Detection of COVID-19: A Metaheuristic-Optimized Maximally Stable Extremal Regions Approach
Abstract
1. Introduction
- The interpretation of the parameter selection problem in MSER-based image segmentation as an optimization problem (including the definition of an appropriate objective function);
- Proposing metaheuristic algorithms for solving the previously mentioned optimization problem;
- The application of both of the points mentioned above to lung X-ray image segmentation as a means to facilitate diagnosis (with a special interest in COVID-19).
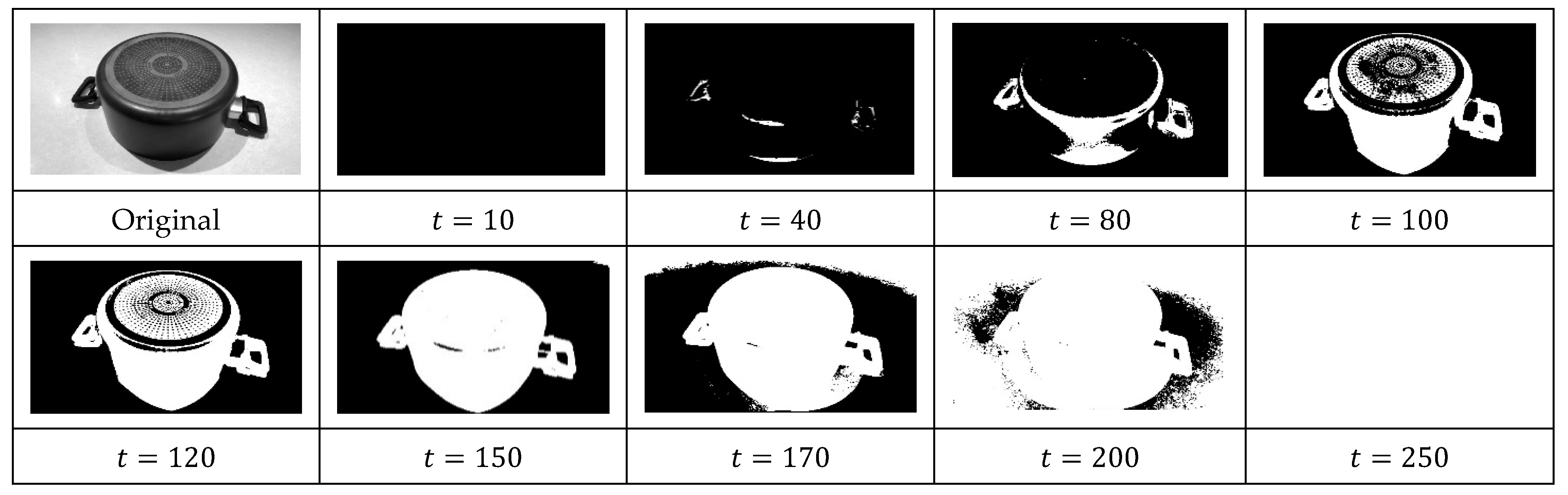
2. Maximally Stable Extremal Regions (MSER)
3. Metaheuristic Methods
3.1. Particle Swarm Optimization (PSO)
3.2. Firefly Method (FF)
3.3. Grey Wolf Optimizer (GWO)
3.4. Genetic Algorithms (GA)
3.5. Artificial Ecosystem Optimization by Means of Fitness–Distance Balance (FDBAEO)
3.6. The Standard Symbiotic Organism Search with the Fitness–Distance Balance Method (FDBSOS)
4. The Proposed Method
4.1. Problem Statement
4.2. Objective Function
4.3. Computational Procedure
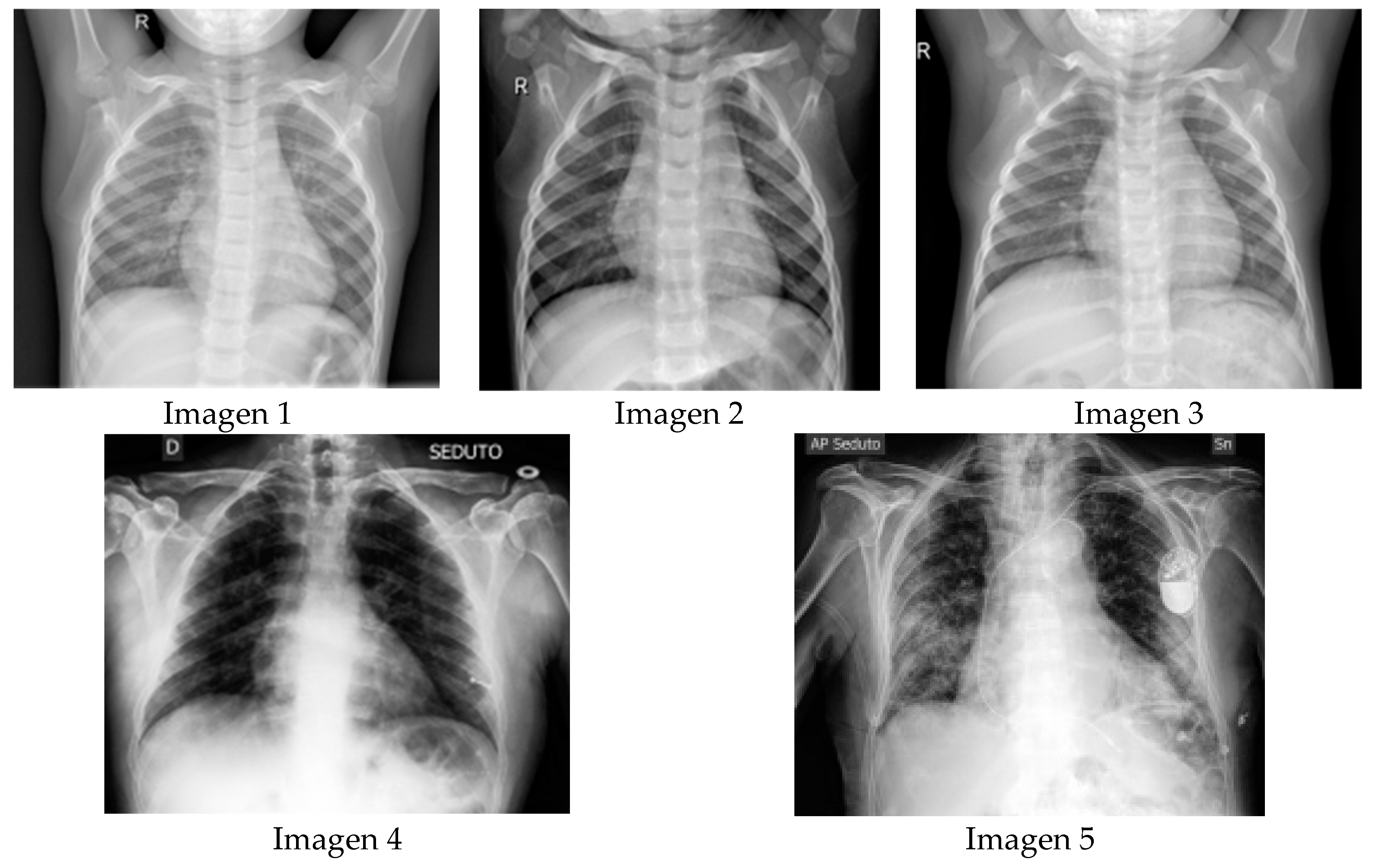
5. Experimental Results
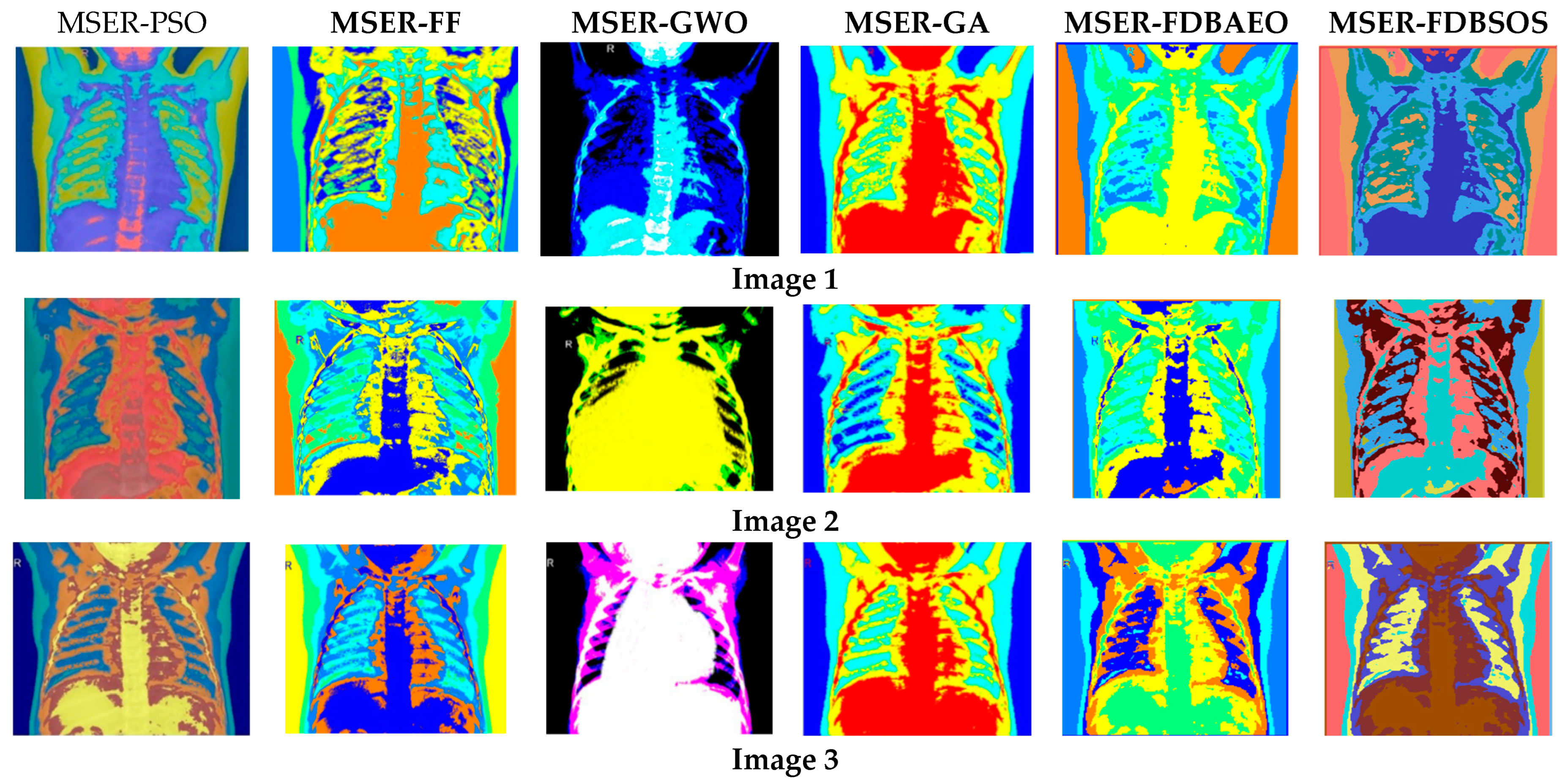
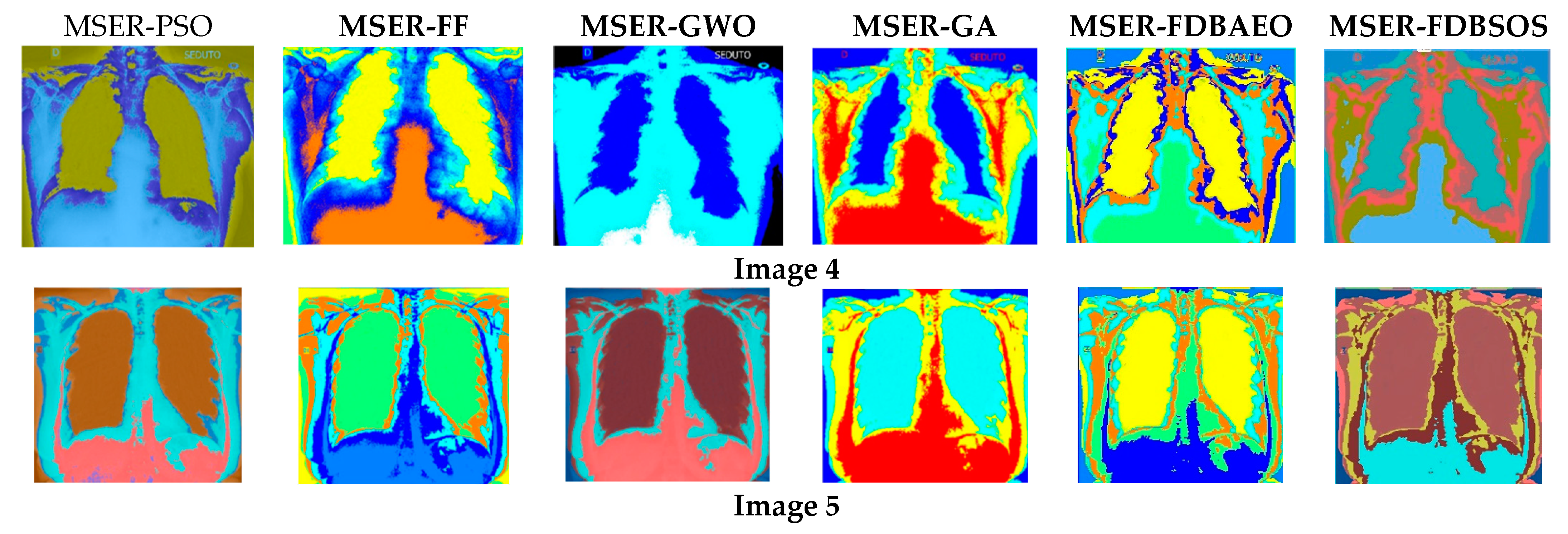
5.1. Visual Evaluation
5.2. Numerical Evaluation
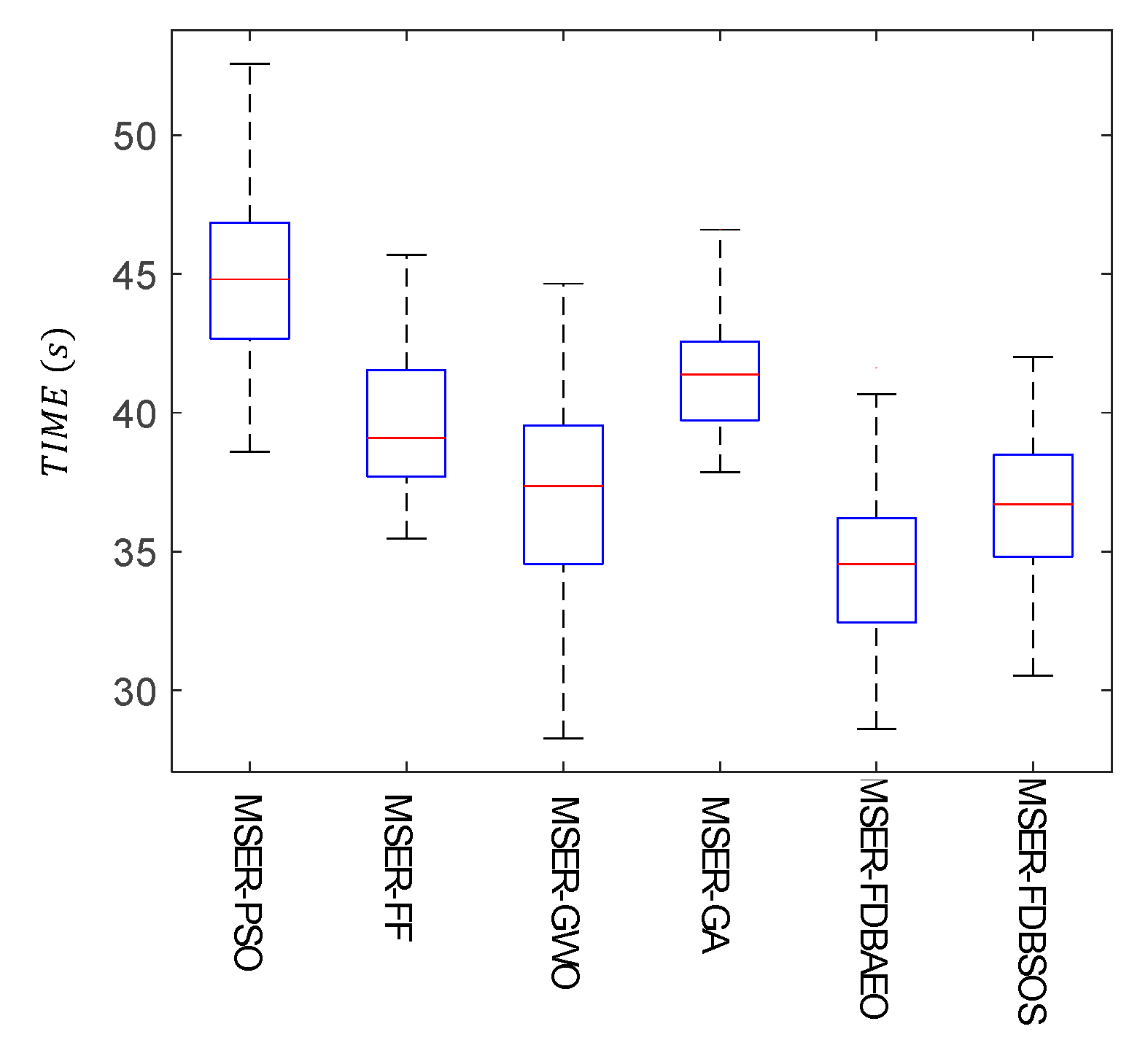
6. Time Analysis and Accuracy
7. Conclusions
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Mishra, N.P.; Das, S.S.; Yadav, S.; Khan, W.; Afzal, M.; Alarifi, A.; Kenawy, E.-R.; Ansari, M.T.; Hasnain, S.; Nayak, A.K. Global impacts of pre- and post-COVID-19 pandemic: Focus on socio-economic consequences. Sens. Int. 2020, 1, 100042. [Google Scholar] [CrossRef] [PubMed]
- van Mulukom, V.; Pummerer, L.J.; Alper, S.; Bai, H.; Čavojová, V.; Farias, J.; Kay, C.S.; Lazarevic, L.B.; Lobato, E.J.; Marinthe, G.; et al. Antecedents and consequences of COVID-19 conspiracy beliefs: A systematic review. Soc. Sci. Med. 2022, 301, 114912. [Google Scholar] [CrossRef] [PubMed]
- Pascarella, G.; Strumia, A.; Piliego, C.; Bruno, F.; Del Buono, R.; Costa, F.; Scarlata, S.; Agrò, F.E. COVID-19 diagnosis and management: A comprehensive review. J. Intern. Med. 2020, 288, 192–206. [Google Scholar] [CrossRef] [PubMed]
- Zhai, P.; Ding, Y.; Wu, X.; Long, J.; Zhong, Y.; Li, Y. The epidemiology, diagnosis and treatment of COVID-19. Int. J. Antimicrob. Agents 2020, 55, 105955. [Google Scholar] [CrossRef] [PubMed]
- Jamshaid, H.; Zahid, F.; Din, I.U.; Zeb, A.; Choi, H.G.; Khan, G.M.; Din, F.U. Diagnostic and Treatment Strategies for COVID-19. AAPS Pharmscitech 2020, 21, 222. [Google Scholar] [CrossRef] [PubMed]
