Statistical Validation Verifies That Enantiomorphic States of Chiral Cells Are Determinant Dictating the Left- or Right-Handed Direction of the Hindgut Rotation in Drosophila
Abstract
1. Introduction
2. Materials and Methods
2.1. Fly Lines
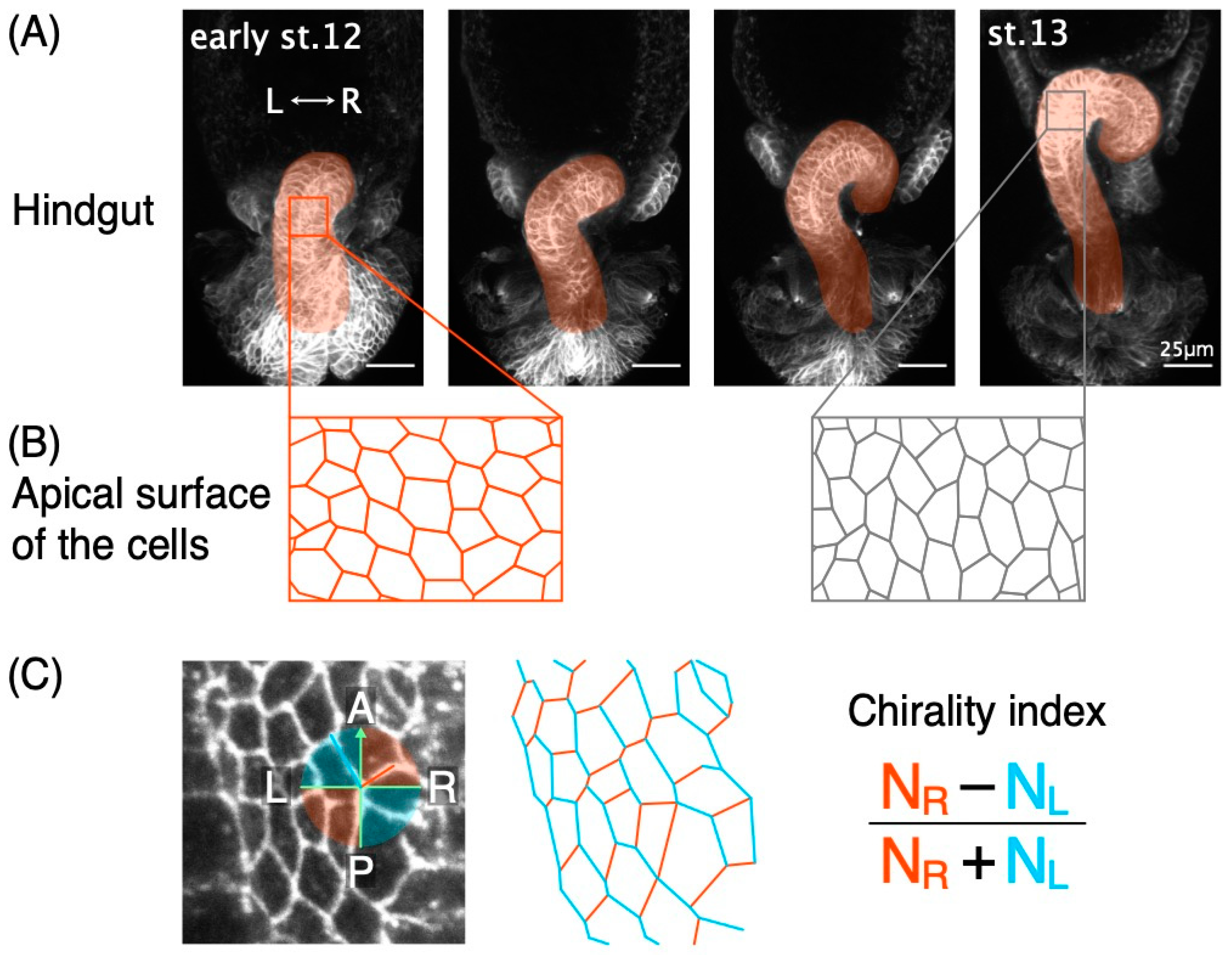
2.2. Live Imaging and Laterality Score
2.3. Analysis of Cell Chirality Index
2.4. Statistical Analysis
3. Results
3.1. A Logistic Model Significantly Explained the Correlation between the Enantiomorphic States of Chiral Cells and the LR-Directionality of Hindgut Rotation
3.2. The Cell Chirality Index Predicts the LR-Directions of Future Hindgut Rotation in Live Embryos
4. Discussion
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Appendix A
| p-Value | ||
|---|---|---|
| Intercept | −4.042 | 0.003723 |
| Chirality Index | −5.992 | 0.000326 |
| p-Value | ||
|---|---|---|
| Intercept | −1.149 | 0.27109 |
| Live Chirality Index | −3.256 | 0.00625 |
| Model | ||
|---|---|---|
| Best fit model | 0.8607872 | |
| Model fitted for Figure 2 | 0.7246853 |
References
- Blum:, M.; Ott, T. Animal left–right asymmetry. Curr. Biol. 2018, 28, R301–R304. [Google Scholar] [CrossRef]
- Hirokawa, N.; Tanaka, Y.; Okada, Y.; Takeda, S. Nodal Flow and the Generation of Left-Right Asymmetry. Cell 2006, 125, 33–45. [Google Scholar] [CrossRef]
- Nakamura, T.; Hamada, H. Left-right patterning: Conserved and divergent mechanisms. Development 2012, 139, 3257–3262. [Google Scholar] [CrossRef] [PubMed]
- Levin, M. Left-right asymmetry in embryonic development: A comprehensive review. Mech. Dev. 2005, 122, 3–25. [Google Scholar] [CrossRef] [PubMed]
- Yoshiba, S.; Hamada, H. Roles of cilia, fluid flow, and Ca2+signaling in breaking of left-right symmetry. Trends Genet. 2014, 30, 10–17. [Google Scholar] [CrossRef] [PubMed]
- Nonaka, S.; Tanaka, Y.; Okada, Y.; Takeda, S.; Harada, A.; Kanai, Y.; Kido, M.; Hirokawa, N. Randomization of left-right asymmetry due to loss of nodal cilia generating leftward flow of extraembryonic fluid in mice lacking KIF3B motor protein. Cell 1998, 95, 829–837. [Google Scholar] [CrossRef]
- Nonaka, S.; Shiratori, H.; Saijoh, Y.; Hamada, H. Determination of left-right patterning of the mouse embryo by artificial nodal flow. Nature 2002, 418, 96–99. [Google Scholar] [CrossRef]
- Takeda, S.; Yonekawa, Y.; Tanaka, Y.; Okada, Y.; Nonaka, S.; Hirokawa, N. Left-right asymmetry and kinesin superfamily protein KIF3a: New insights in determination of laterality and mesoderm induction by KIF3A(-/-) mice analysis. J. Cell Biol. 1999, 145, 825–836. [Google Scholar] [CrossRef]
- Kajikawa, E.; Horo, U.; Ide, T.; Mizuno, K.; Minegishi, K.; Hara, Y.; Ikawa, Y.; Nishimura, H.; Uchikawa, M.; Kiyonari, H.; et al. Nodal paralogues underlie distinct mechanisms for visceral left–right asymmetry in reptiles and mammals. Nat. Ecol. Evol. 2020, 4, 261–269. [Google Scholar] [CrossRef]
