Abstract
COVID-19 is a disease that can be spread easily with minimal physical contact. Currently, the World Health Organization (WHO) has endorsed the reverse transcription-polymerase chain reaction swab test as a diagnostic tool to confirm COVID-19 cases. This test requires at least a day for the results to come out depending on the available facilities. Many countries have adopted a targeted approach in screening potential patients due to the cost. However, there is a need for a fast and accurate screening test to complement this targeted approach, so that the potential virus carriers can be quarantined as early as possible. The X-ray is a good screening modality; it is quick at capturing, cheap, and widely available, even in third world countries. Therefore, a deep learning approach has been proposed to automate the screening process by introducing LightCovidNet, a lightweight deep learning model that is suitable for the mobile platform. It is important to have a lightweight model so that it can be used all over the world even on a standard mobile phone. The model has been trained with additional synthetic data that were generated from the conditional deep convolutional generative adversarial network. LightCovidNet consists of three components, which are entry, middle, and exit flows. The middle flow comprises five units of feed-forward convolutional neural networks that are built using separable convolution operators. The exit flow is designed to improve the multi-scale capability of the network through a simplified spatial pyramid pooling module. It is a symmetrical architecture with three parallel pooling branches that enable the network to learn multi-scale features, which is suitable for cases wherein the X-ray images were captured from all over the world independently. Besides, the usage of separable convolution has managed to reduce the memory usage without affecting the classification accuracy. The proposed method managed to get the best mean accuracy of 0.9697 with a low memory requirement of just 841,771 parameters. Moreover, the symmetrical spatial pyramid pooling module is the most crucial component; the absence of this module will reduce the screening accuracy to just 0.9237. Hence, the developed model is suitable to be implemented for mass COVID-19 screening.
1. Introduction
As of 10th August 2020, COVID-19 disease has infected more than 20 million people all over the world and caused more than 700,000 deaths (https://www.worldometers.info/coronavirus/). Generally, the elderly are more affected by the disease, especially the ones with prior health conditions, whereas younger people rarely show any symptoms even if they have been infected. COVID-19 is a type of respiratory illness that is caused by SARS-CoV-2 strain with three main symptoms, which are fever, dry cough, and tiredness [1]. The disease is believed to have zoonotic origins; that means the virus originated from an animal before being transmitted to humans; the first case was reported in Wuhan, China [2]. At the moment, the vaccines are still in the development and testing stages; none of them has passed through the Phase IV clinical trials (approval phase), which is a phase wherein a vaccine is certified to be safe for public usage.
The average mortality rate of the disease is low, less than , but it can be transmitted easily from one person to others. Bampoe et al. [3] reported that COVID-19 spreads through respiratory droplets from coughing and sneezing, while the risk of airborne transmission is very low. Hence, most governments have mandated their citizens to wear masks in public spaces such as shopping malls, public transportation hubs, schools, universities, worship places, etc. The term “social distancing” has also been popularized; it is advisable for people to maintain a certain distance to the others to reduce the infection rate of the disease. It is a crucial step in controlling the spread of the disease; some cases in Japan have been attributed to sport activities that have not observed strict social distancing measures [4]. Thus, the prevention method is needed so that the hospitals will not be burdened with overcapacity and lack of respiratory-related equipment.
Apart from that, accurate and efficient screening methods to detect COVID-19 are also needed to limit the disease’s spread. Sometimes, a person does not know that he is already infected by the disease; consider cases of those who are returning home after traveling abroad. By with a good detection protocol, any patient can be identified and isolated early, so that the treatment can be administered immediately. In many countries, the reverse transcription-polymerase chain reaction (RT-PCR) swab test is the most common test to confirm COVID-19 as endorsed by the World Health Organization [5]. Due to the constraint of laboratory capacity and the large number of samples, the results rarely come out on the same day. Thus, a good screening method that can give immediate–early results should be developed to identify the potential COVID-19 patients so that they can be quarantined before the confirmation results come out.
Since a severe case of COVID-19 will usually cause pneumonia [6], chest X-rays are a good screening modality, which has been widely used to detect other types of viral pneumonia cases [7]. In [8], Moncada et al. found that the smallest details on the chest X-ray image are important to identifying the pneumonia cases. They also found that an inexperienced observer will have a higher misdetection rate of spotting the fine-details. Hence, these weaknesses in observation can be mitigated through a deep learning-based automatic system. The work in [9] has also explained the sensitivity relationship between pneumonia and chest X-ray images; it is the primary display of prevalence of COVID-19 according to [10]. Moreover, Pereira et al. [11] has also stated that the chest X-ray is a faster and cheaper approach to screening pneumonia cases compared to the CT-scan. Besides, it is widely available in most hospitals, even in third world countries. The cost of X-ray imaging is also relatively cheap; it costs only RM 10 in Malaysia compared to RM 350 for an RT-PCR test [12]. Hence, this paper focuses on the development of an accurate deep learning algorithm to screen COVID-19 using chest X-ray images. Figure 1 shows some samples of the X-ray images used in this study. A lightweight deep learning model was the focus of our development, so that it could be implemented even on a mobile platform. Therefore, the proposed model can be applied all over the world using just a standard mobile phone without using any dedicated hardware. It will be useful for the countries that have chosen targeted diagnoses, especially underdeveloped countries where they cannot bear the cost of mass testing via RT-PCR. A lightweight screening model can be defined as a deep learning network that requires low memory requirements for making the inference. Generally, there is no global definition of a hard boundary that separates a lightweight model from others. Therefore, a maximum number of 20,000,000 parameters was defined to be the upper limit for a lightweight model. This value is in line with most of the networks that have been purposely built for mobile applications [13,14].
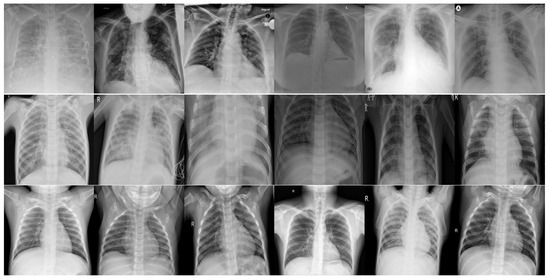
Figure 1.
Image samples of the frontal chest X-ray scans. The first row shows X-ray images of COVID-19 patients, the second row shows X-ray images of patients with other types of viral pneumonia, and the third row shows X-ray images of normal cases, which represents the control group.