- Rahman, T.; Khandakar, A.; Qiblawey, Y.; Tahir, A.; Kiranyaz, S.; Kashem, S.B.A.; Islam, M.T.; Al Maadeed, S.; Zughaier, S.M.; Khan, M.S.; et al. Exploring the effect of image enhancement techniques on COVID-19 detection using chest X-ray images. Comput. Biol. Med. 2021, 132, 104319. [Google Scholar] [CrossRef] [PubMed]
- Feki, I.; Ammar, S.; Kessentini, Y.; Muhammad, K. Federated learning for COVID-19 screening from Chest X-ray images. Appl. Soft Comput. 2021, 106, 107330. [Google Scholar] [CrossRef] [PubMed]
- López-Cabrera, J.D.; Orozco-Morales, R.; Portal-Diaz, J.A.; Lovelle-Enríquez, O.; Pérez-Díaz, M. Current limitations to identify COVID-19 using artificial intelligence with chest X-ray imaging. Health Technol. 2021, 11, 411–424. [Google Scholar] [CrossRef] [PubMed]
- Ahishali, M.; Degerli, A.; Yamac, M.; Kiranyaz, S.; Chowdhury, M.E.H.; Hameed, K.; Hamid, T.; Mazhar, R.; Gabbouj, M. Advance Warning Methodologies for COVID-19 Using Chest X-Ray Images. IEEE Access 2021, 9, 41052–41065. [Google Scholar] [CrossRef]
- Benmalek, E.; Elmhamdi, J.; Jilbab, A. Comparing CT scan and chest X-ray imaging for COVID-19 diagnosis. Biomed. Eng. Adv. 2021, 1, 100003. [Google Scholar] [CrossRef]
- Shen, J.; Zhang, C.J.P.; Jiang, B.; Chen, J.; Song, J.; Liu, Z.; He, Z.; Wong, S.Y.; Fang, P.-H.; Ming, W.-K. Artificial Intelligence Versus Clinicians in Disease Diagnosis: Systematic Review. JMIR Public Heal. Surveill. 2019, 7, e10010. [Google Scholar] [CrossRef] [PubMed]
- Enshaei, N.; Oikonomou, A.; Rafiee, M.J.; Afshar, P.; Heidarian, S.; Mohammadi, A.; Plataniotis, K.N.; Naderkhani, F. COVID-rate: An automated framework for segmentation of COVID-19 lesions from chest CT images. Sci. Rep. 2022, 12, 3212. [Google Scholar] [CrossRef] [PubMed]
- Arun Prakash, J.; Asswin, C.R.; Dharshan Kumar, K.S.; Dora, A.; Ravi, V.; Sowmya, V.; Gopalakrishnan, E.A.; Soman, K.P. Transfer learning approach for pediatric pneumonia diagnosis using channel attention deep CNN architectures. Eng. Appl. Artif. Intell. 2023, 123, 106416. [Google Scholar] [CrossRef]
- Kaya, Y.; Gürsoy, E. A MobileNet-based CNN model with a novel fine-tuning mechanism for COVID-19 infection detection. Soft Comput. 2023, 27, 5521–5535. [Google Scholar]
- Chen, S.; Ren, S.; Wang, G.; Huang, M.; Xue, C. Interpretable CNN-Multilevel Attention Transformer for Rapid Recognition of Pneumonia From Chest X-Ray Images. IEEE J. Biomed. Health Informatics 2023, 28, 753–764. [Google Scholar] [CrossRef] [PubMed]
