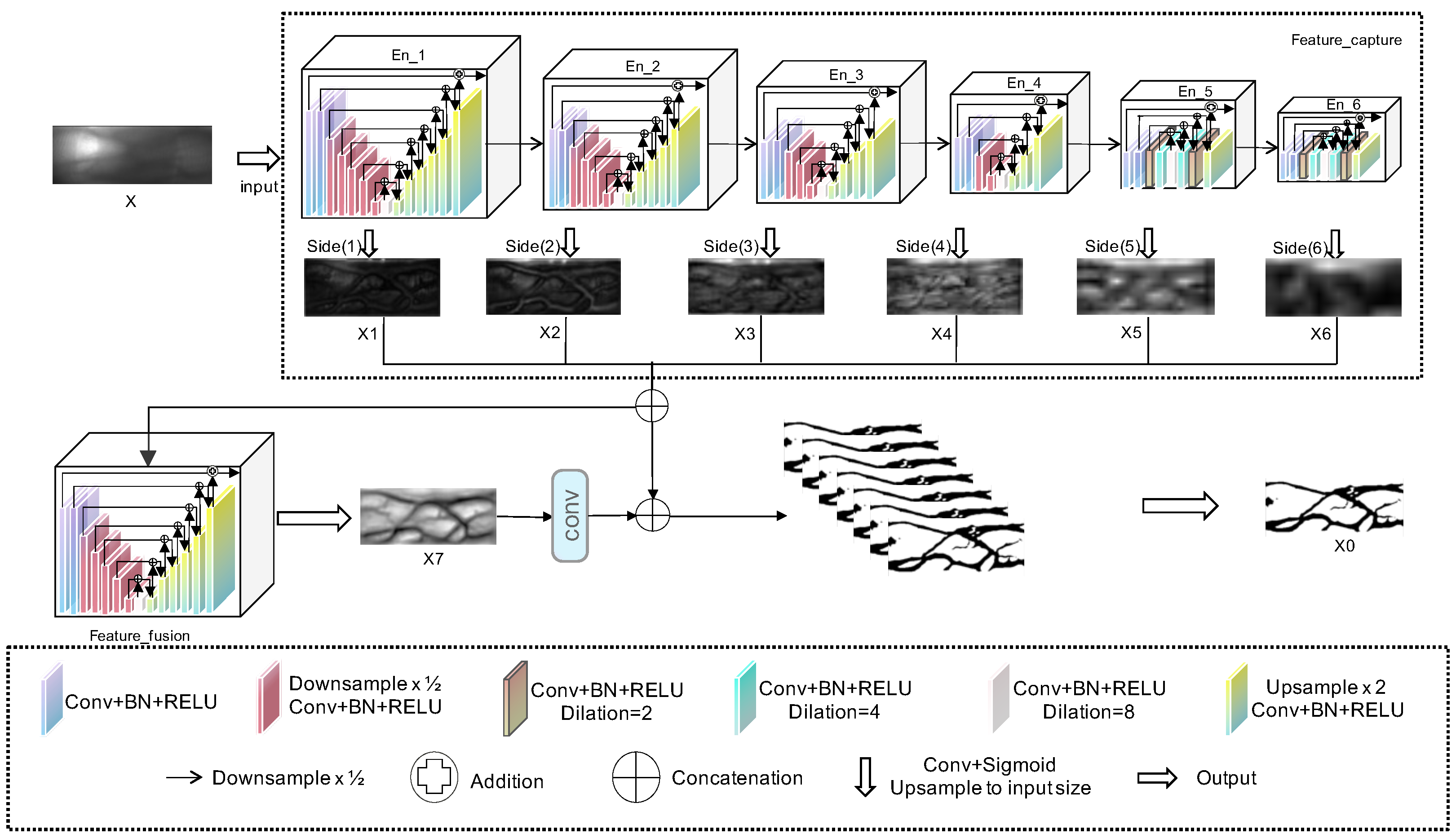
As illustrated in
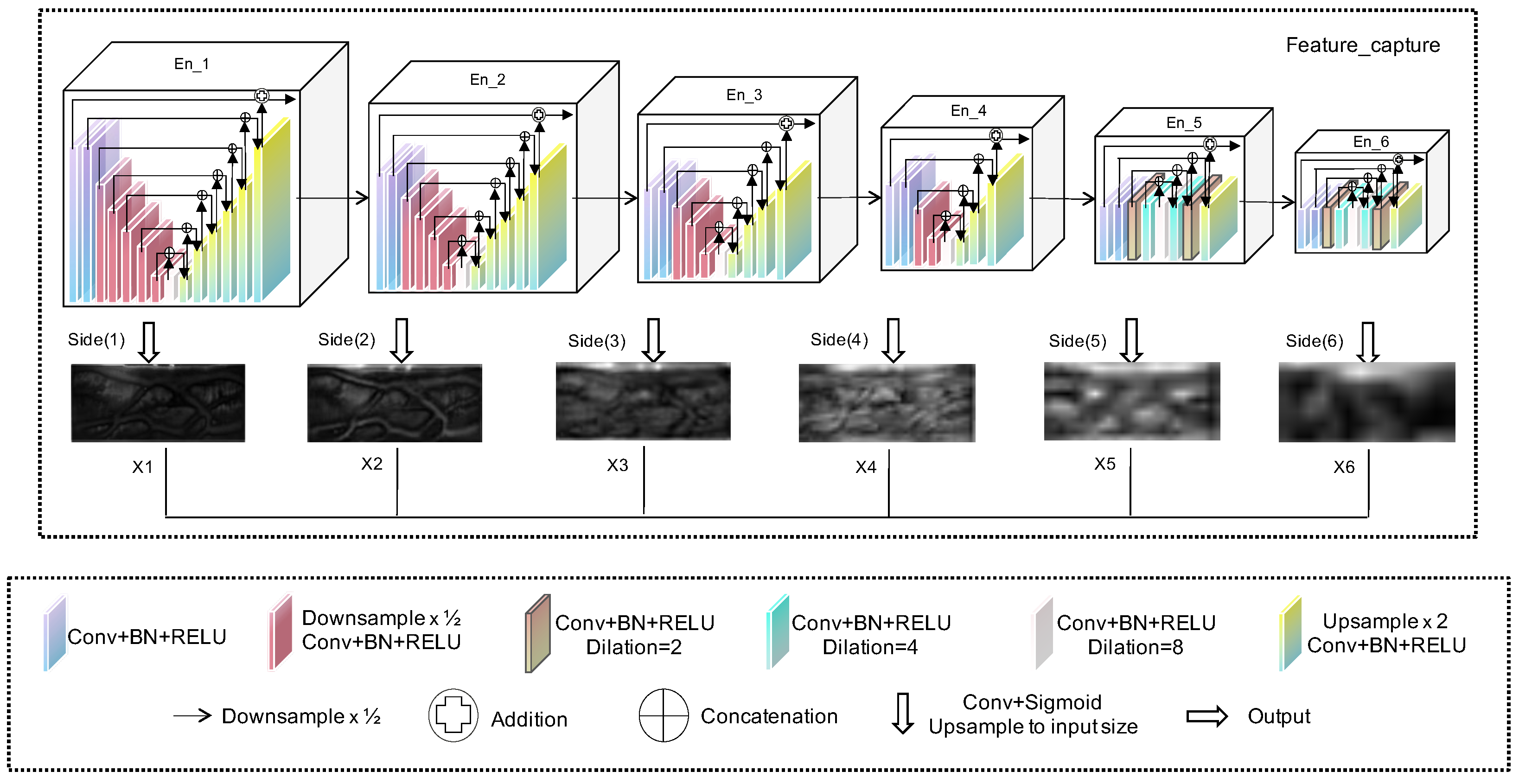
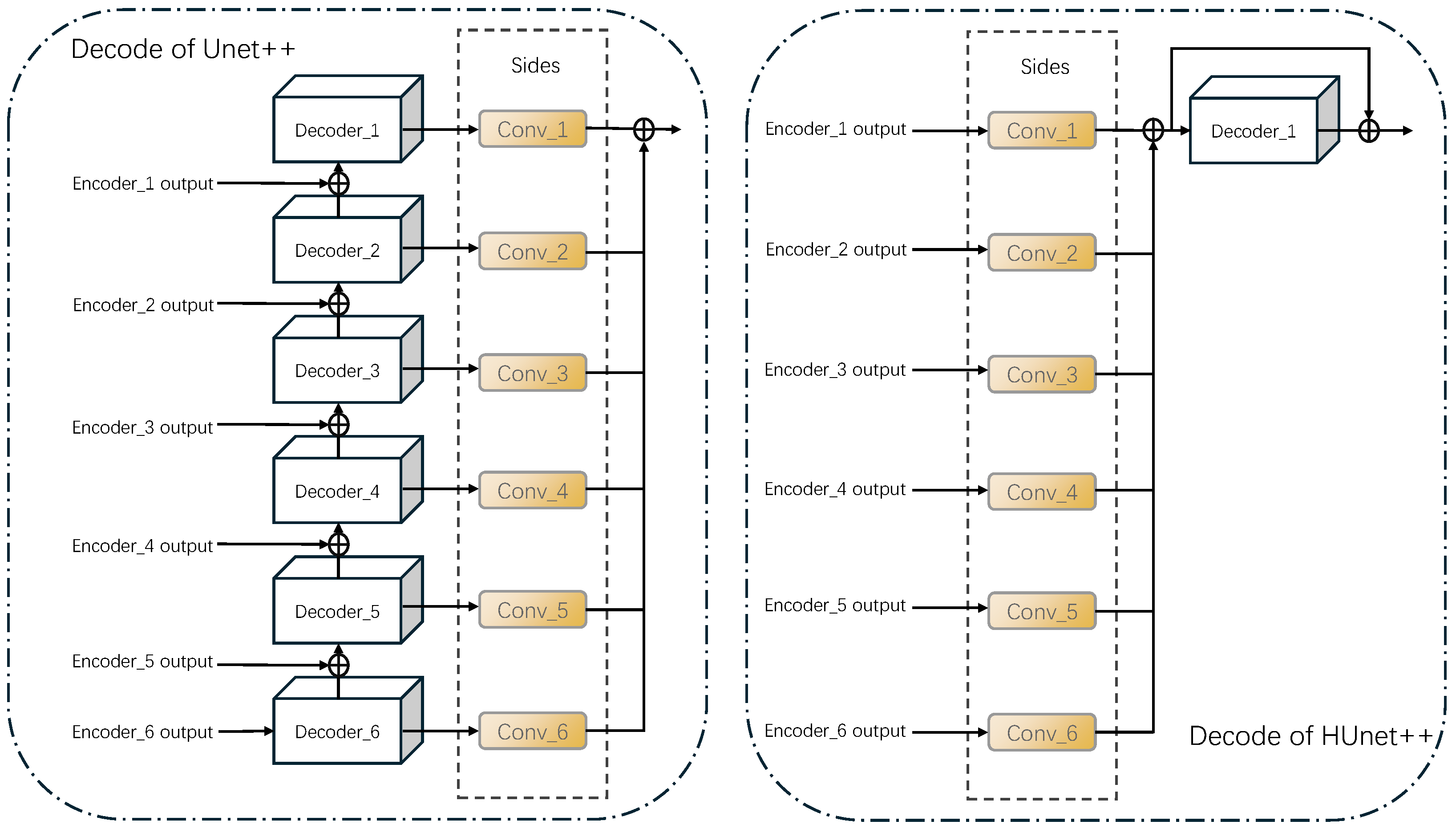
Table 2, this study compared the parameter configurations of four models: Unet++, HUnet_32m, HUnet_36m, and HUnet_40m. In these models, the “hid, in, out” column represents the “hidden, input, output” of each module, and the “n” of En_n is the depth of each module. Specifically, in each of these models, the encoder component consisted of six En_n units. Regarding the decoder architecture, the Unet++ model included five De_n units, whereas the other three models contained only one De_5 unit.
According to the data presented in
Table 2, each En_n unit corresponds to the En_n shown in
Figure 2. The De_5 unit corresponds to the Feature Fusion structure depicted in the same figure. Additionally, the structure of De_m (where m ranged from 1 to 4) was similar to that of its corresponding En_m (also ranging from 1 to 4), although their functions differed. Notably, in the three HUnet models, the decoder consisted solely of a De_5 unit. In contrast to the original Unet++ model, where the De_5 has an input channel of 128, in these models, the De_5 had an input channel of 6. This design was intended to integrate the finger vein features extracted by decoders at six different levels. Due to space constraints, this article uses the En_1 structure of the encoder in the HUnet++ model from
