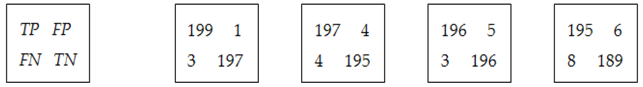1. Introduction
Eye disorders can lead partial or even total loss of vision, significantly affecting life quality [
1]. In order to ensure timely medical intervention and treatment, their early detection relies on the analysis of retinal components such as optic disc (
OD), macula (
MA), blood vessels (
BVs), exudates (
EXs), and hemorrhages (
HEs). Only intelligent processing of a retinal image can provide real measures of these specific ophthalmological zones. Adding complex image processing to dedicated neural networks and efficient descriptors and classifiers, the system for determining the RoIs in retinal images can be considered as an effective support for decision-making in early disease diagnosis. The system’s input is the retinal image and the output is the degree of retinal RoI damage. Automatic detection based on specific imaging devices and real-time image processing can be used for the early-stage decision of an ophthalmologist. Computer-aided diagnosis in an intelligent system also has the advantages of computational power, speed, and continuous development. Many different approaches for retinal RoI identification and evaluation are found in the literature. However, accurate and simultaneous detection, localization, and evaluation of
OD,
MA,
BV,
EX, and
HE is a difficult task for such a system. The general difficulty for simultaneous detection of retinal RoIs is the symmetries between them, like between
OD and
EX (color and, sometimes, segmented shapes) and between
EX and
HE (segmented shapes). It must have intelligent algorithms for image processing and interpretation and sufficient computational power. Usually, it must exclude the similarities (symmetries) and point out the differences.
The first RoI, the
OD, is the easiest to detect, due to its shape, colors, size, and structure. Therefore, many papers are related to its detection and localization [
2,
3,
4,
5,
6,
7,
8,
9,
10,
11,
12,
13,
14]. For example, the maximum number of decisions from a set of detectors can be used to detect the center of the
OD [
7]. By selecting some textural features and image decomposition in overlapping boxes,
OD detection becomes more simple and efficient [
4]. Taking into account the density of large blood vessels in the
OD region, the authors in [
6,
8] proposed fast and accurate methods for
OD detection. Considering that the
OD has the highest radial symmetry, a methodology based on the radial symmetry detector and separation of vascular information is used in [
14] for
OD localization. Sometimes, the
OD and
EX are similar in color, intensity, and shape. In pathological images, a large
EX can be similar to the
OD. To overcome this inconvenience, an automatic
OD detection is proposed in [
13], based on the symmetry axis of blood vessel distribution in this region. Some authors remove the
OD before the detection and localization of
EXs. To this end, the authors in [
15] proposed a methodology to differentiate
EXs from
ODs by considering the features on different color components. The optic disc is identified as a region where the majority of pixels have a great difference on R and G channels. Next, a thresholding method is applied in order to extract the exudates from the obtained binary image. Some features like mean intensity, entropy, uniformity, standard deviation, and smoothness are computed for obtaining an improved accuracy. The mentioned characteristics have different values for
EXs and
ODs; so,
EXs can be identified by removing the areas that do not have matching values of the previous characteristics. For the optic disc detection, in [
16], an algorithm based on the 2D Gaussian filter and blood vessel segmentation was implemented. Also for
OD detection Pereira et al. [
17] applied a modern algorithm based on ant colony optimization and anisotropic diffusion. More recently, the authors in [
18] proposed a supervised model for
OD abnormality detection for different datasets, taking into account a deep learning approach from global retinal images. The results show a good accuracy and indicate potential applications. Automatic image analysis methods aiming to detect and segment
MA and fovea are currently based on support vector machine classification [
19], hierarchical image decomposition [
20,
21], statistical segmentation methods [
22], deep learning [
23], and pixel intensity characteristics [
24]. The authors in [
7] presented a method which combines different
OD and
MA classifiers to benefit from their strong points (high accuracy and low computation time). With the purpose of detection of macular degeneration, Veras et al. [
25] proposed some algorithms to localize and segment the
MA. Sekhar et al. [
26] addresses blood vessels localization, segmentation, and their spatial relations in order to detect the optic disc and macula. Based on pixel intensity (low), the authors in [
27] also proposed the
MA detection technique. They also proposed an algorithm for the
OD diameter estimation. Akram et al. [
28] presented a system based on artificial intelligence for evaluation of macular edema to support the ophthalmologists in early detection of the disease. They used a comprehensive characteristics group and a Gaussian-mixture-model-based classifier for accurate detection of
MA. Chaudhry et al. [
29] detect macula from an autofluorescence imaging device in connection with
OD position. This automatic positioning is based on a top-down approach using vascular tree segmentation and skeletonization. In [
30] the authors proposed a method to detect and localize the macula center using two steps: first, the context - based information is aggregated with a statistical type model in order to obtain the approximate localization of the macula; second, the aggregation of seeded mode tracking is used for final localization of the macula center.
Over time, many methods and approaches have been used for the detection of
EXs (e.g., thresholding methods considering the green channel [
31,
32,
33]). As a consequence that the illumination conditions are different, image preprocessing algorithms for correction are necessary. Kaur and Mittal [
34] proposed an algorithm for
EX detection which first identifies and rejects the healthy areas based on their characteristics and second segments the
EXs from the remaining regions using a region growing method. Bu et al. [
35] presented a hierarchical procedure for
EX detection in eye fundus images to solve some difficulties such as the recognition of similar-colored objects (e.g. cotton wool spots, optic disc, and the normal macular reflection). Using an artificial intelligence method, an automated identification of
EXs is obtained in [
36]. After an initial primary processing phase, an image segmentation based on fuzzy clustering is used and a dataset of regions of interest is obtained. For each area, characteristics such as size, color, edge, and texture are extracted. The authors proposed a deep neural network, based on the extracted characteristics for the detection of areas containing
EXs. The images are firstly filtered by applying a fuzzy clustering technique. Furthermore, the method in [
37] localizes and evaluates the exudates, taking into account different image processing techniques, morphological filters, and image features.