In this work, synthetic data that were generated from a conditional deep convolutional generative adversarial network (conditional DC-GAN) were used to augment the training dataset for the COVID-19 class. This step was done to balance out the numbers of training data among the three classes of COVID-19, viral pneumonia, and normal—around 1300 images per class. In this paper, the viral pneumonia class refers to the pneumonia cases that are caused by viruses other than SARS-CoV-2, which are all grouped into a single class. The proposed method implements three main novel features to produce a lightweight classification model for an accurate COVID-19 screening algorithm. Hence, the main novelties of the proposed model are three fold: data augmentation through conditional DC-GAN, a multi-scale feature extractor with a symmetrical spatial pyramid pooling module and three mirroring branches, and a lightweight model with separable convolution. It uses less than 1,000,000 parameters with a low computational burden of just 1,679,643 floating-point operations (FLOPs). The proposed algorithm, which we termed LightCovidNet can be categorized as a compact architecture compared to other deep networks such as [15,16] that have more than 100 layers of convolutional neural networks (CNN). Compact networks such as in [17,18] are suitable for the situations wherein the training data are limited, such as in the case of chest X-ray images of the COVID-19 patients.
This paper is presented in five main sections. Section 2 describes the state-of-the-art deep learning algorithms used for classification. Section 3 explains in detail the dataset used in this study, and Section 4 explains the architecture of LightCovidNet, a lightweight deep learning model for COVID-19 screening. Experimental results and discussion are given in Section 5; a concise conclusion is summarized in the last section.
2. Related Work
2.1. Convolutional Neural Network Classifier
In 2012, the introduction of AlexNet popularized the usage of CNN for classification, which won the ImageNet Large Scale Visual Recognition Challenge. The model is relatively compact compared to other recent networks with five convolutional and four dense layers. However, it uses a total of 56,059,833 parameters, which is relatively a heavyweight model considering its compact structure. SqueezeNet [19] reduces the memory requirement by introducing fire modules that employ a 1 × 1 convolution kernel, which reduces the total number of parameters to just 736,963. A compact network with transfer learning was also introduced in [20] with just three convolutional and three dense layers. Then, a deeper model was introduced by Simonyan and Zisserman [21] that comprises of 16 convolutional and three dense layers. GoogleNet [22] then introduced embedding inception modules, which consist of parallel convolution layers but with different kernel sizes. As the networks became deeper, the zero diminishing gradient problem led to the introduction of ResNet that uses residual or skip connections between the convolutional layers. DenseNet then introduced feed-forward connections—layers are combined through a concatenate operator rather than an addition operator in ResNet. The spatial pyramid pooling is embedded in [23] to replace the last layer of DenseNet to improve the multi-scale capability of the network.
MobileNet V1 [13] is the first classifier model that was specifically designed for a mobile platform. It utilizes separable convolution that reduces the memory usage of the network; a standard convolution is replaced with a set of depthwise and pointwise convolutions. Similarly, Xception is also built based on separable convolution with the addition of residual connections. It has a much bigger memory requirement with a total number of parameters of 20,867,627 compared to the Mobilenet V1 with just 3,231,939 parameters. Sandler et al. [14] then introduced an inverted residual module to MobileNet, wherein the residual connection is passed through a pointwise convolution first before a depthwise convolution. In [24], the MobileNet architecture was made more efficient by implementing a hardware network architecture search that reduces the total number of parameters to just 1,665,501. Another efficient network was introduced in [25] for the mobile platform by adding shuffle and group convolution to the separable module. A variation of this work, ShuffleNet V2 [26], uses channel splitting before performing a skip connection, instead of using channel shuffling between the separable convolution components.
2.2. COVID-19 Classification Using Deep Learning Models
Generally, there are two approaches that have been used by researchers in using convolutional neural networks (CNN)—either making a fixed non-trainable feature extractor [27] or trainable end-to-end networks [28]. The work by Apostolopoulos and Mpesiana [29] has used a transfer learning approach on VGG-19 architecture to classify chest X-ray images into one of the three classes: COVID-19, other types of viral pneumonia, and normal. Panwar et al. [30] have also modified a variant of the VGG network to reduce the number of parameters by altering the top layers. Global average pooling is used to down-sample the feature maps before being passed to a two-layer dense classification network. Apart from VGG-family network, ResNet architecture [31] has also been implemented by several works [32,33] to identify the X-ray images as either belong ingto COVID-19 patients or not. Narin et al. [32] used a standard ResNet-50 that was pre-trained using the ImageNet database. Sethy and Behera [33] utilized the pre-trained parameters of ResNet-50 as a feature extractor module, which was then classified using a support vector machine (SVM). Using the same approach, the work in [11] extracted the features using a pre-trained Inception-V3 network. Instead of using a single model as a feature extraction module, the work in [34] combined both MobileNet V2 and SqueezeNet bottom layers and applied SVM as the classifier network to identify the COVID-19 cases.
An optimal SqueezeNet architecture was used in [35] to identify COVID-19 cases using optimized hyperparameters through the Bayesian method. Since COVID-19 is a new type of virus, the number of chest X-ray images for this case is still relatively low. The authors augmented the experimental data with synthetic images that were generated through additive noise, a shearing process, and brightness variations. Another compact architecture, DarkCovidNet, was proposed in [36]; it uses object detection architecture YOLO V1 [37] as the basis. The network uses a low number of parameters of just 1,167,363 but has relatively high accuracy. Besides, CapsuleNet has also been used to design a compact classifier network for screening COVID-19 cases with just three standard convolutional layers [38]. Since X-ray images were acquired from all over the world, the scale of the images is not consistent, and some images do include more background information compared to the others. Hence, SPP-Covid-Net was introduced in [2] to include spatial pyramid pooling to replace the top layers in DarkCovidNet architecture. The network size is smaller but it reported higher accuracy than DarkCovidNet. Another work in [7] also used parallel branches of CNN to capture multi-scale information. The method, which is called CovXNet, uses several convolutional layers with different dilation rates to capture different scales of features in the X-ray images. Apart from that, DCGAN has also been used to augment the limited number of training data for COVID-19 cases [39]. Table 1 shows the strengths and weaknesses of the reviewed COVID-19 classification methods.

Table 1.
Strengths and weaknesses of the selected COVID-19 classification methods.