- Sanghvi, H.A.; Patel, R.H.; Agarwal, A.; Gupta, S.; Sawhney, V.; Pandya, A.S. A deep learning approach for classification of COVID and pneumonia using DenseNet-201. Int. J. Imaging Syst. Technol. 2022, 33, 18–38. [Google Scholar] [CrossRef] [PubMed]
- AnbuDevi, M.K.A.; Suganthi, K. Review of Semantic Segmentation of Medical Images Using Modified Architectures of UNET. Diagnostics 2022, 12, 3064. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Z.; Siddiquee, M.M.R.; Tajbakhsh, N.; Liang, J. UNet++: Redesigning Skip Connections to Exploit Multiscale Features in Image Segmentation. IEEE Trans. Med. Imaging 2019, 39, 1856–1867. [Google Scholar] [CrossRef] [PubMed]
- Diakogiannis, F.I.; Waldner, F.; Caccetta, P.; Wu, C. ResUNet-a: A deep learning framework for semantic segmentation of remotely sensed data. ISPRS J. Photogramm. Remote Sens. 2020, 162, 94–114. [Google Scholar] [CrossRef]
- Li, X.; Chen, H.; Qi, X.; Dou, Q.; Fu, C.W.; Heng, P.A. H-DenseUNet: Hybrid densely connected UNet for liver and tumor segmentation from CT volumes. IEEE Trans. Med. Imaging 2018, 37, 2663–2674. [Google Scholar] [CrossRef]
- Aiadi, O.; Khaldi, B. A fast lightweight network for the discrimination of COVID-19 and pulmonary diseases. Biomed. Signal Process. Control. 2022, 78, 103925. [Google Scholar] [CrossRef]
- Basu, A.; Sheikh, K.H.; Cuevas, E.; Sarkar, R. COVID-19 detection from CT scans using a two-stage framework. Expert Syst. Appl. 2021, 193, 116377. [Google Scholar] [CrossRef] [PubMed]
- Xue, X.; Chinnaperumal, S.; Abdulsahib, G.M.; Manyam, R.R.; Marappan, R.; Raju, S.K.; Khalaf, O.I. Design and Analysis of a Deep Learning Ensemble Framework Model for the Detection of COVID-19 and Pneumonia Using Large-Scale CT Scan and X-ray Image Datasets. Bioengineering 2023, 10, 363. [Google Scholar] [CrossRef] [PubMed]
- Cuevas, E.; Rodríguez, A. Metaheuristic Computation with MATLAB®; Informa UK Limited: London, UK, 2020. [Google Scholar]
- Ztürk, H.T.; Kahraman, H.T. Meta-heuristic search algorithms in truss optimization: Research on stability and complexity analyses. Appl. Soft Comput. 2023, 145, 110573. [Google Scholar] [CrossRef]
- Riaz, M.; Bashir, M.; Younas, I. Metaheuristics based COVID-19 detection using medical images: A review. Comput. Biol. Med. 2022, 144, 105344. [Google Scholar] [CrossRef] [PubMed]
- Júnior, D.A.D.; da Cruz, L.B.; Diniz, J.O.B.; da Silva, G.L.F.; Junior, G.B.; Silva, A.C.; de Paiva, A.C.; Nunes, R.A.; Gattass, M. Automatic method for classifying COVID-19 patients based on chest X-ray images, using deep features and PSO-optimized XGBoost. Expert Syst. Appl. 2021, 183, 115452. [Google Scholar] [CrossRef] [PubMed]