Table 2 as an example to illustrate the significance of the En_n and De_n structure parameters detailed in
Table 2.
Table 3 elaborates on the parameter configuration of the En_1 unit within the encoder structure of the HUnet_32m model, as referenced in
Table 2. Specifically, within the encoder structure of the HUnet_32m model in
Table 2, the hidden, input, and output parameters of the En_1 unit are specified as (32, 1, 64). The input channels count corresponds to the input channels of the conv_in module within the encoder structure in
Table 3. The output channels’ count aligns with the output channels of the last module, decoder_6, in the decoder structure of
Table 3. The hidden layer parameters are consistent with the output channel numbers of all encoder_n units and all decoder_n units, except for decoder_6. Additionally, in
Table 2, the depth associated with the En_1 unit in the encoder structure of HUnet_32m is 7, which corresponds to the highest number, encoder_7, in the encoder_n structure in
Table 3. Across all En_n structures, the total number of decoder_n units is always one less than the total number of encoder_n units. Note that in all En_n structures, the convolutional layer in the final encoder_n module employs dilated convolution.
In the experiments involving the HUnet++ and Unet++ model, AdamW was adopted as the optimizer used throughout, with a weight-decay value of 1
. After training the models, the one with the best performance was compressed using the method of structural reparameterization. Structural reparameterization is a technique that enables the compression of model parameters while preserving accuracy, thus enhancing the model's prediction speed. This method typically involves merging convolution branches or convolution+BN blocks into a single convolution with an added bias term. To implement this, a new compressed model must be constructed, which, compared to the original model, only lacks the BN or convolution branch components. Afterward, the compressed weight file is loaded into the new model. The primary goal of this approach is to accelerate prediction, enabling real-time performance. In the experiments conducted in this study, the convolution+BN blocks were primarily compressed. The output of the matrix after passing through both the convolution block
and the BN block is equivalent to the output of a newly created convolution block
with an added bias term
. As explained by Ding et al. [
27], the value of the new convolution block
can be represented by Formula (
5):
The value of bias
is Formula (
6):
By substituting the original and the weights of the BN layer with and , the resulting model was compressed. Subsequently, the compressed model was evaluated against the original Unet++ (a lightweight variant of the Unet++ architecture) model for vein mask extraction on the test set, with a focus on comparing the extraction speed of vein masks between the two models.