3. Material
3.1. Chest X-ray Dataset
This work focuses on a ternary classification problem, wherein the X-ray images are classified into one of these classes: COVID-19, other types of viral pneumonia, and normal cases. This study only considers frontal chest area X-ray images. The images of COVID-19 cases consist of a set of X-ray images that were captured from COVID-19 patients that exhibited pneumonia conditions of various stages. The images for other types of viral pneumonia cases consist of X-ray images that were captured from patients with pneumonia caused by viruses other than SARS-CoV-2. All COVID-19 X-ray images were obtained from the COVID Chest X-ray dataset [40], which is still being continually updated with new X-ray images. The dataset consists of X-ray images of COVID-19 patients from various countries and ethnicities. We would like to clarify that the goal of this research was not to produce a diagnostic method, as we focused only on the screening stage, which is a preliminary step before a diagnosis is made [41]. Screening is done to reduce the number of mass screening tests of RT-PCR so that the available equipment can be directed towards targeted patients, as has been practiced in Malaysia [42]. The screening method, which is based on an X-ray image is used to screen for the possibility of COVID-19—since it is a cheap modality and is readily available in many countries—before diagnosis through RT-PCR test [43]. The dataset has been verified by medical practitioners, as stated in [11], as being suitable for COVID-19 screening of various severity levels.
The images were captured using a portable radiography unit to reduce the infection rate; they were stored in either 8-bit or 24-bit depth formats. They were then cropped into several sizes with a maximum resolution of 5623 × 4757; the minimum resolution was 331 × 324. The image resolution depends on the machine used to capture the X-ray images; it will vary a lot since the data are collected from various sources all over the world. The dataset has been annotated by several medical practitioners, as stated in [40]. The subjects comprise males and females that cover an age range of 11–94 years old. Several images came from the same patients but were captured at different timestamps; the images were sampled at times up to the 12th week of the COVID-19 disease. As of 1st August 2020, there are 446 cleaned X-ray images of COVID-19 patients; the side chest X-ray images and computed tomography (CT) scan images were removed from this study. Some of the removed samples are shown in Figure 2. This removal was done to ensure that a fair comparison can be made with other types of viral pneumonia and normal cases whose data have been captured from the frontal view only. Otherwise, COVID-19 detection will become easier, as any side view X-ray and CT scan images will be easily spotted as COVID-19 cases. The X-ray images for other types of viral pneumonia and normal cases were taken from Kaggle platform [44]. Most of the data were extracted from their training folder; we have selected 1345 and 1341 images of pneumonia and normal cases. Only 1345 images out of 3875 pneumonia cases were considered so that training data among the classes were uniformly distributed. On the other hand, conditional DC-GAN was used to buff up the number of training data in the COVID-19 class. All X-ray images were then saved in the Portable Network Graphics (PNG) format with a standard size of 1024 × 1024.
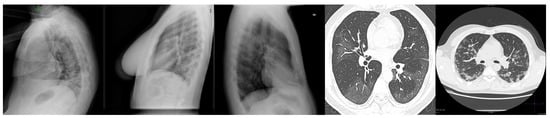
Figure 2.
Some samples of X-ray images that were removed from this study. The first three images are chest X-ray images that were captured from the side view, while the last two images were captured using CT-scan modality.
3.2. Synthetic Data Generation through Conditional DC-GAN
Since the number of COVID-19 X-ray images was less than those for other classes, synthetic data were generated such that the numbers of data in each class were almost equal during the training process. Hence, conditional DC-GAN [45] has been designed to produce a set of synthetic data to augment COVID-19 chest X-ray images. The conditional DC-GAN was trained using all data from the three classes, but only the COVID-19 class were augmented with the synthetic data to reduce training bias by balancing the number of samples among the classes. However, no synthetic data were utilized during the testing phase. Contrary to the method in [39], no condition has been applied and thus the data were generated solely using certain class information. However, we trained our network through conditioning the network with the label information to ensure that the generated images were unique to the targeted class. The proposed architecture of the conditional DC-GAN is shown in Figure 3, where the discriminator and generator networks are symmetrical in design with five sets of convolution and pooling operators each. Each of the generated samples had an output size of 224 × 224 pixels, which is closed enough to the required input size for most of the deep learning models.
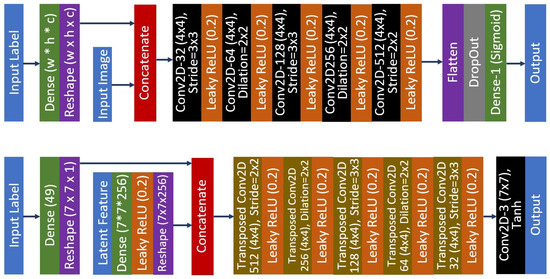
Figure 3.
The proposed conditional DC-GAN architecture that produces 224 × 224 synthetic images. The top image is the architecture for the discriminator network, while the bottom image is the architecture of the generator network.
Let define be an input X of layer i with a size of , where W, H, and C represent the width, height, and channel, respectively. for the discriminator network is a concatenation of the input image I and input label L that has been reshaped to match the I size.
where ⊕ represents the concatenate operator. Thus, the proposed discriminator network is a composite function of five convolution units with a dense classification layer.
where is a composite function convolution layer and leaky ReLU activation function, while is a composite function of a DropOut unit, a dense layer, and softmax activation function. On the other hand, the generator network takes input from a concatenated layer of L and the latent variables V.
The size of the random latent features is set to 100, where both discriminator and generator networks are trained by using binary cross-entropy loss function. Latent features are then upsampled using dense layers to the size of 7 × 7 × 256. The generator network comprises of five transposed convolution units and one exit convolution layer.
where is a composite function transposed convolution layer and leaky ReLU activation function, while is an exit network flow of a convolution layer with a tanh activation function. The parameters of the discriminator network are fixed when the combined generator-discriminator network is trained. The Leaky ReLU activation function is used in both generator and discriminator networks. It allows small non-negative gradient values to pass through the network, which provides stronger gradient flows from the discriminator to the generator. Atrous convolution is also used to increase the kernel reach while maintaining the same kernel size. Feature map upsampling in the generator network is done through transposed convolution, while downsampling operation in the discriminator network is done through a strided convolution operator, rather than a down-pooling operator. An Adam optimizer [46] with a learning rate of 0.0002 and a momentum of 0.5 was used to train both of the networks. The intensity values of the images were normalized to the floating-point representation from −1.0 to 1.0 so that contrast variation among the X-ray images could be reduced. The networks were trained in batches of 256 for 100 epochs using the NVIDIA Titan RTX graphics processing unit. Figure 4 depicts some of the generated frontal chest X-ray images that were used to train our lightweight deep learning classification model.