The approaches for
HE identification commonly used can also be grouped into different categories such as thresholding, segmentation, mathematical morphology, classification, clustering, neural network, etc. For example, Seoud et al. [
38] described and validated a method for the identification of micro-aneurysms and
HEs in retinal images, which can be used in the diagnosis and grading of diabetic retinopathy. Their contribution is a set of dynamic shape features which show the evolution of shapes under a processed image and allow differentiation between lesions and blood vessels. The method was tested against six image datasets, both public and private. A splat-based feature classification algorithm with applications to large, irregular
HE detection in retinal images was proposed in [
39].
For detection and segmentation of retinal RoIs, different classifiers were used: model-based (template matching), minimum distance between vector features, voting scheme, and, recently, neural network (NN). Garcia et al. [
40] proposed an algorithm for computer - based identification of the lesions due to the diabetic retinopathy. The algorithm is integrated into a system in order to support the ophthalmologist decision in the detection and monitoring of the disease. The authors analyze the performance of three different NN classifiers in automated detection of lesions or exudates: support vector machine, radial basis function, and multilayer perceptron. By applying different criteria and testing on 67 images, they conclude that the multilayer NN classifier based on image criterion has the best results. This research was extended in [
41], where new criteria and classifiers (e.g., logistic regression) were tested against those in the previous paper. Now, the relationship between the extracted set of characteristics and the class of objects is highlighted. Again, the multilayer perceptron performed the best out of the selected methods, but only by a small margin in comparison with logistic regression. Moreover, in [
42] a new neural network supervised approach for blood vessel detection was proposed. The proposed algorithm uses an NN arhitecture for pixel classification and creates a vector of moment-invariants-based and gray-level features for pixel representation. The performance of the algorithm was tested on STARE [
43] and DRIVE [
44] datasets, with better performance than other existing solutions. The authors in [
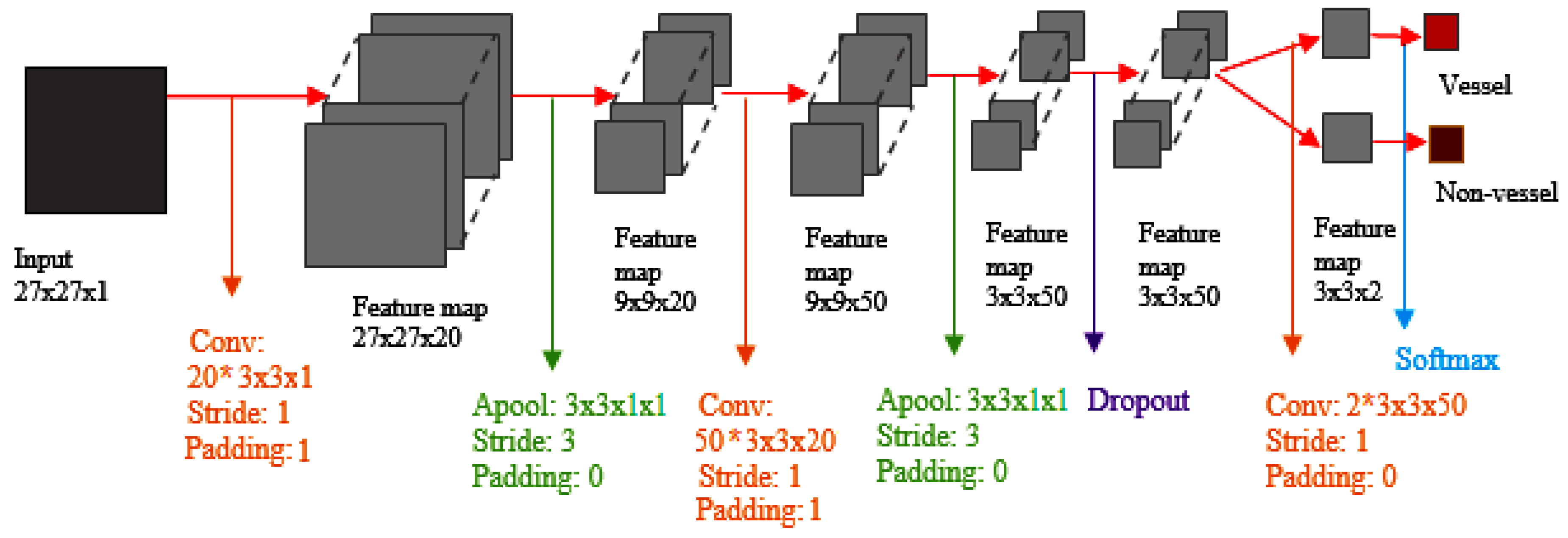
45] used CNNs (Convolutional NNs) to detect hemorrhages. They tried to simplify and speed it up by heuristically classifying samples of the training phase.
The authors in [
46] presented a methodology and their implementation which is based on a voting procedure with the aim of raising the decision accuracy. They proposed the technique of majority voting and probability approach to evaluate the correctness of decision. The important goal of the work consisted in solving detection problems in image processing algorithms which are geometrically constrained. For example, the algorithm performance was tested with good accuracy for
OD identification on the MESSIDOR dataset [
47]. In order to increase the performances of classifications, the voting scheme and NN approaches were combined in [
48]. In this case, two connected NNs were considered: one for feature selection and the other for classification, based on local detectors. Finally, a voting scheme with different weights for local classifiers was used, taking into account the results from associated confusion matrices.