- Tabin, C. Do we know anything about how left-right asymmetry is first established in the vertebrate embryo? J. Mol. Histol. 2005, 36, 317–323. [Google Scholar] [CrossRef]
- Inaki, M.; Liu, J.; Matsuno, K. Cell chirality: Its origin and roles in left-right asymmetric development. Philos. Trans. R. Soc. B Biol. Sci. 2016, 371, 20150403. [Google Scholar] [CrossRef]
- Inaki, M.; Sasamura, T.; Matsuno, K. Cell chirality drives left-right asymmetric morphogenesis. Front. Cell Dev. Biol. 2018, 6. [Google Scholar] [CrossRef]
- Utsunomiya, S.; Sakamura, S.; Sasamura, T.; Ishibashi, T.; Maeda, C.; Inaki, M.; Matsuno, K. Cells with Broken Left–Right Symmetry: Roles of Intrinsic Cell Chirality in Left–Right Asymmetric Epithelial Morphogenesis. Symmetry 2019, 11, 505. [Google Scholar] [CrossRef]
- Kuroda, R.; Endo, B.; Abe, M.; Shimizu, M. Chiral blastomere arrangement dictates zygotic left-right asymmetry pathway in snails. Nature 2009, 462, 790–794. [Google Scholar] [CrossRef]
- Naganathan, S.R.; Fürthauer, S.; Nishikawa, M.; Jülicher, F.; Grill, S.W. Active torque generation by the actomyosin cell cortex drives left-right symmetry breaking. Elife 2014, 3, e04165. [Google Scholar] [CrossRef]
- Hayashi, T.; Murakami, R. Left-right asymmetry in Drosophila melanogaster gut development. Dev. Growth Differ. 2001, 43, 239–246. [Google Scholar] [CrossRef]
- Hozumi, S.; Maeda, R.; Taniguchi, K.; Kanai, M.; Shirakabe, S.; Sasamura, T.; Spéder, P.; Noselli, S.; Aigaki, T.; Murakami, R.; et al. An unconventional myosin in Drosophila reverses the default handedness in visceral organs. Nature 2006, 440, 798–802. [Google Scholar] [CrossRef]
- Ligoxygakis, P.; Strigini, M.; Averof, M. Specification of left-right asymmetry in the embryonic gut of Drosophila. Development 2001, 128, 1171–1174. [Google Scholar]
- Pascual, A.; Huang, K.L.; Neveu, J.; Préat, T. Brain asymmetry and long-term memory: Fruitflies that have structurally similar brain hemispheres forget within a matter of hours. Nature 2004, 427, 605–606. [Google Scholar] [CrossRef]
- Spéder, P.; Ádám, G.; Noselli, S. Type ID unconventional myosin controls left-right asymmetry in Drosophila. Nature 2006, 440, 803–807. [Google Scholar] [CrossRef]
- Maeda, R.; Hozumi, S.; Taniguchi, K.; Sasamura, T.; Murakami, R.; Matsuno, K. Roles of single-minded in the left-right asymmetric development of the Drosophila embryonic gut. Mech. Dev. 2007, 124, 204–217. [Google Scholar] [CrossRef]
- Nakamura, M.; Matsumoto, K.; Iwamoto, Y.; Muguruma, T.; Nakazawa, N.; Hatori, R.; Taniguchi, K.; Maeda, R.; Matsuno, K. Reduced cell number in the hindgut epithelium disrupts hindgut left-right asymmetry in a mutant of pebble, encoding a RhoGEF, in Drosophila embryos. Mech. Dev. 2013, 130, 169–180. [Google Scholar] [CrossRef]
- Taniguchi, K.; Hozumi, S.; Maeda, R.; Ooike, M.; Sasamura, T.; Aigaki, T.; Matsuno, K. D-JNK signaling in visceral muscle cells controls the laterality of the Drosophila gut. Dev. Biol. 2007, 311, 251–263. [Google Scholar] [CrossRef]