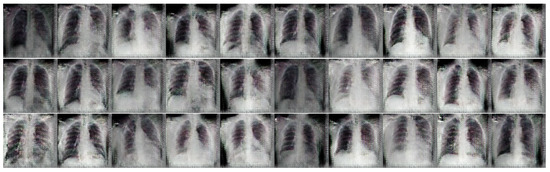
Figure 4.
Samples of the generated frontal chest X-ray images for COVID-19 class using conditional DC-GAN.
4. LightCovidNet: A Lightweight Deep Learning Model
The proposed network, LightCovidNet is a lightweight deep learning model with only 841,771 parameters. For comparison, a widely used deep learning model such as ResNet-50 uses 23,567,299 parameters, which requires 27 times more memory storage compared to the proposed model. Due to the lightweight nature of the LightCovidNet, it can be processed at 1,679,643 FLOPs, whereas ResNet-50 requires 47,028,459 FLOPs. The full architecture of the LightCovidNet is shown in Figure 5. It comprises of three parts, which are the entry (), middle (), and exit () flows with a total of only 14 convolutional layers. Let us define the LightCovidNet classification network as , and thus:
where is the input image with a size of 224 × 224 pixels. The selection of this input resolution was inspired by many state-of-the-art lightweight models, such as those in [13,14,24,25,26]. It is small enough to be implemented on a mobile platform but big enough to identify the pattern of COVID-19 cases. For the entry flow, LightCovidNet consists of two units of standard convolution unit, B. Let us define B to be a composite function of a convolution layer, batch normalization, leaky ReLU activation function, and maximum down-pooling operator. Note that we maintain the usage of leaky ReLU activation function from the conditional DC-GAN in this because it provides training stability when a small gradient value of 0.2 is used.
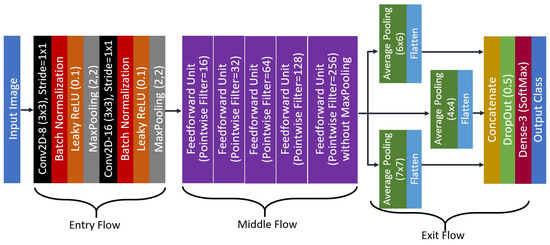
Figure 5.
The proposed architecture of the LightCovidNet.
The convolution stride is maintained at 1 × 1 and the down-pooling step is performed using a maximum pooling operator at the end of each CNN layer. The kernel size of the down-pooling operator is to 2 × 2 with a stride step of 2. All convolution layers in the LightCovidNet are set to 3 × 3 kernel, which is the optimized kernel speed in TensorFlow. For the middle flow, the network comprises of five layers of the feed-forward unit, K, which is inspired by DenseNet [15]. To be specific, only a single feedforward layer is used for each K. However, a separable convolution S approach has been adopted to reduce the memory usage while maintaining the 3 × 3 convolution layer. Therefore, a single unit of K can be written as
In each K, the number of filters in B is twice the number of filters in . The separable unit is a composite function of a depthwise and pointwise convolution, where batch normalization and leaky ReLU are applied after each operation. A maximum pooling operator is added at the end of K so that the feature maps will be downsampled for four times except for the last unit. Thus, the feature map at the end of the middle flow will have a reduced size by a factor 16 compared to its initial input size. Figure 6 shows the architecture of a single middle unit, where the total number of filters used can be found in Figure 5. Therefore, the middle flow can be represented by Equation (8).
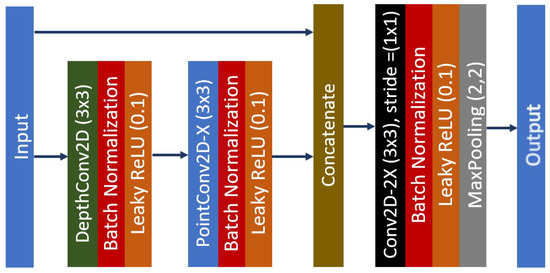
Figure 6.
The feed-forward unit network with X representing the total number of the convolution filters.
The exit flow is designed such that the features are extracted symmetrically from compact multi-scale layers that require low numbers of parameters. It is a simplified version of the spatial pyramid pooling module with three symmetrical down-pooling branches that follows a similar network flow as in the work by Abdani et al. [23]. It is a crucial step in improving the accuracy, which we proved later, as shown in the ablation study subsection—the features are sampled from three scales which provides better extracted features, knowing that the images are captured from various X-ray machines. The multi-scale down-pooling unit is used through a symmetrical network of average pooling operations with three different kernel sizes. Thus, the exit flow can be written as
where follows the same definition in the discriminator network with a dense classification layer. The resultant feature maps from M are then flattened and concatenated, which are then fed to the dense classifier as the input features. DropOut [47] scheme is also added to reduce the probability of network over-fitting, followed by a SoftMax activation function to decide the final likelihood of each output class.
5. Results and Discussion
A five-fold cross-validation method was used to verify the LightCovidNet performance, wherein the synthetic data were only added to the COVID-19 class to reduce bias during the training process. The same images in each training fold were used for training and testing of all benchmarked methods for a fair comparison. Similarly, the same set of synthetic images was also added while training the other models. LightCovidNet was trained using an Adam optimizer for 200 epochs with a learning rate of 0.0001. In general, all models were trained until convergence, as shown by the graphs of training accuracy and loss in Figure 7 and Figure 8. The loss function used in this study was categorical cross-entropy and the network was processed in batches of 64 input images. Tensorflow back-end with Keras front-end was used to model all benchmarked methods. Since the objective of LightCovidNet development is to produce an accurate, lightweight COVID-19 screening method, there were three performance measures used to rank the methods—mean accuracy (), total parameters, and floating-point operations (FLOPs). Mean accuracy is a metric that measures the number of predictions that are correctly classified according to the ground truth label.
where is the output class by predicted the deep learning network and is the ground truth label that has been annotated by the medical practitioners; is the total number of tested samples. Thus, the only situation where the and will be similar is for the case of a true positive for each class. Then, the accuracy metric is obtained through the ratio between the summation of all true positive classes and the total number of samples. The total parameters metric refers to the number of memory units used to store the deep learning model; a larger number of total parameters indicates that the model requires bigger memory storage and vice versa. On the other hand, FLOPs is a metric that measures the computational burden of processing the whole deep learning model. In this study, several state-of-the-art deep learning models which have less than 20 millions parameters were selected as the benchmarks, including MobileNet V1 [13], MobileNet V2 [14], MobileNet V3 [24], Xception-41 [48], ShuffleNet V1 [25], and ShuffleNet V2 [26]. These selected benchmarked methods have been specifically designed to be lightweight models, in which separable convolution units have been applied to reduce the memory usage. Besides, several recent deep learning methods that have been developed specifically for COVID-19 classification were also tested for performance comparisons, including the methods by Ucar et al. [35], Oztruk et al. [36], Abdani et al. [2], Loey et al. [39], Mahmud et al. [7], Narin et al. [32], and Panwar et al. [30].
