Abstract
Early detection of pancreatic cancer is difficult, and thus many cases of pancreatic cancer are diagnosed late. When pancreatic cancer is detected, the cancer is usually well developed. Machine learning is an approach that is part of artificial intelligence and can detect pancreatic cancer early. This paper proposes a machine learning approach with the twin support vector machine (TWSVM) method as a new approach to detecting pancreatic cancer early. TWSVM aims to find two symmetry planes such that each plane has a distance close to one data class and as far as possible from another data class. TWSVM is fast in building a model and has good generalizations. However, TWSVM requires kernel functions to operate in the feature space. The kernel functions commonly used are the linear kernel, polynomial kernel, and radial basis function (RBF) kernel. This paper uses the TWSVM method with these kernels and compares the best kernel for use by TWSVM to detect pancreatic cancer early. In this paper, the TWSVM model with each kernel is evaluated using a 10-fold cross validation. The results obtained are that TWSVM based on the kernel is able to detect pancreatic cancer with good performance. However, the best kernel obtained is the RBF kernel, which produces an accuracy of 98%, a sensitivity of 97%, a specificity of 100%, and a running time of around 1.3408 s.
1. Introduction
According to the World Health Organization (WHO), the second leading cause of death in the world is cancer [1]. In 2018, around 17 million new cases of cancer occurred in the world and 9.6 million of the world’s population died from cancer [2]. Cancer has many types depending on location. If cancer occurs in the pancreas, then the cancer is called pancreatic cancer. Pancreatic cancer is the seventh leading cause of cancer deaths in the world and ranks as the 14th most common cancer [3]. Based on the Global Cancer Observatory in 2018, the estimated number of diagnoses of this cancer in the world is 458,918 and the estimated number of deaths is 432,242 [4]. This cancer is expected to be the second leading cause of cancer deaths in the world in 2030 [5].
Pancreatic cancer is a type of cancer that is difficult to detect physically. This is because the pancreas is an organ deep in the body; there are no external lumps or external skin changes such as in cases of breast lesions [6]. In addition, non-specific symptoms such as nausea, anorexia, jaundice, weight loss, and abdominal pain are also factors in the difficulty of detecting pancreatic cancer early [7]. The difficulty in detecting pancreatic cancer early results in many cases of pancreatic cancer being diagnosed late. When pancreatic cancer is detected, cancer is usually well developed, whereas in one study, only 7% of pancreatic cancer was diagnosed as localized disease [6]. If pancreatic cancer is detected too late, then the cancer can develop properly and spread to other body parts so that the cancer is difficult to treat. Therefore, early detection of pancreatic cancer is an important problem.
The problem of detecting pancreatic cancer early is a classification problem in machine learning. Machine learning has been accepted for various classification problems in various fields, one of which is in the field of medicine. In the field of medicine, several machine learning methods are used to detect several types of cancer, namely breast cancer [8,9,10,11], cervical cancer [12,13], ovarian cancer [14,15], colon cancer [16], prostate cancer [17], and lung cancer [18].
Machine learning has also been implemented to detect pancreatic cancer. Qiu et al. [19] implemented several methods of machine learning, namely the decision tree, the -nearest neighbor, and the support vector machine. From the results of the implementation, the support vector machine method is the method that produces the best performance, which is 70% sensitivity and 70% specificity.
The support vector machine (SVM) is one of the well-known machine learning methods; it uses the concept of “maximum margin”, and this concept reduces generalization errors by maximizing margins between two half-separated planes [20]. Many researchers have proposed the development of SVM to obtain better performance. One such development is the twin support vector machine proposed by Jayadeva et al. in 2007.
The twin support vector machine (TWSVM) aims to find two symmetry planes such that each plane has a distance close to one data class and as far as possible from another data class [20]. On several benchmark data sets, TWSVM is not only fast, but shows good generalization [20]. At present, TWSVM has become one of the popular methods because of its excellent learning performance [21]. However, both SVM and TWSVM indirectly need a kernel method to classify data. The kernel method is a method that uses a kernel function that allows an algorithm to operate in a feature space that has a higher dimension by using product operations between images of all data pairs in the feature space [22]. The kernel functions commonly used for SVM methods are the linear kernel, polynomial kernel, and radial basis function (RBF) kernel.