- Kuroda, J.; Nakamura, M.; Yoshida, M.; Yamamoto, H.; Maeda, T.; Taniguchi, K.; Nakazawa, N.; Hatori, R.; Ishio, A.; Ozaki, A.; et al. Canonical Wnt signaling in the visceral muscle is required for left-right asymmetric development of the Drosophila midgut. Mech. Dev. 2012, 128, 625–639. [Google Scholar] [CrossRef]
- Inaki, M.; Hatori, R.; Nakazawa, N.; Okumura, T.; Ishibashi, T.; Kikuta, J.; Ishii, M.; Matsuno, K.; Honda, H. Chiral cell sliding drives left-right asymmetric organ twisting. Elife 2018, 7, e32506. [Google Scholar] [CrossRef]
- Hozumi, S.; Maeda, R.; Taniguchi-Kanai, M.; Okumura, T.; Taniguchi, K.; Kawakatsu, Y.; Nakazawa, N.; Hatori, R.; Matsuno, K. Head region of unconventional myosin I family members is responsible for the organ-specificity of their roles in left-right polarity in Drosophila. Dev. Dyn. 2008, 237, 3528–3537. [Google Scholar] [CrossRef]
- Taniguchi, K.; Maeda, R.; Ando, T.; Okumura, T.; Nakazawa, N.; Hatori, R.; Nakamura, M.; Hozumi, S.; Fujiwara, H.; Matsuno, K. Chirality in planar cell shape contributes to left-right asymmetric epithelial morphogenesis. Science 2011, 333, 339–341. [Google Scholar] [CrossRef]
- Hatori, R.; Ando, T.; Sasamura, T.; Nakazawa, N.; Nakamura, M.; Taniguchi, K.; Hozumi, S.; Kikuta, J.; Ishii, M.; Matsuno, K. Left-right asymmetry is formed in individual cells by intrinsic cell chirality. Mech. Dev. 2014, 133, 146–162. [Google Scholar] [CrossRef]
- Ishibashi, T.; Hatori, R.; Maeda, R.; Nakamura, M.; Taguchi, T.; Matsuyama, Y.; Matsuno, K. E and ID proteins regulate cell chirality and left–right asymmetric development in Drosophila. Genes Cells 2019, 24, 214–230. [Google Scholar] [CrossRef]
- Sato, K.; Hiraiwa, T.; Maekawa, E.; Isomura, A.; Shibata, T.; Kuranaga, E. Left-right asymmetric cell intercalation drives directional collective cell movement in epithelial morphogenesis. Nat. Commun. 2015, 6. [Google Scholar] [CrossRef]
- Coutelis, J.B.; Géminard, C.; Spéder, P.; Suzanne, M.; Petzoldt, A.G.; Noselli, S. Drosophila Left/Right Asymmetry Establishment Is Controlled by the Hox Gene Abdominal-B. Dev. Cell 2013, 24, 89–97. [Google Scholar] [CrossRef] [PubMed]
- González-Morales, N.; Géminard, C.; Lebreton, G.; Cerezo, D.; Coutelis, J.B.; Noselli, S. The Atypical Cadherin Dachsous Controls Left-Right Asymmetry in Drosophila. Dev. Cell 2015, 33, 675–689. [Google Scholar] [CrossRef]
- Ray, P.; Chin, A.S.; Worley, K.E.; Fan, J.; Kaur, G.; Wu, M.; Wan, L.Q. Intrinsic cellular chirality regulates left–right symmetry breaking during cardiac looping. Proc. Natl. Acad. Sci. USA 2018, 115, E11568–E11577. [Google Scholar] [CrossRef]
- Onuma, T.A.; Hayashi, M.; Gyoja, F.; Kishi, K.; Wang, K.; Nishida, H. A chordate species lacking Nodal utilizes calcium oscillation and Bmp for left–right patterning. Proc. Natl. Acad. Sci. USA 2020, 117, 4188–4198. [Google Scholar] [CrossRef]