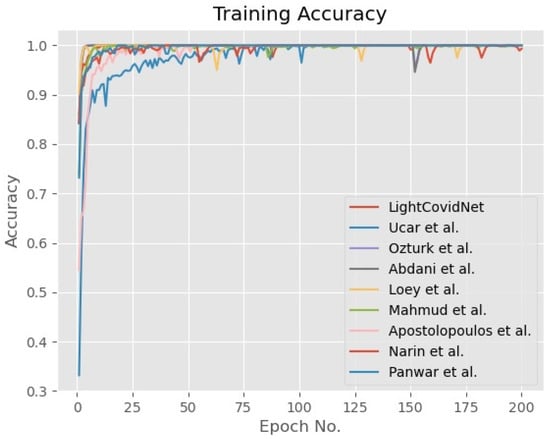
Figure 7.
Graph of the training accuracy of the LightCovidNet and its benchmarked models.
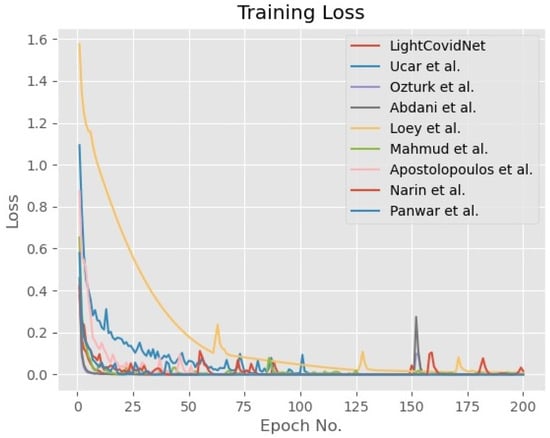
Figure 8.
Graph of the training loss of the LightCovidNet and its benchmarked models.
Table 2 shows the performance measures of the proposed LightCovidNet and its benchmarked methods. LightCovidNet returned the best of 0.9697, followed by Xception-41, Abdani et al. method, and Ozturk et al. method. All top three methods have utilized separable convolutions with skip connections to improve their models’ accuracies. Xception-41 also uniquely uses three separable convolution schemes followed by a residual skip connection; its composite operators start with a ReLU activation function, depthwise convolution, pointwise convolution, and batch normalization. It also uses the same entry network as used in LightCovidNet with two CNN layers, but the feedforward layers are combined through an addition operator, rather than a concatenation operator. The downside of using the addition operator is the size of the feedforward network must be similar to the intended combined layer. Hence, it limits the network’s flexibility as it grows deeper, which will not be a problem if a concatenation operator is used. However, the total number of parameters used in the concatenation operation is bigger compared to the addition operation, given that the network flow is similar. The method by Ozturk et al. is the best-performing model that does not apply any skip connection scheme, where all the convolution layers use the standard single network flow. However, they apply squeeze and expansion schemes to better extract the latent variables, as used in SqueezeNet [19]. Therefore, their model size is still relatively low compared to the other methods with just 1,167,363 parameters. On the other hand, the proposed LightCovidNet is also the second lightest model with just 841,771 parameters, compared to Xception-41 with 20,867,627 parameters, which requires more than 24 times memory storage. Our low memory usage can be attributed to the simplified spatial pyramid pooling module, where various parallel branches are down-sampled without applying any CNN layers; as such, the concatenated features are directly fed to the dense classification layers.

Table 2.
Experimental results of the LightCovidNet and its benchmarked methods.
The most lightweight model was produced by Ucar et al.’s method with 736,963 parameters but the performance was low with just 0.8633. It implemented eight layers of fire modules, which are closely similar to the squeeze and expansion schemes used in the Ozturk et al. method. Their entry network is also simple in design with just one CNN layer, while their exit network consists only of one dense classification layer. There is no skip or residual connection is implemented; the widest number of CNN filters applied was 64. This model is relatively thin compared to the LightCovidNet; the latter uses 256 CNN filters in its last layer. Hence, with a slightly larger total number of parameters, LightCovidNet manages to screen COVID-19 cases better, which is a crucial step in preventing the disease from spreading. Apart from that, the network by Narin et al. used the biggest number of parameters with the highest number of FLOPs, but still achieved a slightly lower accuracy compared to the LightCovidNet. It is interesting to note that the first version of MobileNet returnd the best performance with 0.9330, which can be attributed to the number of parameters used compared to its other versions. It is also worth noting that the inverted residual module in the MobileNet family did not perform well; the performances for all three of them were relatively low compared to the standard separable module. Their inverted scheme applies a pointwise convolution first, followed by a depthwise convolution and another pointwise convolution, which are added together with a skip connection.
The network by Loey et al. performed the worst with of just 0.8528, even though the memory usage was quite big with 11,192,003 parameters. Their method is based on ResNet with 18 layers of CNN, which is still a deeper network compared to ours with just 14 CNN layers. Thus, the dense feed-forward layer allowed LightCovidNet to reuse previous layers’ information efficiently and reduce the likelihood of network overfitting. Moreover, the usage of separable convolution also managed to reduce the memory usage without sacrificing much accuracy performance. Apart from the top three models, the best-performing model that uses separable convolution and combines a depthwise operator with a following pointwise operator was ShuffleNet V2 with an of 0.9387. This model shuffles the channels information, which is then processed through group convolutions. The smaller number of group convolutions in ShuffleNet V2 performed better compared to the bigger number of group convolutions in ShuffleNet V1.
A compact LightCovidNet performed relatively stably in identifying all the three classes, as shown in the confusion matrix in Figure 9. This matrix details the experimental performance of the LightCovidNet; the true positive, false positive, and false negative cases are shown according to each respected class. The largest error happened when the model misclassified the normal cases to be the other types of viral pneumonia, and the second largest error occurred when the model misclassified the other types of viral pneumonia cases to be normal cases. Generally, LightCovidNet is a good classifier for COVID-19 screening; it correctly identified 429 cases out of 446, which is an accuracy of 96.19%. Only seven cases of the COVID-19 images were wrongly identified as the other types of viral pneumonia cases, while only five cases were wrongly identified as normal cases. Thus, the screening rate of LightCovidNet is very high, which is 97.28%, which makes it suitable to be implemented as a mass screening tool.