In order to increase the efficiency of the classifiers, primary processing of retinal images is necessary. Therefore, Nayak et al. [
49,
50] presented algorithms for identification and monitoring glaucoma and diabetic retinopathy, using primary processing l (histogram equalization, filtering, and morphological operations). For a particular disease, specific characteristics are established. Based on these features, an artificial NN classifier identifies the presence of a particular disease and its evolution. The authors in [
51] introduce an algorithm based on morphological operations for the identification of micro-aneurysms and
HEs in retinal images. The image is firstly primary processed and then, important regions of interest such as
OD, blood vessels, and fovea are successively removed from the retinal image. Only the
HEs and micro-aneurysms remain. The obtained results are similar to those of other papers as sensitivity and specificity. The method for computer aided detection of the diabetic retinopathy, developed in [
52], consists of the following phases: image enhancement by local histogram equalization, Gabor filters, SVM (support vector machine) classifier,
HE detection, fovea localization, and diabetic retinopathy identification. The algorithms were implemented and tested on free database HRF [
53] with acceptable sensitivity and specificity. The authors in [
54] presented a method and algorithms for successive detection of important RoIs in retinal images using an artificial neural network (ANN) and
k-means clustering. The ANN was simulated on a common PC and it created time-consuming difficulties for real implementation. The application was developed only for the MESSIDOR database.
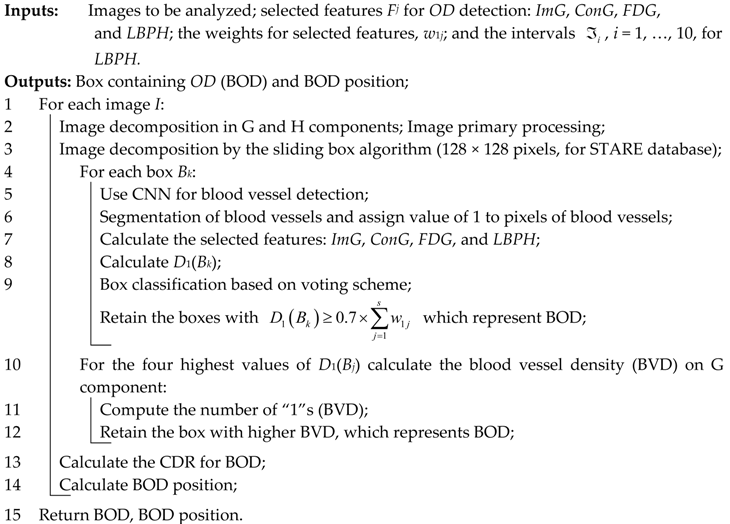
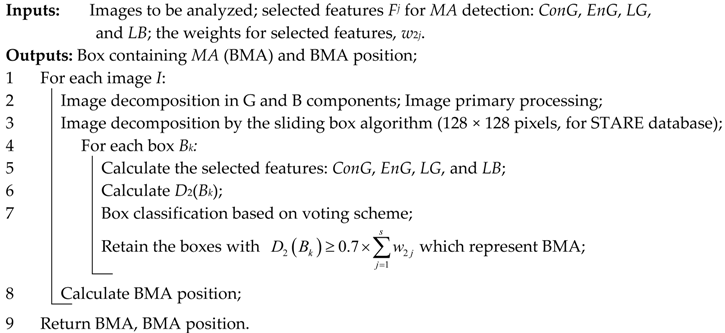
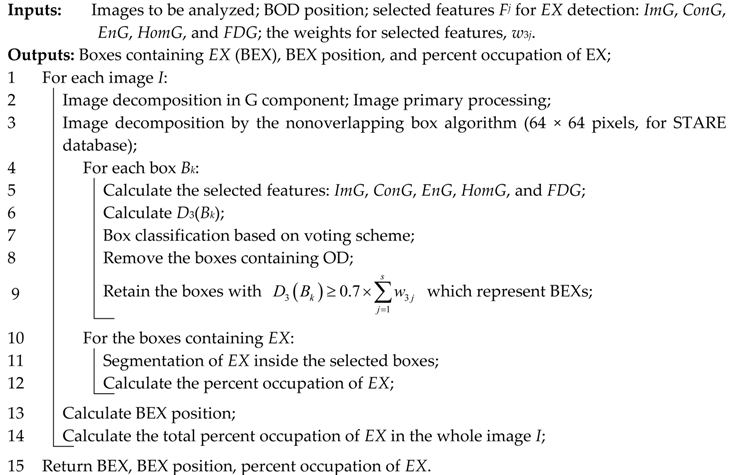
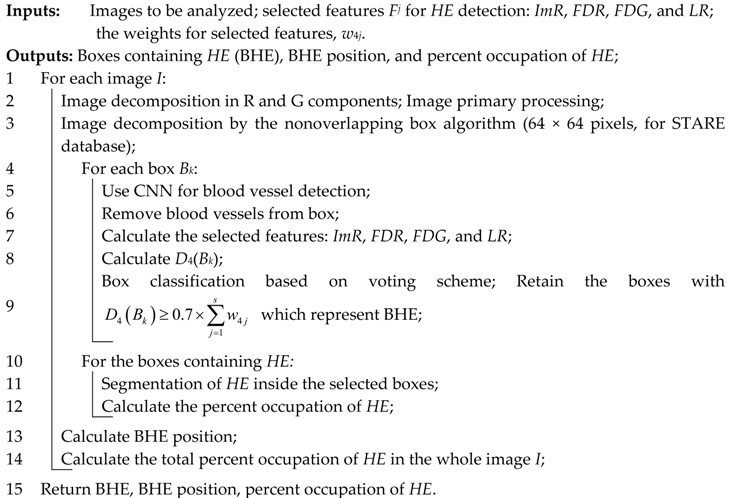
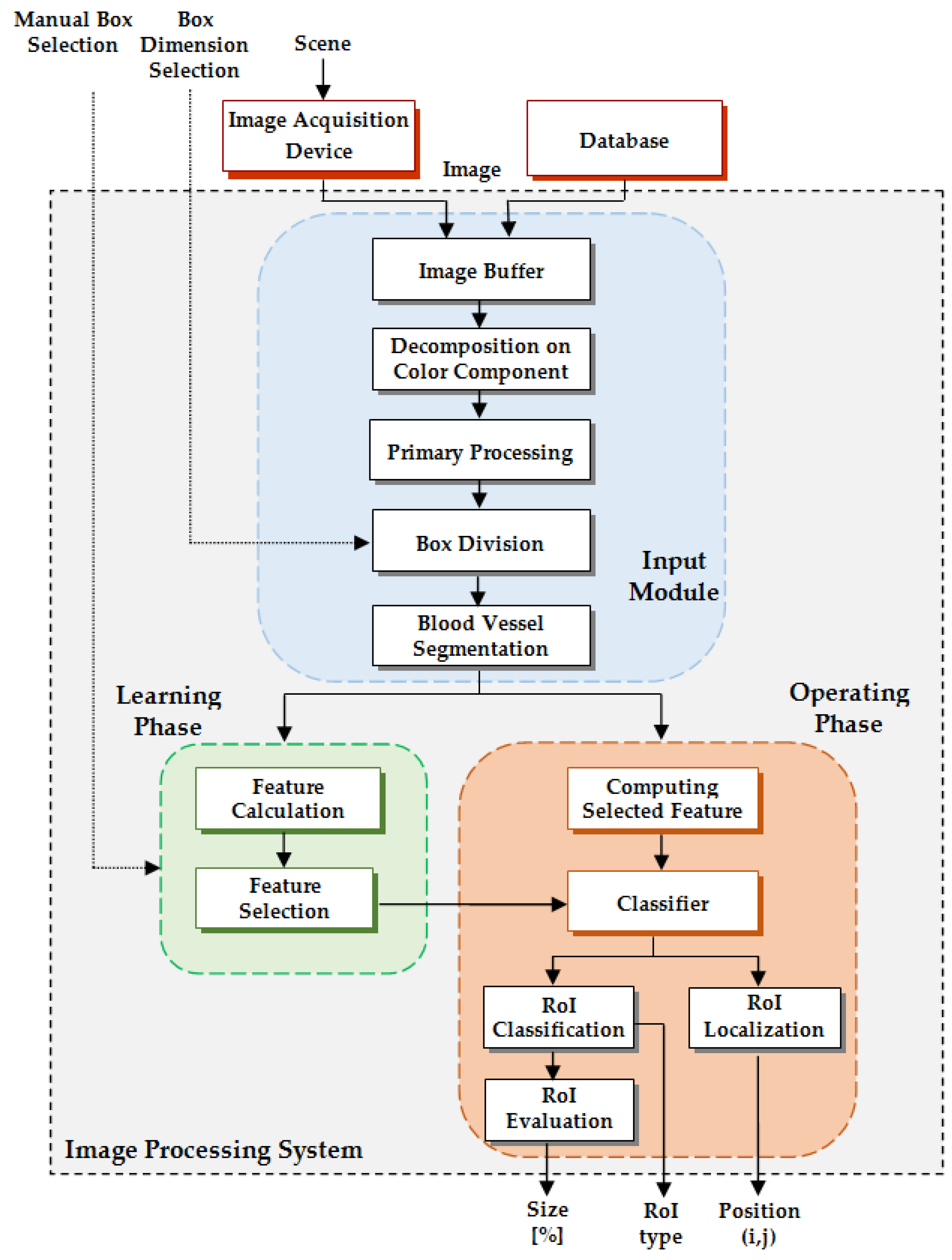
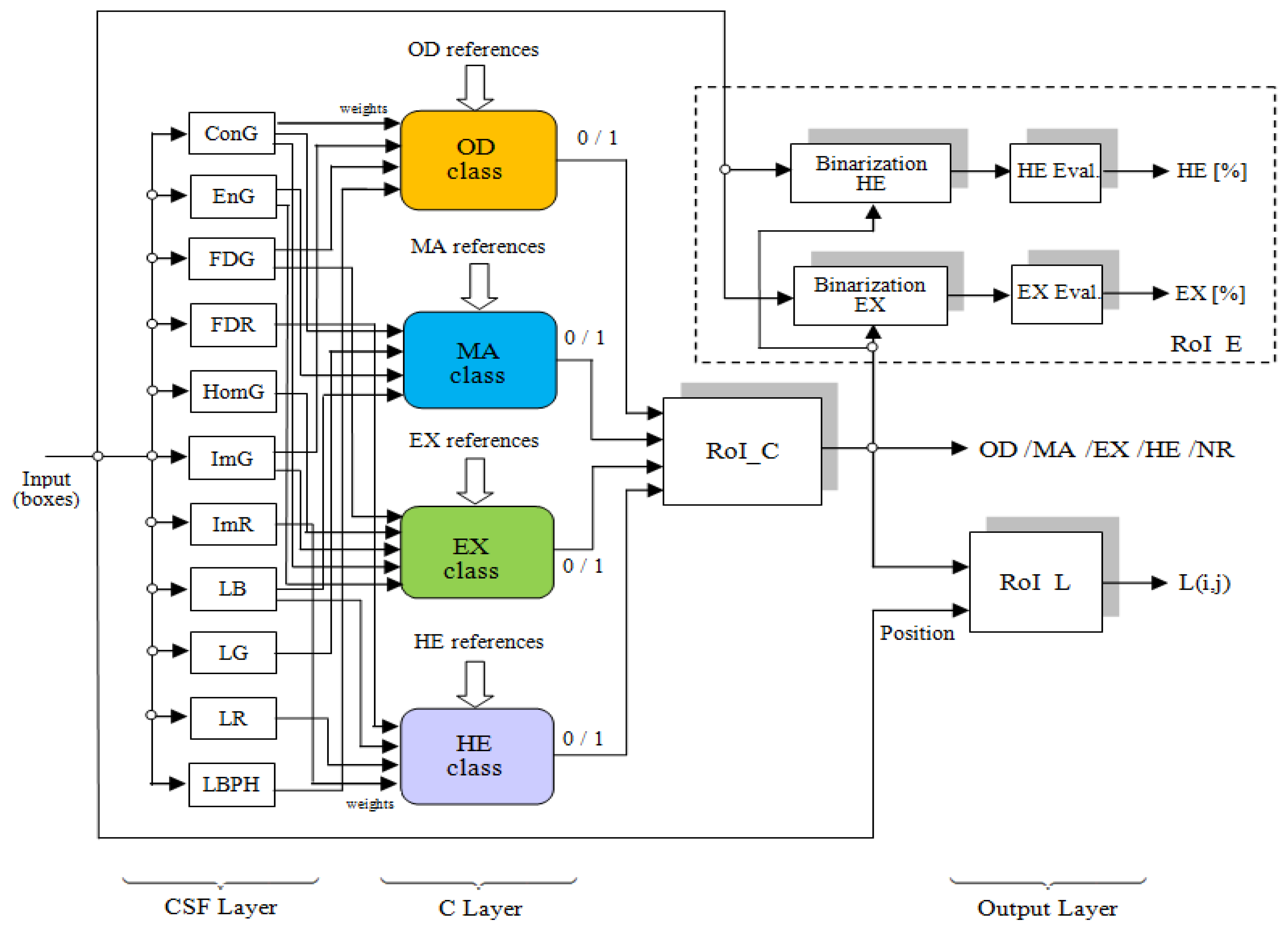
As a technical novelty, this paper proposes an Intelligent System for diagnosis and evaluation of regions of interest from Retinal Images (ISRI), with multiple functions: F1,
OD detection; F2,
MA detection; F3,
EX detection and size evaluation; and F4,
HE detection and size evaluation. All of these four functions are addressed. Two main contributions of the authors can be mentioned: (a) proper feature selection based on statistical analysis in confusion matrices for different feature type, and (b) the weight-based fusion of the local classifiers (associated with features) into global classifiers (corresponding to the RoIs). The paper is organized as follows: In
Section 2, the methodology and algorithms for primary processing of retinal images with the goal of RoI detection, segmentation, and size estimation are described and implemented. In
Section 3 the experimental results obtained using images from two databases (STARE and MESSIDOR) are reported and analyzed. Finally, the discussions and conclusions are presented in
Section 4 and
Section 5, respectively.