This paper proposes the TWSVM method as a novel approach for early detection of pancreatic cancer. The kernel functions used are the linear kernel, the polynomial kernel, and the RBF kernel. This paper compares the performance of TWSVM with each kernel to get the best kernel for early detection of pancreatic cancer.
2. Materials and Methods
2.1. Kernel Method
The kernel method uses a kernel function that allows an algorithm to operate in a feature space that has a higher dimension by using product operations between images of all data pairs in the feature space [22].
Let be an input space; is a feature space, and . Kernel function is defined by
where .
Kernel functions that are often used are the linear kernel, the polynomial kernel, and the radial basis function (RBF) kernel. The formulas for the kernel functions are listed below.
2.1.1. Linear Kernel
Linear kernels are the simplest kernel functions, represented by the product [23]. This kernel is the basic kernel that is most often used by SVM because with this kernel, SVM divides data linearly. The linear kernel formula is presented in Equation (2) [24].
2.1.2. Polynomial Kernel
The polynomial kernel is a kernel that is suitable for problems where training data is normalized [24]. From Equation (3), is the parameter that must be settled. Variable is the constant that is set and variable is the degree of polynomial that is set.
2.1.3. Radial Basis Function (RBF) Kernel
The RBF kernel is a kernel family where distance measurements are smoothed by radials function (exponential function) [23]. The RBF kernel is denoted as in Equation (4),
where is the adjustable parameter.
2.2. Twin Support Vector Machine (TWSVM)
The twin support vector machine (TWSVM) is one of the developments of the support vector machine proposed by Jayadeva et al. in 2007. TWSVM aims to find two symmetry planes such that each plane has a distance close to one data class and as far as possible from another data class. [20]. Figure 1 shows an illustration of TWSVM.
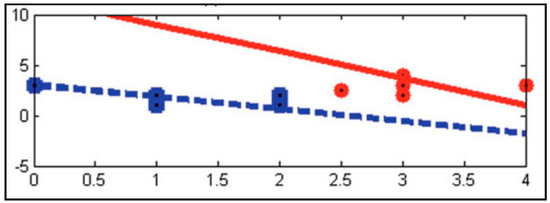
Figure 1.
Illustration of the twin support vector machine (TWSVM) [20].
Let is the training data. Then let there be data points in class +1 and data points in class −1 such that . We form matrix A, which contains the data points in class +1, and matrix B, which contains the data points in class −1. The two non-parallel hyperplanes are [20]:
where is data vector, is weight parameter for first hyperplane, is bias parameter for first hyperplane, is weight parameter for second hyperplane, and is bias parameter for second hyperplane.
The TWSVM method is obtained by solving the following pair of quadratic programming problems [20]:
TWSVM 1:
subject to
and
TWSVM 2:
subject to
where and are penalty parameters, and are slack variables, and and are vectors of ‘ones’, i.e., each component is ‘one’ only [20].
The two hyperplanes of TWSVM with kernel [20]:
where , , and is the kernel matrix corresponding to an appropriately chosen kernel function [11].
The kernel TWSVM can be obtained by solving the optimization problems [20]:
KTWSVM 1:
subject to
and
KTWSVM 2:
subject to
where and are penalty parameters, and are slack variables, and are vectors of ‘ones’, i.e., each component is ‘one’ only, , , and is the kernel matrix corresponding to an appropriately chosen kernel function [20].
2.3. k-Fold Cross Validation
In this paper, to obtain a model and evaluate the model obtained, the dataset was divided into training data and testing data. Training data are data used by machines to recognize and study pancreatic cancer data patterns, while testing data are data used to evaluate models obtained after a machine learns data patterns. The dataset was divided into training data and testing data using the -fold cross validation method. -fold cross validation is a method for selecting training data samples. The -fold cross validation method divided the dataset into sections of equal size [25]. Each subsample was taken as validation data to test the model and repeat the process times [25]. The advantage of this method is the repetition of random samples as training data and validation [25].