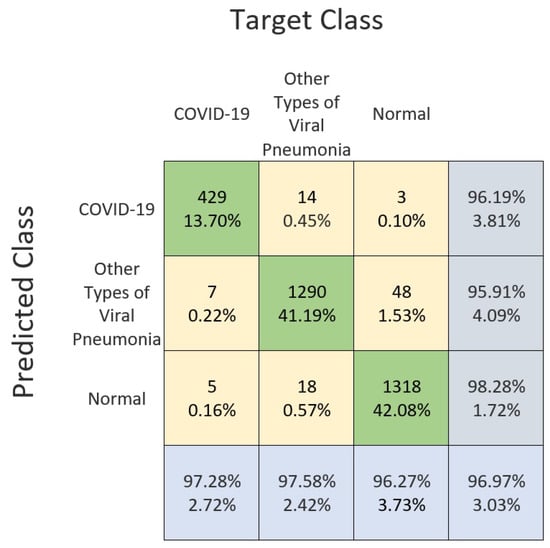
Figure 9.
Confusion matrix of the LightCovidNet classification performance using frontal chest X-ray images.
Table 3 shows the ablation study on the LightCovidNet. There are five components that have been tested to quantify the extents of their performance contributions to the LightCovidNet, which are a spatial pyramid pooling module, synthetic data generated by conditional DC-GAN, concatenated feedforward layers, separable convolution, and a DropOut unit. The results indicate that the addition of exit flow with spatial pyramid pooling improved the accuracy significantly compared to the standard global average pooling; has increased from 0.9237 to 0.9697. By having parallel branches with different pooling kernels, the features were extracted from multiple scales of feature maps, which fits the situation of our COVID-19 dataset that was collected from various sources. Note that the number of parameters in LightCovidNet without the pyramid module was the highest because of a denser connection in the classification layers. Concatenated feed-forward layers also improved the mean accuracy performance; a single network flow without any residual connection returned of just 0.9493. On the other hand, the memory usage of a single network flow was significantly reduced to just 450,091 parameters. However, a two percent improvement in is important for the COVID-19 situation, and then LightCovidNet is still a lightweight model with less than one million parameters. The lightweight nature of the proposed method can be attributed to the usage of separable convolution that reduces the memory usage by more than 20% compared to the standard convolution operator. Even more, the mean accuracy also slightly improved when separable convolution was utilized, which fits the situation of our small number of COVID-19 X-ray images. The usage of synthetic data through conditional DC-GAN also improved the network’s accuracy by a 2% margin. Similarly to the previous reasoning, the smaller number of datasets in COVID-19 resulted in imbalanced data distribution among the classes, which made the training process more biased towards the other classes. Finally, the performance improvement due to the DropOut unit was minimal with just 0.5% accuracy increment. However, the role of DropOut is important in avoiding overfitting issues due to the lightweight nature of the proposed method.

Table 3.
Performance results of the LightCovidNet ablation study.
6. Conclusions
In conclusion, the proposed LightCovidNet managed to achieve the best mean classification accuracy of 0.9697 with a low memory requirement of just 841,771 parameters. The model manages to achieve high accuracy because of its compact feed-forward architecture that utilizes separable convolution. A simplified spatial pyramid pooling module also improved the network’s capability regarding extracting features from X-ray images of various conditions—some images were taken while leaning towards the left or right side, while some images were zoomed into the chest area. The experimental results show that LightCovidNet without a spatial pyramid pooling module managed to achieve only 0.9237 accuracy. Besides, our synthetic data that were generated from conditional DC-GAN further improved classification accuracy from 0.9493 to 0.9697. Hence, LightCovidNet is a fast and accurate mobile-based deep learning model for COVID-19 screening, where the input relies on X-ray images that are readily available in most countries, even in the third world countries. This work has not considered any network pruning techniques in reducing the number of redundant features due to the limited number of COVID-19 X-ray images. The proposed model also requires input images with good contrast values, such that the air pockets are visible—the main pneumonia symptom. For future works, network pruning and more variants of the conditional DC-GAN will be explored to better classify the COVID-19 cases.
Author Contributions
Conceptualization, M.A.Z., S.R.A. and N.H.Z.; Formal analysis, M.A.Z., S.R.A. and N.H.Z.; Methodology, M.A.Z., S.R.A. and N.H.Z.; Software, M.A.Z., S.R.A. and N.H.Z.; Writing—original draft, M.A.Z., S.R.A. and N.H.Z.; Writing—review and editing, M.A.Z., S.R.A. and N.H.Z. All authors contributed equally to this work. All authors have read and agreed to the published version of the manuscript.
Funding
This work was supported in part by the Universiti Kebangsaan Malaysia through the Research University Grant Scheme under grant GUP-2019-008, and in part by the Ministry of Higher Education Malaysia through the Fundamental Research Grant Scheme under grant FRGS/1/2019/ICT02/UKM/02/1.
Conflicts of Interest
The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript, or in the decision to publish the results.