- Gopatoti, A.; Vijayalakshmi, P. CXGNet: A tri-phase chest X-ray image classification for COVID-19 diagnosis using deep CNN with enhanced grey-wolf optimizer. Biomed. Signal Process. Control 2022, 77, 103860. [Google Scholar] [CrossRef] [PubMed]
- Khadija, B. Automatic detection of COVID-19 using CNN model combined with Firefly algorithm. In Proceedings of the 2022 8th International Conference on Optimization and Applications (ICOA), Sestri Levante, Italy, 6–7 October 2022; pp. 1–4. [Google Scholar]
- Gülmez, B. A novel deep neural network model based Xception and genetic algorithm for detection of COVID-19 from X-ray images. Ann. Oper. Res. 2023, 328, 617–641. [Google Scholar] [CrossRef]
- Naik, M.K.; Swain, M.; Panda, R.; Abraham, A. Novel Square Error Minimization-Based Multilevel Thresholding Method for COVID-19 X-Ray Image Analysis Using Fast Cuckoo Search. Int. J. Image Graph. 2024, 24, 2450004. [Google Scholar] [CrossRef]
- Ramkumar, M.; Gowtham, M.; Jamaesha, S.S.; Vigenesh, M. Attention induced multi-head convolutional neural network organization with MobileNetv1 transfer learning and COVID-19 diagnosis using jellyfish search optimization process on chest X-ray images. Biomed. Signal Process. Control. 2024, 93, 106133. [Google Scholar] [CrossRef]
- Liu, B.; Nie, X.; Li, Z.; Yang, S.; Tian, Y. Evolving deep convolutional neural networks by IP-based marine predator algorithm for COVID-19 diagnosis using chest CT scans. J. Ambient. Intell. Humaniz. Comput. 2024, 15, 451–464. [Google Scholar] [CrossRef] [PubMed]
- Rachel, R.B.; Nehemiah, H.K.; Singh, V.K.; Manoharan, R.M.V. Diagnosis of Covid-19 from CT slices using Whale Optimization Algorithm, Support Vector Machine and Multi-Layer Perceptron. J. X-ray Sci. Technol. 2024, 32, 253–269. [Google Scholar] [CrossRef]
- Mahdy, A.; Shaheen, A.; El-Sehiemy, R.; Ginidi, A. Artificial ecosystem optimization by means of fitness distance balance model for engineering design optimization. J. Supercomput. 2023, 79, 18021–18052. [Google Scholar] [CrossRef]
- Kahraman, H.T.; Aras, S.; Gedikli, E. Fitness-distance balance (FDB): A new selection method for meta-heuristic search algorithms. Knowledge-Based Syst. 2020, 190, 105169. [Google Scholar] [CrossRef]
- Nistér, D.; Stewénius, H. Linear time maximally stable extremal regions. In Proceedings of the Computer Vision–ECCV 2008: 10th European Conference on Computer Vision, Marseille, France, 12–18 October 2008; Proceedings, Part II 10. Springer: Berlin/Heidelberg, Germany, 2008; pp. 183–196. [Google Scholar]