3. Experimental Results
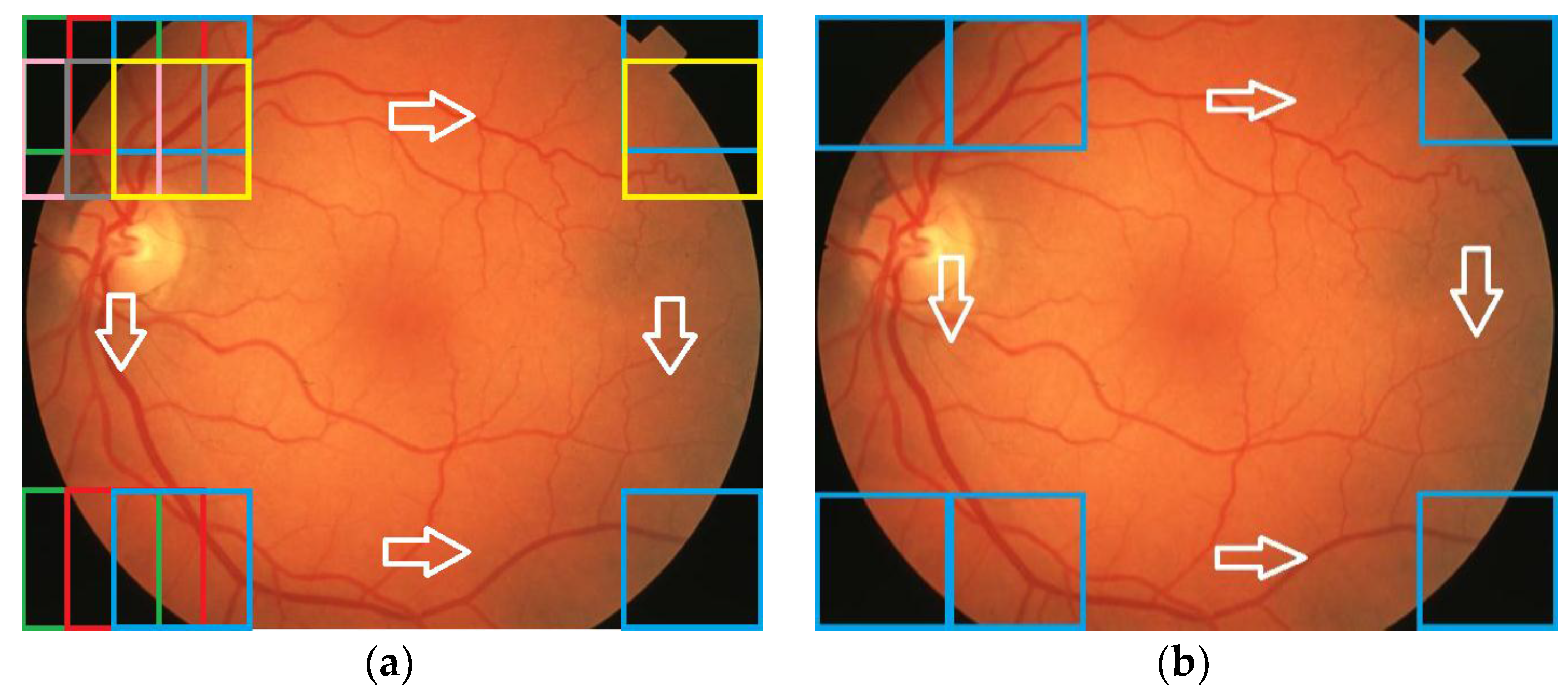
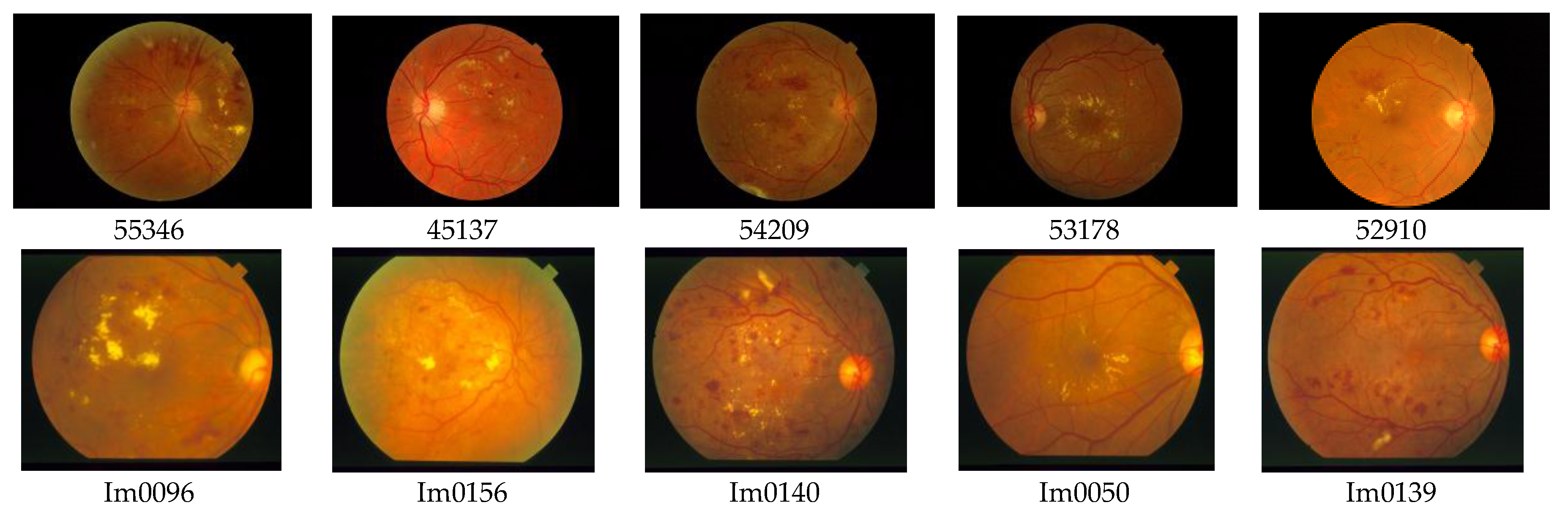
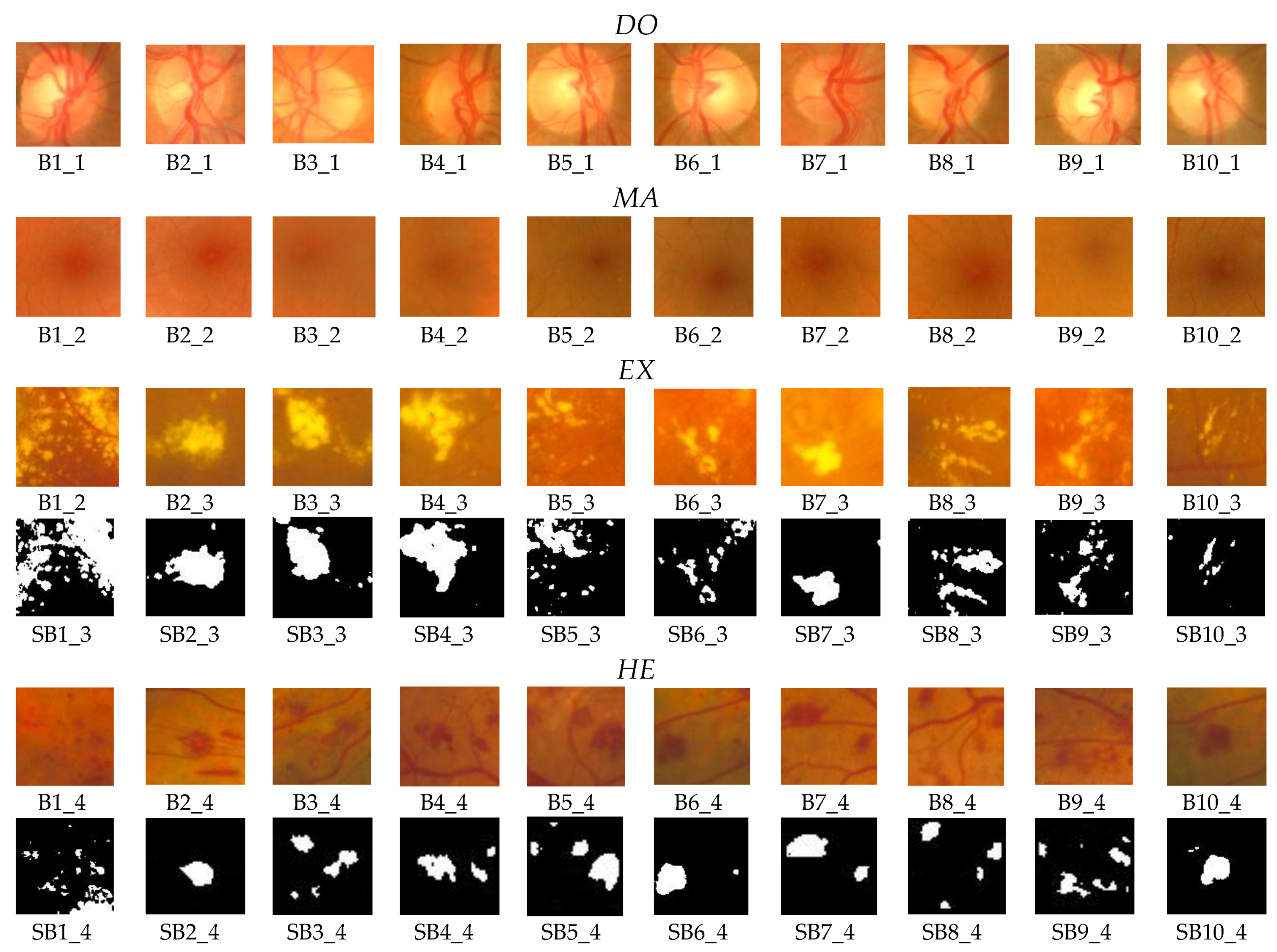
For the testing and validation of the proposed sensor we used 100 boxes from 60 images (
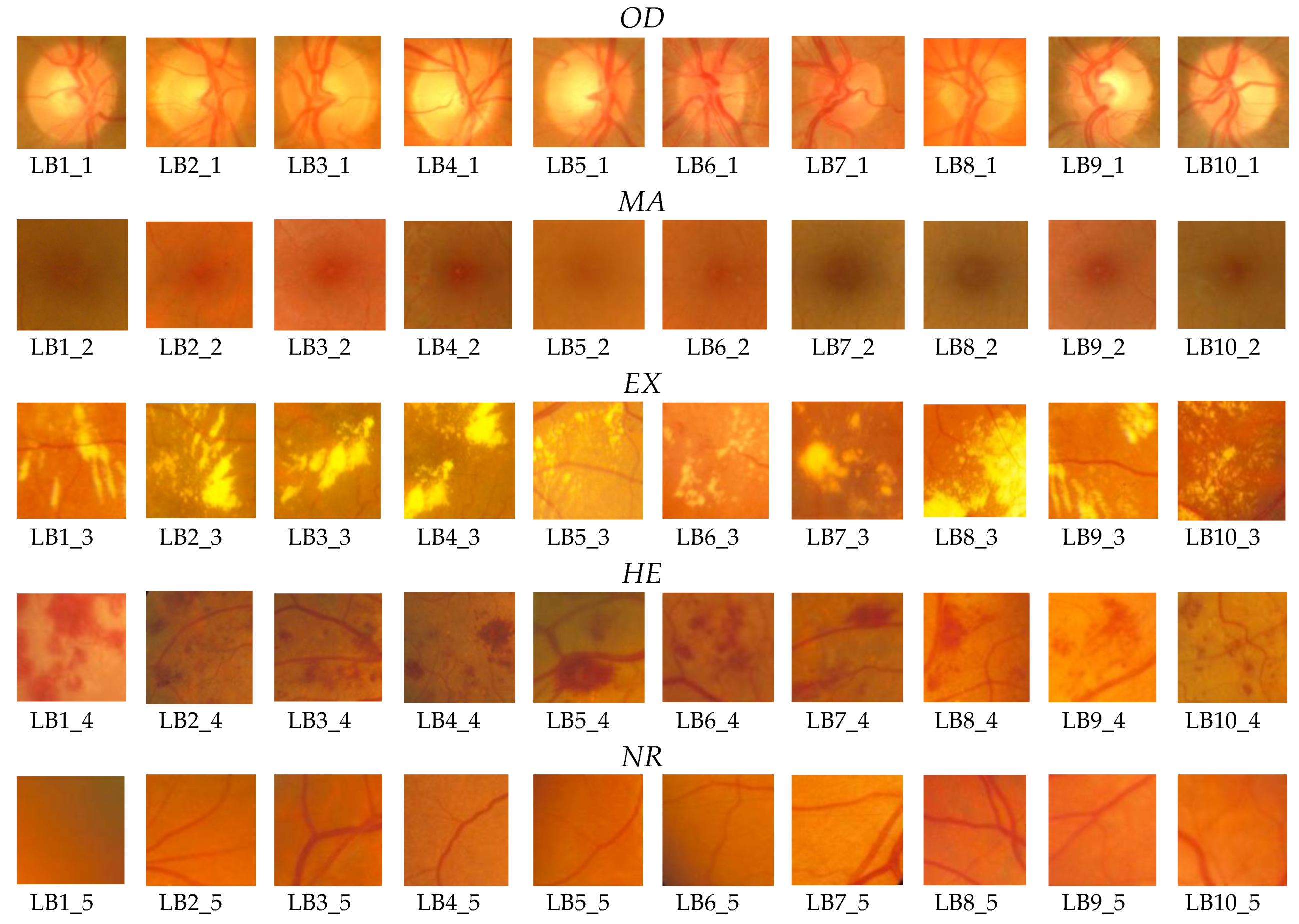
Figure 7) for the learning phase and 200 images for the operating phase. As mentioned in