2.4. Proposed Method
In this paper, the method proposed for early detection of pancreatic cancer consisted of four stages. In the first stage, the data were divided into training data and testing data using k-fold cross validation. The k value chosen was 10. This means that the dataset was divided into 10 samples of the same size. In this dataset, 9 samples were used as training data, and 1 sample was used as testing data. In the second stage, the training data were used by the TWSVM method based on linear kernels, polynomial kernels, and RBF kernels to study data patterns and build classification models. In the third stage, the classification model obtained was evaluated based on the parameters of accuracy, sensitivity, specificity, and required running time. This evaluation used testing data. After that, evaluation parameters generated by each kernel were compared to find out the best kernel to detect pancreatic cancer early using the TWSVM method. The stages carried out in this paper used the Python 3 programming language.
3. Results and Discussions
3.1. Data
In this paper, the data used were pancreatic cancer data obtained from Al Islam Hospital Bandung, Indonesia. The data consisted of 203 samples and six features. The six features were the diagnosis of patient, namely pancreatic cancer (Y) and not pancreatic cancer (N), and blood tests which consisted of cancer antigens, hemoglobin, leukocytes, hematocrit, and platelets. The diagnosis feature became a target feature in detecting pancreatic cancer. Table 1 shows part of the data.

Table 1.
Part of the pancreatic data.
In the data shown in Table 1, the diagnosis feature is a categorical feature. This feature must be changed into a numeric feature for the proposed method to work. Therefore, based on the TWSVM method, the Y category in the diagnostic feature was transformed into +1 and the N category in the diagnostic feature was transformed into −1.
3.2. Confusion Matrix
In this paper, a confusion matrix was used to assist in calculating the evaluation parameters of the classification model. Table 2 shows the confusion matrix used to evaluate the TWSVM classification model based on the kernel for early detection of pancreatic cancer.

Table 2.
Confusion matrix.
Explanation:
- TP = Many cases of pancreatic cancer are predicted to be correct
- TN = Many cases of not pancreatic cancer are predicted to be correct
- FP = Many cases of not pancreatic cancer are predicted to be wrong (predicted as pancreatic cancer)
- FN = Many pancreatic cancer cases are predicted to be wrong (predicted as not pancreatic cancer)
3.3. Evaluation Parameters
The parameters to evaluate the performance of the TWSVM classification model were accuracy, sensitivity, specificity, and required running time. Table 3 shows the formula for accuracy, sensitivity, and specificity.

Table 3.
Parameter formulae to evaluate the classification model.
3.4. Results
In this section, we discuss the performance evaluation of the TWSVM classification model with a linear kernel, polynomial kernel, and RBF kernel. The TWSVM classification model based on kernel that is proposed in this paper refers to research conducted by [20] which detects hepatitis using TWSVM with a linear kernel and a RBF kernel. In research conducted by the authors of [20], the accuracy produced by the RBF kernel is superior to that of a linear kernel. This indicates that the RBF kernel is the appropriate kernel in detecting hepatitis using TWSVM.
In this paper, we have built the TWSVM classification model with a linear kernel, a polynomial kernel, and a RBF kernel in detecting pancreatic cancer. Table 4 presents a comparison of TWSVM performance with linear kernels, polynomial kernels with , and RBF kernels with . The performance evaluation parameters compared are accuracy, sensitivity, specificity, and running time.

Table 4.
Results of TWSVM classification model based on kernel.
From Table 4, it can be seen that for accuracy, TWSVM models with RBF kernel have the most superior accuracy compared to the linear kernel and the polynomial kernel, reaching 98%. That is, the TWSVM model with the RBF kernel correctly detected 98% of the total cases. The lowest accuracy is the TWSVM model with a polynomial kernel, with a resulting percentage of 80%. In addition, for consideration of sensitivity and specificity, TWSVM models with RBF kernel have percentages that are also superior than the linear kernel and polynomial kernel, which are 97% sensitivity and 100% specificity. This means that the TWSVM model with the RBF kernel is able to detect 98% of cases correctly, with a truth of 97% of all cases of pancreatic cancer, and 100% of all cases of non-pancreatic cancer.