- Lee, M.H.; Park, I.K. Performance evaluation of local descriptors for maximally stable extremal regions. J. Vis. Commun. Image Represent. 2017, 47, 62–72. [Google Scholar] [CrossRef]
- Matas, J.; Chum, O.; Urban, M.; Pajdla, T. Robust wide-baseline stereo from maximally stable extremal regions. Image Vis. Comput. 2004, 22, 761–767. [Google Scholar] [CrossRef]
- Martins, P.; Carvalho, P.; Gatta, C. On the completeness of feature-driven maximally stable extremal regions. Pattern Recognit. Lett. 2016, 74, 9–16. [Google Scholar] [CrossRef]
- Chang, H.; Li, D.; Zhang, X.; Cui, X.; Fu, Z.; Chen, X.; Song, Y. Real-time height measurement with a line-structured-light based imaging system. Sensors Actuators A Phys. 2024, 368, 115164. [Google Scholar] [CrossRef]
- Arul, U.; Arun, V.; Rao, T.P.; Baskaran, R.; Kirubakaran, S.; Hussan, M.I.T. Effective Anomaly Identification in Surveillance Videos Based on Adaptive Recurrent Neural Network. J. Electr. Eng. Technol. 2024, 19, 1793–1805. [Google Scholar] [CrossRef]
- Atlas, L.G.; Arjun, K.; Kumar, K.S.; Dhanaraj, R.K.; Nayyar, A. Deep learning enabled hemorrhage detection in retina with DPFE and splat segmentation in fundus images. Biomed. Signal Process. Control. 2024, 88, 105692. [Google Scholar] [CrossRef]
- Tohidi, N.; Rustamov, R.B. Short overview of advanced metaheuristic methods. Int. J. Tech. Phys. Probl. Eng. (IJTPE) 2022, 14, 84–97. [Google Scholar]
- Joyce, T.; Herrmann, J.M. A review of no free lunch theorems, and their implications for metaheuristic optimisation. Nat. -Inspired Algorithms Appl. Optim. 2018, 744, 27–51. [Google Scholar]
- Eberhart, R.C.; Kennedy, J. A new optimizer using particle swarm theory. In Proceedings of the 6th International Symposium on Micro Machine and Human Science 1995, Nagoya, Japan, 13–16 March 1995; pp. 39–43. [Google Scholar]
- Johari, N.F.; Zain, A.M.; Noorfa, M.H.; Udin, A. Firefly Algorithm for Optimization Problem. Appl. Mech. Mater. 2013, 421, 512–517. [Google Scholar] [CrossRef]
- Mirjalili, S.; Mirjalili, S.M.; Lewis, A. Grey wolf optimizer. Adv. Eng. Softw. 2014, 69, 46–61. [Google Scholar] [CrossRef]
- Lambora, A.; Gupta, K.; Chopra, K. Genetic algorithm-A literature review. In Proceedings of the 2019 International Conference on Machine Learning, Big Data, Cloud and Parallel Computing (COMITCon), Faridabad, India, 14–16 February 2019; pp. 380–384. [Google Scholar]
- Zhang, H.; Fritts, J.E.; Goldman, S.A. Entropy-based objective evaluation method for image segmentation. In Storage and Retrieval Methods and Applications for Multimedia 2004; SPIE: Bellingham, WA, USA, 2003; Volume 5307, pp. 38–49. [Google Scholar]