Section 2, 50 boxes (10 for
OD, 10 for
MA, 10 for
EX, 10 for
HE, and 10 for
NR;
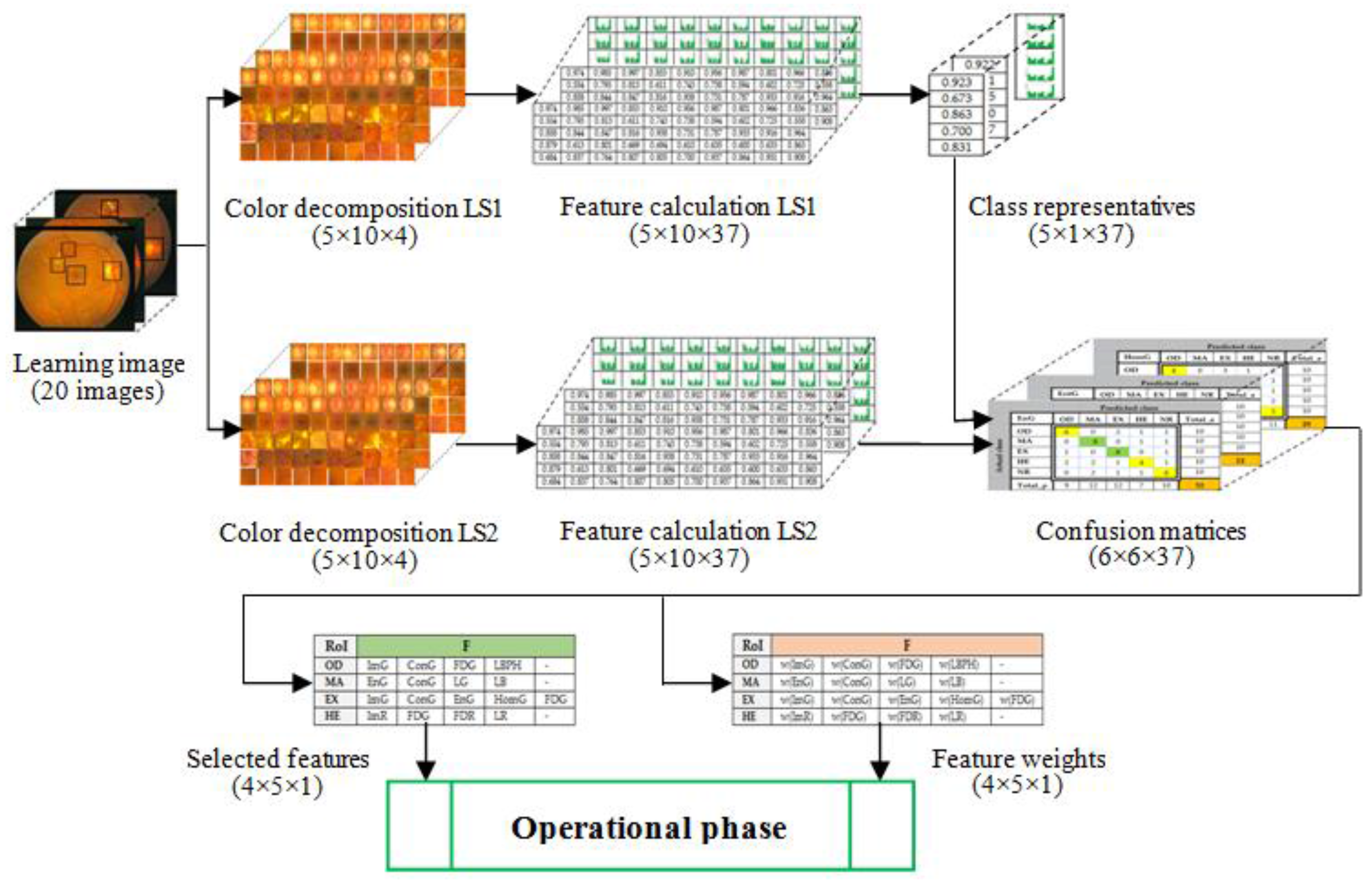
Figure 8) were used in feature selection and another 50 boxes were used in classifier implementation. A box dimension of 128 × 128 pixels was used for STARE and a box dimension of 256 × 256 pixels was used for MESSIDOR. The sliding step sizes (BDS) were 