Based on consideration of accuracy, sensitivity, and specificity produced by the TWVM model with a linear kernel, a polynomial kernel, and a RBF kernel, overall the TWSVM model with RBF kernel is the most superior kernel. This means that the pancreatic cancer dataset can be separated almost precisely by RBF function. However, for consideration of running time, the TWSVM model with RBF kernel has the longest running time compared to linear and polynomial kernels, which is around 1.3408 s. The TWSVM model with the polynomial kernel actually produces the fastest running time, which is around 1.2040 s. Even so, the running time produced by the RBF kernel is quite good and acceptable for detecting pancreatic cancer early. Thus, the RBF kernel is the best kernel for TWSVM in detecting pancreatic cancer early.
4. Conclusions
Early detection of pancreatic cancer is very important so that the handling of pancreatic cancer does not occur too late, before the cancer spreads to other organs in the body. However, early detection of pancreatic cancer is difficult because this cancer has non-specific symptoms. The twin support vector machine method based on the kernel can help detect pancreatic cancer early, based on blood tests. The most appropriate kernel for the TWSVM method in detecting pancreatic cancer is the RBF kernel which produces an accuracy of 98%, sensitivity of 97%, and 100% specificity, and the required running time is 1.3408 s.
Author Contributions
Conceptualization, W.S. and Z.R.; methodology, W.S. and H.H.; software, H.H. and Z.R.; validation, W.S., H.H., Z.R., and A.R.C.; formal analysis, W.S., H.H., and Z.R.; investigation, W.S. and Z.R.; resources, W.S.; data curation, W.S. and Z.R.; writing—original draft preparation, H.H.; writing—review and editing, A.R.C.; visualization, W.S.; supervision, Z.R.; project administration, Z.R.; funding acquisition, W.S. and Z.R. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by Universitas Indonesia with a DRPM PUTI Q2 2020 research grant scheme and The APC received no external funding.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Cancer. Available online: https://www.who.int/cancer/en/ (accessed on 19 January 2020).
- Worldwide Cancer Statistics. Available online: https://www.cancerresearchuk.org/health-professional/cancer-statistics/worldwide-cancer (accessed on 25 January 2020).
- McGuigan, A.; Kelly, P.; Turkington, R.C.; Jones, C.; Coleman, H.G.; MacCain, R.S. Pancreatic cancer: A review of clinical diagnosis, epidemiology, treatment and outcomes. World J. Gastroenterol. 2018, 24, 4846–4861. [Google Scholar] [CrossRef] [PubMed]
- Global Cancer Observatory 2018. Available online: http://gco.iarc.fr/ (accessed on 23 December 2019).
- Rahib, L.; Smith, B.D.; Aizenberg, R.; Rosenzweig, A.B.; Fleshman, J.M.; Matrisian, L.M. Projecting cancer incidence and deaths to 2030: The unexpected burden of thyroid, liver, and pancreas cancers in the United States. Cancer Res. 2014, 74, 2913–2921. [Google Scholar] [CrossRef] [PubMed]
- Kim, V.M.; Ahuja, N. Early Detection of Pancreatic Cancer. Chin. J. Cancer Res. 2015, 27, 321–331. [Google Scholar] [PubMed]
- Badger, S.A.; Brant, J.L.; Jones, C.; McClements, J.; Loughrey, M.B.; Taylor, M.A.; Diamond, T.; McKie, L.D. The role of surgery for pancreatic cancer: A 12-year review of patient outcome. Ulster Med. J. 2010, 79, 70–75. [Google Scholar] [PubMed]
- Octaviani, T.L.; Rustam, Z. Random Forest for Breast Cancer Prediction. In Proceedings of the 4th International Symposium on Current Progress in Mathematics and Sciences (ISCPMS2018), Depok, Indonesia, 30–31 October 2018. [Google Scholar]
- Rustam, Z.; Putri, R.A. Comparison between stochastic support vector machine (stochastic SVM) and Fuzzy Kernel Robust C-Means (FKRCM) in breast cancer classification. In Proceedings of the 4th International Symposium on Current Progress in Mathematics and Sciences (ISCPMS2018), Depok, Indonesia, 30–31 October 2018. [Google Scholar]