- Ben-David, S.; Ackerman, M. Measures of clustering quality: A working set of axioms for clustering. In Proceedings of the Advances in Neural Information Processing Systems 21, Proceedings of the Twenty-Second Annual Conference on Neural Information Processing Systems, Vancouver, BC, Canada, 8–11 December 2008; pp. 121–128. [Google Scholar]
- Müller, D.; Soto-Rey, I.; Kramer, F. Towards a guideline for evaluation metrics in medical image segmentation. BMC Res. Notes 2022, 15, 210. [Google Scholar] [CrossRef]
- Taha, A.A.; Hanbury, A.; del Toro, O.A.J. A formal method for selecting evaluation metrics for image segmentation. In Proceedings of the 2014 IEEE International Conference on Image Processing (ICIP), Paris, France, 27–30 October 2014; pp. 932–936. [Google Scholar]
- Kumar, S.N.; Lenin Fred, A.; Ajay Kumar, H.; Sebastin Varghese, P. Performance metric evaluation of segmentation algorithms for gold standard medical images. In Recent Findings in Intelligent Computing Techniques: Proceedings of the 5th ICACNI 2017, Goa, India, 1–3 June 2017; Springer: Singapore, 2018; Volume 3, pp. 457–469. [Google Scholar]
- Friedman, M. A Comparison of Alternative Tests of Significance for the Problem of m Rankings. Ann. Math. Stat. 1940, 11, 86–92. Available online: http://www.jstor.org/stable/2235971 (accessed on 1 June 2024). [CrossRef]



| Algorithm | Parameters |
|---|---|
| PSO [47] | , , , |
| FF [48] | , , , |
| GWO [49] | , |
| GA [50] | , , , selection method = roulette |
| FDBAEO [36] | The parameter values have been configured according to [r1] |
| FDBSOS [37] | The parameter values have been configured according to [r1] |
| Image 1 | Image 2 | Image 3 | Image 4 | Image 5 | ||
|---|---|---|---|---|---|---|
| FF | AVG | 2.78 × 10−3 | 2.74 × 10−3 | 2.79 × 10−3 | 2.73 × 10−3 | 2.72 × 10−3 |
| STD | 2.76 × 10−3 | 2.74 × 10−3 | 2.79 × 10−3 | 2.73 × 10−3 | 2.72 × 10−3 | |
| MEDIAN | 1.20 × 10−3 | 1.10 × 10−3 | 1.40 × 10−3 | 1.00 × 10−3 | 1.90 × 10−3 | |
| GA | AVG | 2.28 × 10−3 | 2.17 × 10−3 | 2.28 × 10−3 | 2.28 × 10−3 | 2.17 × 10−3 |
| STD | 1.76 × 10−4 | 8.82 × 10−4 | 1.76 × 10−4 | 1.76 × 10−4 | 8.82 × 10−4 | |
| MEDIAN | 2.20 × 10−3 | 2.10 × 10−3 | 2.20 × 10−3 | 2.20 × 10−3 | 2.10 × 10−3 | |
| PSO | AVG | 2.78 × 10−3 | 2.74 × 10−3 | 2.79 × 10−3 | 2.73 × 10−3 | 2.72 × 10−3 |
| STD | 2.76 × 10−3 | 2.74 × 10−3 | 2.79 × 10−3 | 2.73 × 10−3 | 2.72 × 10−3 | |
| MEDIAN | 1.20 × 10−3 | 1.10 × 10−3 | 1.40 × 10−3 | 1.00 × 10−3 | 1.90 × 10−3 | |
| GWO | AVG | 2.28 × 10−3 | 2.17 × 10−3 | 2.28 × 10−3 | 2.28 × 10−3 | 2.17 × 10−3 |
| STD | 1.76 × 10−4 | 8.82 × 10−4 | 1.76 × 10−4 | 1.76 × 10−4 | 8.82 × 10−4 | |