- Chen, T.H.; Hsu, J.J.; Zhao, X.; Guo, C.; Wong, M.N.; Huang, Y.; Li, Z.; Garfinkel, A.; Ho, C.M.; Tintut, Y.; et al. Left-right symmetry breaking in tissue morphogenesis via cytoskeletal mechanics. Circ. Res. 2012, 110, 551–559. [Google Scholar] [CrossRef]
- Tamada, A.; Kawase, S.; Murakami, F.; Kamiguchi, H. Autonomous right-screw rotation of growth cone filopodia drives neurite turning. J. Cell Biol. 2010, 188, 429–441. [Google Scholar] [CrossRef]
- Tee, Y.H.; Shemesh, T.; Thiagarajan, V.; Hariadi, R.F.; Anderson, K.L.; Page, C.; Volkmann, N.; Hanein, D.; Sivaramakrishnan, S.; Kozlov, M.M.; et al. Cellular chirality arising from the self-organization of the actin cytoskeleton. Nat. Cell Biol. 2015, 17, 445–457. [Google Scholar] [CrossRef]
- Wan, L.Q.; Ronaldson, K.; Park, M.; Taylor, G.; Zhang, Y.; Gimble, J.M.; Vunjak-Novakovic, G. Micropatterned mammalian cells exhibit phenotype-specific left-right asymmetry. Proc. Natl. Acad. Sci. USA 2011, 108, 12295–12300. [Google Scholar] [CrossRef]
- Xu, J.; Van Keymeulen, A.; Wakida, N.M.; Carlton, P.; Berns, M.W.; Bourne, H.R. Polarity reveals intrinsic cell chirality. Proc. Natl. Acad. Sci. USA 2007, 104, 9296–9300. [Google Scholar] [CrossRef]
- Lebreton, G.; Géminard, C.; Lapraz, F.; Pyrpassopoulos, S.; Cerezo, D.; Spéder, P.; Ostap, E.M.; Noselli, S. Molecular to organismal chirality is induced by the conserved myosin 1D. Science 2018, 362, 949–952. [Google Scholar] [CrossRef]
- Okumura, T.; Sasamura, T.; Inatomi, M.; Hozumi, S.; Nakamura, M.; Hatori, R.; Taniguchi, K.; Nakazawa, N.; Suzuki, E.; Maeda, R.; et al. Class I myosins have overlapping and specialized functions in left-right asymmetric development in Drosophila. Genetics 2015, 199, 1183–1199. [Google Scholar] [CrossRef]
- Wang, L.H.; Baker, N.E. E Proteins and ID Proteins: Helix-Loop-Helix Partners in Development and Disease. Dev. Cell 2015, 35, 269–280. [Google Scholar] [CrossRef]
- Ellis, H.M. Embryonic expression and function of the Drosophila helix-loop-helix gene, extramacrochaetae. Mech. Dev. 1994, 47, 65–72. [Google Scholar] [CrossRef]
- Wülbeck, C.; Fromental-Ramain, C.; Campos-Ortega, J.A. The HLH domain of a zebrafish hE12 homologue can partially substitute for functions of the HLH domain of Drosophila daughterless. Mech. Dev. 1994, 46, 73–85. [Google Scholar] [CrossRef]
- Petzoldt, A.G.; Coutelis, J.-B.; Géminard, C.; Spéder, P.; Suzanne, M.; Cerezo, D.; Noselli, S. DE-Cadherin regulates unconventional Myosin ID and Myosin IC in Drosophila left-right asymmetry establishment. Development 2012, 139, 1874–1884. [Google Scholar] [CrossRef]
- Popova, M.K.; He, W.; Korenjak, M.; Dyson, N.J.; Moon, N.-S. Rb deficiency during Drosophila eye development deregulates EMC, causing defects in the development of photoreceptors and cone cells. J. Cell Sci. 2011, 124, 4203–4212. [Google Scholar] [CrossRef]