- Rustam, Z.; Hartini, S. Classification of Breast Cancer using Fast Fuzzy Clustering based on Kernel. In Proceedings of the 9th Annual Basic Science International Conference 2019 (BaSIC 2019), Malang, Indonesia, 20–21 March 2019. [Google Scholar]
- Fijri, A.L.; Rustam, Z. Comparison between Fuzzy Kernel C-Means and Sparse Learning Fuzzy C-Means for Breast Cancer Clustering. In Proceedings of the ICAITI 2018—1st International Conference on Applied Information Technology and Innovation: Toward A New Paradigm for the Design of Assistive Technology in Smart Home Care, Padang, Indonesia, 4–5 September 2018. [Google Scholar]
- Rustam, Z.; Hapsari, V.A.W.; Solihin, M.R. Optimal cervical cancer classification using Gauss-Newton representation based algorithm. In Proceedings of the 4th International Symposium on Current Progress in Mathematics and Sciences (ISCPMS2018), Depok, Indonesia, 30–31 October 2018. [Google Scholar]
- Zahras, D.; Rustam, Z. Cervical Cancer Risk Classification Based on Deep Convolutional Neural Network. In Proceedings of the ICAITI 2018—1st International Conference on Applied Information Technology and Innovation: Toward A New Paradigm for the Design of Assistive Technology in Smart Home Care, Padang, Indonesia, 4–5 September 2018. [Google Scholar]
- Arfiani, A.; Rustam, Z. Ovarian cancer data classification using bagging and random forest. In Proceedings of the 4th International Symposium on Current Progress in Mathematics and Sciences (ISCPMS2018), Depok, Indonesia, 30–31 October 2018. [Google Scholar]
- Octaviani, T.L.; Rustam, Z.; Siswantining, T. Ovarian Cancer Classification using Bayesian Logistic Regression. In Proceedings of the 9th Annual Basic Science International Conference 2019 (BaSIC 2019), Malang, Indonesia, 20–21 March 2019. [Google Scholar]
- Salmi, N.; Rustam, Z. Naïve Bayes Classifier Models for Predicting the Colon Cancer. In Proceedings of the 9th Annual Basic Science International Conference 2019 (BaSIC 2019), Malang, Indonesia, 20–21 March 2019. [Google Scholar]
- Huljanah, M.; Rustam, Z.; Utama, S.; Siswantining, T. Feature Selection using Random Forest Classifier for Predicting Prostate Cancer. In Proceedings of the 9th Annual Basic Science International Conference 2019 (BaSIC 2019), Malang, Indonesia, 20–21 March 2019. [Google Scholar]
- Rustam, Z.; Kharis, S.A.A. Comparison of Support Vector Machine Recursive Feature Elimination and Kernel Function as feature selection using Support Vector Machine for lung cancer classification. In Proceedings of the Basic and Applied Sciences Interdisciplinary Conference, Depok, Indonesia, 18–19 August 2017. [Google Scholar]
- Qiu, Y.; Jiang, H.; Shimada, K.; Hiraoka, N.; Maeshiro, K.; Ching, W.K.; Aoki-Kinoshita, K.; Furuta, K. Towards Prediction of Pancreatic Cancer Using SVM Study Model. J. Clin. Oncol. Res. 2014, 2, 1031. [Google Scholar]
- Jayadeva; Khemchandani, R.; Chandra, S. Twin Support Vector Machines for Pattern Classification. IEEE Trans. Pattern Anal. Mach. Intell. 2007, 29, 905–910. [Google Scholar] [CrossRef] [PubMed]
- Huang, H.; Wei, X.; Zhou, Y. Twin support vector machines: A survey. Neurocomputing 2018, 300, 34–43. [Google Scholar] [CrossRef]
- Shawe-Taylor, J.; Cristianini, N. Kernel Methods for Pattern Analysis; Cambridge University Press: Cambridge, UK, 2004. [Google Scholar]
- Suo, H.; Li, M.; Lu, P.; Yan, Y. Using SVM as Back-End Classifier for Language Identification. EURASIP J. Audio Speech Music Process. 2008, 1, 674859. [Google Scholar] [CrossRef][Green Version]
- Chidambaram, S.; Srinivasagan, K.G. Performance evaluation of support vector machine classification approaches in data mining. Clust. Comput. 2018, 22, S189–S196. [Google Scholar]
- Raju, K.S.; Murty, M.R.; Rao, M.V.; Satapathy, S.C. Support Vector Machine with K-fold Cross Validation Model for Software Fault Prediction. Int. J. Pure Appl. Math. 2018, 118, 321–334. [Google Scholar]
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).