| MEDIAN | 2.20 × 10−3 | 2.10 × 10−3 | 2.20 × 10−3 | 2.20 × 10−3 | 2.10 × 10−3 | |
| FDBAEO | AVG | 1.08 × 10−3 | 1.08 × 10−3 | 1.31 × 10−3 | 1.31 × 10−3 | 1.10 × 10−3 |
| STD | 1.10 × 10−4 | 1.10 × 10−4 | 2.79 × 10−5 | 2.79 × 10−5 | 1.10 × 10−4 | |
| MEDIAN | 1.00 × 10−3 | 1.00 × 10−3 | 1.30 × 10−3 | 1.30 × 10−3 | 1.10 × 10−3 | |
| FDBSOS | AVG | 1.08 × 10−3 | 1.31 × 10−3 | 1.31 × 10−3 | 1.31 × 10−3 | 1.31 × 10−3 |
| STD | 1.10 × 10−4 | 2.79 × 10−5 | 2.79 × 10−5 | 2.79 × 10−5 | 2.79 × 10−5 | |
| MEDIAN | 1.00 × 10−3 | 1.30 × 10−3 | 1.30 × 10−3 | 1.30 × 10−3 | 1.30 × 10−3 |
| Image | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| MSER-PSO | Image 1 | 22.40 | 0.3972 | 0.8998 | 0.8320 | 0.8711 | 0.7884 | 0.6090 | 0.8420 |
| Image 2 | 20.20 | 0.3940 | 0.8888 | 0.8173 | 0.8412 | 0.7379 | 0.6830 | 0.8810 | |
| Image 3 | 21.98 | 0.3820 | 0.7092 | 0.8244 | 0.8310 | 0.7988 | 0.6840 | 0.8822 | |
| Image 4 | 24.16 | 0.2630 | 0.9863 | 0.9644 | 0.9827 | 0.9845 | 0.8988 | 0.4261 | |
| Image 5 | 24.30 | 0.2872 | 0.9940 | 0.9442 | 0.9422 | 0.9884 | 0.8920 | 0.4620 | |
| Averaged | 22.60 | 0.3446 | 0.8956 | 0.8764 | 0.8936 | 0.8596 | 0.7533 | 0.6986 | |
| MSER-FF | Image 1 | 23.18 | 0.3794 | 0.8042 | 0.7920 | 0.8340 | 0.6699 | 0.6742 | 0.8800 |
| Image 2 | 23.99 | 0.4087 | 0.7820 | 0.7830 | 0.8720 | 0.7720 | 0.7640 | 0.8840 | |
| Image 3 | 23.61 | 0.3864 | 0.7037 | 0.8940 | 0.8740 | 0.6637 | 0.7630 | 0.8840 | |
| Image 4 | 23.63 | 0.2400 | 0.9040 | 0.8619 | 0.9199 | 0.8840 | 0.8740 | 0.5820 | |
| Image 5 | 24.50 | 0.2409 | 0.8987 | 0.8849 | 0.9448 | 0.8687 | 0.8698 | 0.5920 | |
| Averaged | 22.68 | 0.3310 | 0.8185 | 0.8431 | 0.8888 | 0.7716 | 0.7890 | 0.7644 | |
| MSER-GWO | Image 1 | 20.46 | 0.7761 | 0.7888 | 0.7220 | 0.7453 | 0.8402 | 0.7998 | 0.7840 |
| Image 2 | 21.99 | 0.7412 | 0.7787 | 0.7440 | 0.7427 | 0.7300 | 0.7993 | 0.7920 | |
| Image 3 | 21.79 | 0.7841 | 0.6740 | 0.7840 | 0.7445 | 0.8000 | 0.7994 | 0.7979 | |
| Image 4 | 22.40 | 0.4027 | 0.8763 | 0.8903 | 0.8930 | 0.9020 | 0.8798 | 0.4244 | |
| Image 5 | 23.79 | 0.4171 | 0.8277 | 0.8906 | 0.9442 | 0.9008 | 0.8977 | 0.4261 | |
| Averaged | 22.08 | 0.6242 | 0.7891 | 0.8061 | 0.8139 | 0.8346 | 0.8352 | 0.6448 | |
| MSER-GA | Image 1 | 19.40 | 0.5710 | 0.6420 | 0.6040 | 0.6390 | 0.6088 | 0.6990 | 0.8840 |
| Image 2 | 19.73 | 0.5900 | 0.5490 | 0.6900 | 0.6440 | 0.6370 | 0.6993 | 0.9920 | |
| Image 3 | 19.21 | 0.5820 | 0.6240 | 0.5720 | 0.6420 | 0.6984 | 0.6883 | 0.8840 | |
| Image 4 | 22.85 | 0.3720 | 0.7120 | 0.7900 | 0.8459 | 0.8084 | 0.8897 | 0.5720 | |
| Image 5 | 22.37 | 0.3000 | 0.7962 | 0.8040 | 0.8666 | 0.8340 | 0.8999 | 0.5261 | |