- Giebel, B.; Stüttem, I.; Hinz, U.; Campos-Ortega, J.A. Lethal of Scute requires overexpression of Daughterless to elicit ectopic neuronal development during embryogenesis in Drosophila. Mech. Dev. 1997, 63, 75–87. [Google Scholar] [CrossRef]
- Pfeiffer, B.D.; Truman, J.W.; Rubin, G.M. Using translational enhancers to increase transgene expression in Drosophila. Proc. Natl. Acad. Sci. USA 2012, 109, 6626–6631. [Google Scholar] [CrossRef]
- Hayashi, S.; Ito, K.; Sado, Y.; Taniguchi, M.; Akimoto, A.; Takeuchi, H.; Aigaki, T.; Matsuzaki, F.; Nakagoshi, H.; Tanimura, T.; et al. GETDB, a database compiling expression patterns and molecular locations of a collection of gal4 enhancer traps. Genesis 2002, 34, 58–61. [Google Scholar] [CrossRef]
- Brand, A.H.; Perrimon, N. Targeted gene expression as a means of altering cell fates and generating dominant phenotypes. Development 1993, 118, 401–415. [Google Scholar]
- Schindelin, J.; Arganda-Carreras, I.; Frise, E.; Kaynig, V.; Longair, M.; Pietzsch, T.; Preibisch, S.; Rueden, C.; Saalfeld, S.; Schmid, B.; et al. Fiji: An open-source platform for biological-image analysis. Nat. Methods 2012, 9, 676–682. [Google Scholar] [CrossRef] [PubMed]
- Campbell, M.J.; Dobson, A.J. An Introduction to Generalized Linear Models. Biometrics 1991, 47, 347. [Google Scholar] [CrossRef]
- R Core Team. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2020. [Google Scholar]
- Van Rossum, G.; Drake, F.L. Python Tutorial, Technical Report CS-R9526. In Proceedings of the Centrum Voor Wiskunde en Informatica (CWI); Academic Press: Cambridge, MA, USA, 1995. [Google Scholar]
- Hunter, J.D. Matplotlib: A 2D graphics environment. Comput. Sci. Eng. 2007, 9, 99–104. [Google Scholar] [CrossRef]


Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ishibashi, T.; Inaki, M.; Matsuno, K. Statistical Validation Verifies That Enantiomorphic States of Chiral Cells Are Determinant Dictating the Left- or Right-Handed Direction of the Hindgut Rotation in Drosophila. Symmetry 2020, 12, 1991. https://doi.org/10.3390/sym12121991
Ishibashi T, Inaki M, Matsuno K. Statistical Validation Verifies That Enantiomorphic States of Chiral Cells Are Determinant Dictating the Left- or Right-Handed Direction of the Hindgut Rotation in Drosophila. Symmetry. 2020; 12(12):1991. https://doi.org/10.3390/sym12121991
Chicago/Turabian StyleIshibashi, Tomoki, Mikiko Inaki, and Kenji Matsuno. 2020. "Statistical Validation Verifies That Enantiomorphic States of Chiral Cells Are Determinant Dictating the Left- or Right-Handed Direction of the Hindgut Rotation in Drosophila" Symmetry 12, no. 12: 1991. https://doi.org/10.3390/sym12121991
APA StyleIshibashi, T., Inaki, M., & Matsuno, K. (2020). Statistical Validation Verifies That Enantiomorphic States of Chiral Cells Are Determinant Dictating the Left- or Right-Handed Direction of the Hindgut Rotation in Drosophila. Symmetry, 12(12), 1991. https://doi.org/10.3390/sym12121991