Abbreviations
The following abbreviations are used in this manuscript:
| CNN | Convolutional neural networks |
| DC-GAN | Deep convolutional generative adversarial network |
| CT | Computed tomography |
| RT-PCR | Reverse transcription polymerase chain reaction |
| FLOPs | Floating points operations |
References
- Park, G.S.; Ku, K.; Baek, S.H.; Kim, S.J.; Kim, S.I.; Kim, B.T.; Maeng, J.S. Development of Reverse Transcription Loop-Mediated Isothermal Amplification Assays Targeting Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2). J. Mol. Diagn. 2020, 22, 729–735. [Google Scholar] [CrossRef] [PubMed]
- Abdani, S.R.; Zulkifley, M.A.; Zulkifley, N.H. A Lightweight Deep Learning Model for Covid-19 Detection. In Proceedings of the IEEE Symposium on Industrial Electronics & Applications (ISIEA), Shah Alam, Selangor, Malaysia, 17–18 July 2020; pp. 1–5. [Google Scholar] [CrossRef]
- Bampoe, S.; Odor, P.; Lucas, D. Novel coronavirus SARS-CoV-2 and COVID-19. Practice recommendations for obstetric anaesthesia: What we have learned thus far. Int. J. Obstet. Anesth. 2020, 43, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Kosaka, H.; Meno, H. 88 affiliated with school soccer club among 91 infected with virus in western Japan city. The Mainichi, 10 August 2020. [Google Scholar]
- Lv, D.F.; Ying, Q.M.; Weng, Y.S.; Shen, C.B.; Chu, J.G.; Kong, J.P.; Sun, D.H.; Gao, X.; Weng, X.B.; Chen, X.Q. Dynamic change process of target genes by RT-PCR testing of SARS-Cov-2 during the course of a Coronavirus Disease 2019 patient. Clin. Chim. Acta 2020, 506, 172–175. [Google Scholar] [CrossRef]
- Neveu, S.; Saab, I.; Dangeard, S.; Bennani, S.; Tordjman, M.; Chassagnon, G.; Revel, M.P. Incidental diagnosis of Covid-19 pneumonia on chest computed tomography. Diagn. Interv. Imaging 2020, 101, 457–461. [Google Scholar] [CrossRef]
- Mahmud, T.; Rahman, M.A.; Fattah, S.A. CovXNet: A multi-dilation convolutional neural network for automatic COVID-19 and other pneumonia detection from chest X-ray images with transferable multi-receptive feature optimization. Comput. Biol. Med. 2020, 122, 103869. [Google Scholar] [CrossRef] [PubMed]
- Moncada, D.C.; Rueda, Z.V.; Macias, A.; Suarez, T.; Ortega, H.; Velez, L.A. Reading and interpretation of chest X-ray in adults with community-acquired pneumonia. Braz. J. Infect. Dis. 2011, 15, 540–546. [Google Scholar] [CrossRef][Green Version]
- Rajpurkar, P.; Irvin, J.; Zhu, K.; Yang, B.; Mehta, H.; Duan, T.; Ding, D.; Bagul, A.; Langlotz, C.; Shpanskaya, K.; et al. Chexnet: Radiologist-level pneumonia detection on chest x-rays with deep learning. arXiv 2017, arXiv:1711.05225. [Google Scholar]
- Hani, C.; Trieu, N.; Saab, I.; Dangeard, S.; Bennani, S.; Chassagnon, G.; Revel, M. COVID-19 pneumonia: A review of typical CT findings and differential diagnosis. Diagn. Interv. Imaging 2020, 101, 263–268. [Google Scholar] [CrossRef] [PubMed]
- Pereira, R.M.; Bertolini, D.; Teixeira, L.O.; Silla, C.N.; Costa, Y.M. COVID-19 identification in chest X-ray images on flat and hierarchical classification scenarios. Comput. Methods Prog. Biomed. 2020, 105532. [Google Scholar] [CrossRef]
- Atif, M.; Sulaiman, S.A.; Shafie, A.A.; Saleem, F.; Ahmad, N. Determination of chest x-ray cost using activity based costing approach at Penang General Hospital, Malaysia. Pan Afr. Med. J. 2012, 12, 1–7. [Google Scholar]
- Howard, A.G.; Zhu, M.; Chen, B.; Kalenichenko, D.; Wang, W.; Weyand, T.; Andreetto, M.; Adam, H. MobileNets: Efficient Convolutional Neural Networks for Mobile Vision Applications. CoRR 2017, arXiv:1704.04861v1. [Google Scholar]
- Sandler, M.; Howard, A.; Zhu, M.; Zhmoginov, A.; Chen, L. MobileNetV2: Inverted Residuals and Linear Bottlenecks. In Proceedings of the 2018 IEEE/CVF Conference on Computer Vision and Pattern Recognition, Salt Lake City, UT, USA, 18–23 June 2018; pp. 4510–4520. [Google Scholar]
- Huang, G.; Liu, Z.; Van Der Maaten, L.; Weinberger, K.Q. Densely Connected Convolutional Networks. In Proceedings of the 2017 IEEE Conference on Computer Vision and Pattern Recognition (CVPR), Honolulu, HI, USA, 21–26 July 2017; pp. 2261–2269. [Google Scholar] [CrossRef]
- Jegou, S.; Drozdzal, M.; Vazquez, D.; Romero, A.; Bengio, Y. The One Hundred Layers Tiramisu: Fully Convolutional DenseNets for Semantic Segmentation. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition Workshops (CVPRW), Honolulu, HI, USA, 21–26 July 2017; pp. 1175–1183. [Google Scholar] [CrossRef]
- Zulkifley, M.A.; Trigoni, N. Multiple-Model Fully Convolutional Neural Networks for Single Object Tracking on Thermal Infrared Video. IEEE Access 2018, 6, 42790–42799. [Google Scholar] [CrossRef]
- Zulkifley, M.A. Two Streams Multiple-Model Object Tracker for Thermal Infrared Video. IEEE Access 2019, 7, 32383–32392. [Google Scholar] [CrossRef]
- Iandola, F.N.; Moskewicz, M.W.; Ashraf, K.; Han, S.; Dally, W.J.; Keutzer, K. SqueezeNet: AlexNet-level accuracy with 50x fewer parameters and <1MB model size. CoRR 2016, arXiv:1602.07360v4. [Google Scholar]
- Zulkifley, M.A.; Abdani, S.R.; Zulkifley, N.H. Pterygium-Net: A deep learning approach to pterygium detection and localization. Multimed. Tools Appl. 2019. [Google Scholar] [CrossRef]
- Simonyan, K.; Zisserman, A. Very deep convolutional networks for large-scale image recognition. Technical Report, University of Oxford. arXiv 2014, arXiv:1409.1556v6. [Google Scholar]
- Szegedy, C.; Liu, W.; Jia, Y.; Sermanet, P.; Reed, S.; Anguelov, D.; Erhan, D.; Vanhoucke, V.; Rabinovich, A. Going deeper with convolutions. In Proceedings of the 2015 IEEE Conference on Computer Vision and Pattern Recognition (CVPR), Boston, MA, USA, 7–12 June 2015; pp. 1–9. [Google Scholar] [CrossRef]