| Averaged | 20.71 | 0.4830 | 0.6646 | 0.6920 | 0.7275 | 0.7173 | 0.7752 | 0.7716 | |
| MSER-FDBAEO | Image 1 | 23.20 | 0.4240 | 0.9087 | 0.9849 | 0.9428 | 0.7697 | 0.8198 | 0.7810 |
| Image 2 | 23.63 | 0.4200 | 0.9740 | 0.9619 | 0.9129 | 0.7840 | 0.8140 | 0.7220 | |
| Image 3 | 23.61 | 0.4286 | 0.9937 | 0.9240 | 0.8720 | 0.8622 | 0.71830 | 0.7910 | |
| Image 4 | 23.99 | 0.4108 | 0.9010 | 0.9430 | 0.9720 | 0.8300 | 0.7640 | 0.2820 | |
| Image 5 | 23.18 | 0.4779 | 0.9042 | 0.9320 | 0.8320 | 0.8402 | 0.7742 | 0.4200 | |
| Averaged | 23.52 | 0.4322 | 0.9363 | 0.9491 | 0.9163 | 0.8172 | 0.7780 | 0.5992 | |
| MSER-FDBSOS | Image 1 | 23.10 | 0.3240 | 0.4787 | 0.8849 | 0.9428 | 0.8397 | 0.7998 | 0.8710 |
| Image 2 | 23.62 | 0.4200 | 0.8440 | 0.8719 | 0.9219 | 0.7242 | 0.7930 | 0.8420 | |
| Image 3 | 23.61 | 0.4286 | 0.9737 | 0.9242 | 0.9740 | 0.8622 | 0.7842 | 0.8310 | |
| Image 4 | 23.99 | 0.4108 | 0.8810 | 0.9830 | 0.9703 | 0.8600 | 0.8340 | 0.8420 | |
| Image 5 | 23.18 | 0.3779 | 0.8042 | 0.8902 | 0.8220 | 0.8902 | 0.8942 | 0.8200 | |
| Averaged | 23.51 | 0.3922 | 0.7963 | 0.9108 | 0.9062 | 0.8352 | 0.8010 | 0.8412 |
| Algorithm | Ranking | Mean Value |
|---|---|---|
| MSER-FDBAEO | 1 | 5.607121 |
| MSER-FDBSOS | 2 | 5.201473 |
| MSER-PSO | 3 | 4.892410 |
| MSER-FF | 4 | 4.552140 |
| MSER-GWO | 5 | 3.788974 |
| MSER-GA | 6 | 3.571401 |
| Algorithm | SR |
|---|---|
| MSER-FDBAEO | 94.25% |
| MSER-FDBSOS | 92.14% |
| MSER-PSO | 89.62% |
| MSER-FF | 85.68% |
| MSER-GWO | 70.11% |
| MSER-GA | 65.74% |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
García-Gutiérrez, V.; González, A.; Cuevas, E.; Fausto, F.; Pérez-Cisneros, M. Detection of COVID-19: A Metaheuristic-Optimized Maximally Stable Extremal Regions Approach. Symmetry 2024, 16, 870. https://doi.org/10.3390/sym16070870
García-Gutiérrez V, González A, Cuevas E, Fausto F, Pérez-Cisneros M. Detection of COVID-19: A Metaheuristic-Optimized Maximally Stable Extremal Regions Approach. Symmetry. 2024; 16(7):870. https://doi.org/10.3390/sym16070870
Chicago/Turabian StyleGarcía-Gutiérrez, Víctor, Adrián González, Erik Cuevas, Fernando Fausto, and Marco Pérez-Cisneros. 2024. "Detection of COVID-19: A Metaheuristic-Optimized Maximally Stable Extremal Regions Approach" Symmetry 16, no. 7: 870. https://doi.org/10.3390/sym16070870
APA StyleGarcía-Gutiérrez, V., González, A., Cuevas, E., Fausto, F., & Pérez-Cisneros, M. (2024). Detection of COVID-19: A Metaheuristic-Optimized Maximally Stable Extremal Regions Approach. Symmetry, 16(7), 870. https://doi.org/10.3390/sym16070870