- Abdani, S.R.; Zulkifley, M.A. DenseNet with Spatial Pyramid Pooling for Industrial Oil Palm Plantation Detection. In Proceedings of the 2019 International Conference on Mechatronics, Robotics and Systems Engineering, Bali, Indonesia, 4–6 December 2019. [Google Scholar]
- Howard, A.; Sandler, M.; Chen, B.; Wang, W.; Chen, L.; Tan, M.; Chu, G.; Vasudevan, V.; Zhu, Y.; Pang, R.; et al. Searching for MobileNetV3. In Proceedings of the 2019 IEEE/CVF International Conference on Computer Vision (ICCV), Seoul, Korea, 27 October–2 November 2019; pp. 1314–1324. [Google Scholar] [CrossRef]
- Zhang, X.; Zhou, X.; Lin, M.; Sun, J. ShuffleNet: An Extremely Efficient Convolutional Neural Network for Mobile Devices. In Proceedings of the 2018 IEEE/CVF Conference on Computer Vision and Pattern Recognition, Salt Lake City, UT, USA, 18–23 June 2018; pp. 6848–6856. [Google Scholar] [CrossRef]
- Ma, N.; Zhang, X.; Zheng, H.T.; Sun, J. Shufflenet v2: Practical guidelines for efficient cnn architecture design. In Proceedings of the European Conference on Computer Vision (ECCV), Munich, Germany, 8–14 September 2018; pp. 116–131. [Google Scholar]
- Zulkifley, M.A.; Mohamed, N.A.; Zulkifley, N.H. Squat Angle Assessment through Tracking Body Movements. IEEE Access 2019, 7, 48635–48644. [Google Scholar] [CrossRef]
- Badrinarayanan, V.; Kendall, A.; Cipolla, R. SegNet: A Deep Convolutional Encoder-Decoder Architecture for Image Segmentation. IEEE Trans. Pattern Anal. Mach. Intell. 2017, 39, 2481–2495. [Google Scholar] [CrossRef]
- Apostolopoulos, I.D.; Mpesiana, T.A. Covid-19: Automatic detection from x-ray images utilizing transfer learning with convolutional neural networks. Phys. Eng. Sci. Med. 2020, 43, 635–640. [Google Scholar] [CrossRef]
- Panwar, H.; Gupta, P.; Siddiqui, M.K.; Morales-Menendez, R.; Singh, V. Application of deep learning for fast detection of COVID-19 in X-Rays using nCOVnet. Chaos Solitons Fractals 2020, 138, 109944. [Google Scholar] [CrossRef]
- He, K.; Zhang, X.; Ren, S.; Sun, J. Deep residual learning for image recognition. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Las Vegas, NV, USA, 27–30 June 2016; pp. 770–778. [Google Scholar]
- Narin, A.; Kaya, C.; Pamuk, Z. Automatic detection of coronavirus disease (covid-19) using X-ray images and deep convolutional neural networks. arXiv 2020, arXiv:2003.10849. [Google Scholar]
- Sethy, P.K.; Behera, S.K. Detection of coronavirus disease (covid-19) based on deep features. Int. J. Math. Eng. Manag. Sci. 2020, 5, 643–651. [Google Scholar]
- Togacar, M.; Ergen, B.; Cömert, Z. COVID-19 detection using deep learning models to exploit Social Mimic Optimization and structured chest X-ray images using fuzzy color and stacking approaches. Comput. Biol. Med. 2020, 121, 103805. [Google Scholar] [CrossRef] [PubMed]
- Ucar, F.; Korkmaz, D. COVIDiagnosis-Net: Deep Bayes-SqueezeNet based diagnosis of the coronavirus disease 2019 (COVID-19) from X-ray images. Med. Hypotheses 2020, 140, 109761. [Google Scholar] [CrossRef] [PubMed]
- Ozturk, T.; Talo, M.; Yildirim, E.A.; Baloglu, U.B.; Yildirim, O.; Acharya, U.R. Automated detection of COVID-19 cases using deep neural networks with X-ray images. Comput. Biol. Med. 2020, 121, 103792. [Google Scholar] [CrossRef]
- Redmon, J.; Divvala, S.; Girshick, R.; Farhadi, A. You Only Look Once: Unified, Real-Time Object Detection. In Proceedings of the 2016 IEEE Conference on Computer Vision and Pattern Recognition (CVPR), Las Vegas, NV, USA, 27–30 June 2016; pp. 779–788. [Google Scholar]
- Toraman, S.; Alakus, T.B.; Turkoglu, I. Convolutional capsnet: A novel artificial neural network approach to detect COVID-19 disease from X-ray images using capsule networks. Chaos Solitons Fractals 2020, 140, 110122. [Google Scholar] [CrossRef]
- Loey, M.; Smarandache, F.; Khalifa, M.N.E. Within the Lack of Chest COVID-19 X-ray Dataset: A Novel Detection Model Based on GAN and Deep Transfer Learning. Symmetry 2020, 12, 651. [Google Scholar] [CrossRef]
- Cohen, J.P.; Morrison, P.; Dao, L. COVID-19 image data collection. arXiv 2020, arXiv:2003.11597. [Google Scholar]
- Adeyemia, H.O.; Nabotha, S.A.; Yusufa, S.O.; Dadaa, O.M.; Alaob, P.O. The Development of Fuzzy Logic-Base Diagnosis Expert System for Typhoid Fever. J. Kejuruteraan 2020, 32, 9–16. [Google Scholar] [CrossRef]
- Rahman, F. The Malaysian Response to COVID-19: Building Preparedness for Surge Capacity, Testing Efficiency and Containment; Russell Publishing Ltd.: Brasted, Kent, UK, 2020. [Google Scholar]
- Shibly, K.H.; Dey, S.K.; Islam, M.T.U.; Rahman, M.M. COVID faster R—CNN: A novel framework to Diagnose Novel Coronavirus Disease (COVID-19) in X-ray images. Inform. Med. Unlocked 2020, 20, 100405. [Google Scholar] [CrossRef]
- Kermany, D.; Zhang, K.; Goldbaum, M. Labeled Optical Coherence Tomography (OCT) and Chest X-Ray Images for Classification. Mendeley Data 2018. [Google Scholar] [CrossRef]
- Radford, A.; Metz, L.; Chintala, S. Unsupervised representation learning with deep convolutional generative adversarial networks. arXiv 2015, arXiv:1511.06434. [Google Scholar]
- Kingma, D.P.; Ba, J. Adam: A Method for Stochastic Optimization. In Proceedings of the 3rd International Conference on Learning Representations. arXiv 2014, arXiv:1412.6980v9. [Google Scholar]
- Srivastava, N.; Hinton, G.; Krizhevsky, A.; Sutskever, I.; Salakhutdinov, R. Dropout: A Simple Way to Prevent Neural Networks from Overfitting. J. Mach. Learn. Res. 2014, 15, 1929–1958. [Google Scholar]
- Chollet, F. Xception: Deep Learning with Depthwise Separable Convolutions. In Proceedings of the 2017 IEEE Conference on Computer Vision and Pattern Recognition (CVPR), Honolulu, HI, USA, 21–26 July 2017; pp. 1800–1807. [Google Scholar]
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).