10 pixels for the STARE DB and 15 pixels for the MESSIDOR DB. In BVS, the sliding step size was 1 pixel for both. The same features were selected both in the STARE case and the MESSIDOR case. Examples of feature calculations for
OD class representations are presented in
Table 2 (36 features) and
Table 3 (
LBP feature). The last column contains the average values of the row. The results concerning the representatives for the classes
OD,
MA,
EX, and
HE are presented in
Table 4. The features from CF (
Table 1) were tested on R, G, B, and H color components and, in concordance with the selection criterion, the selected features and the corresponding color channels are marked with green in
Table 4.
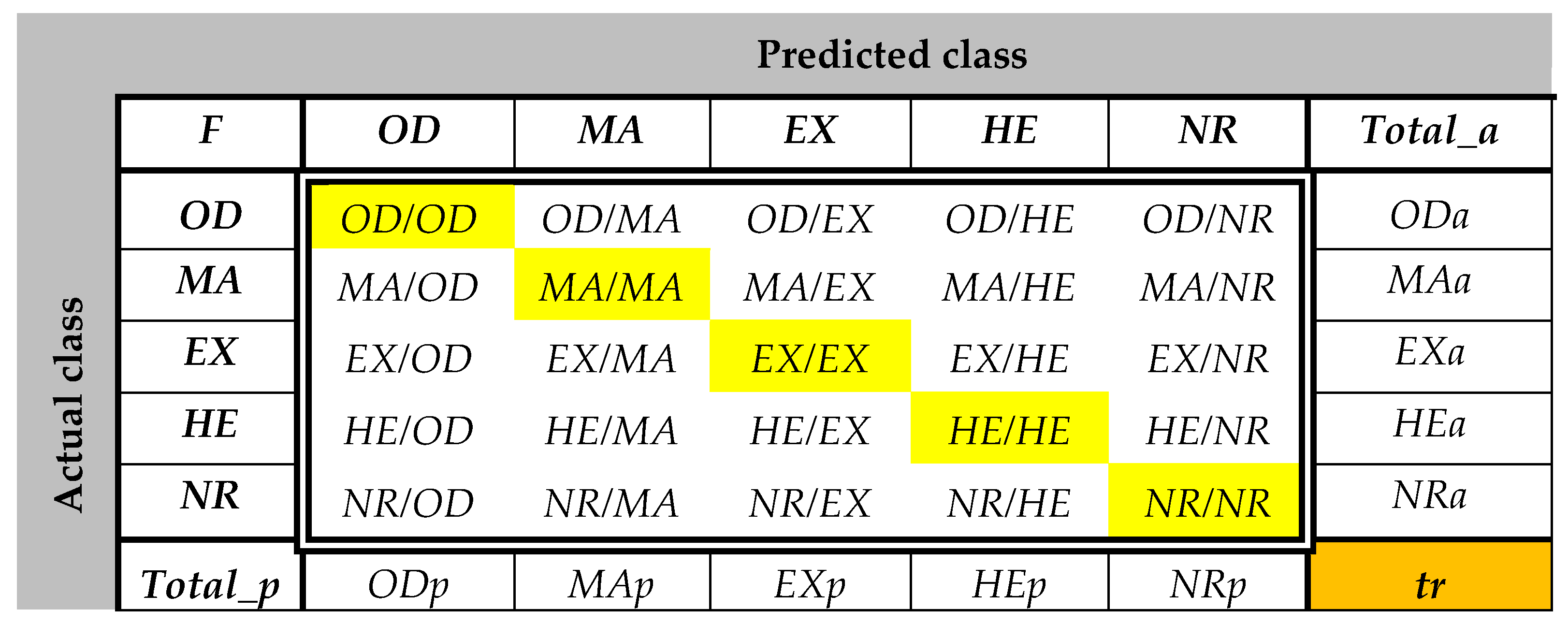
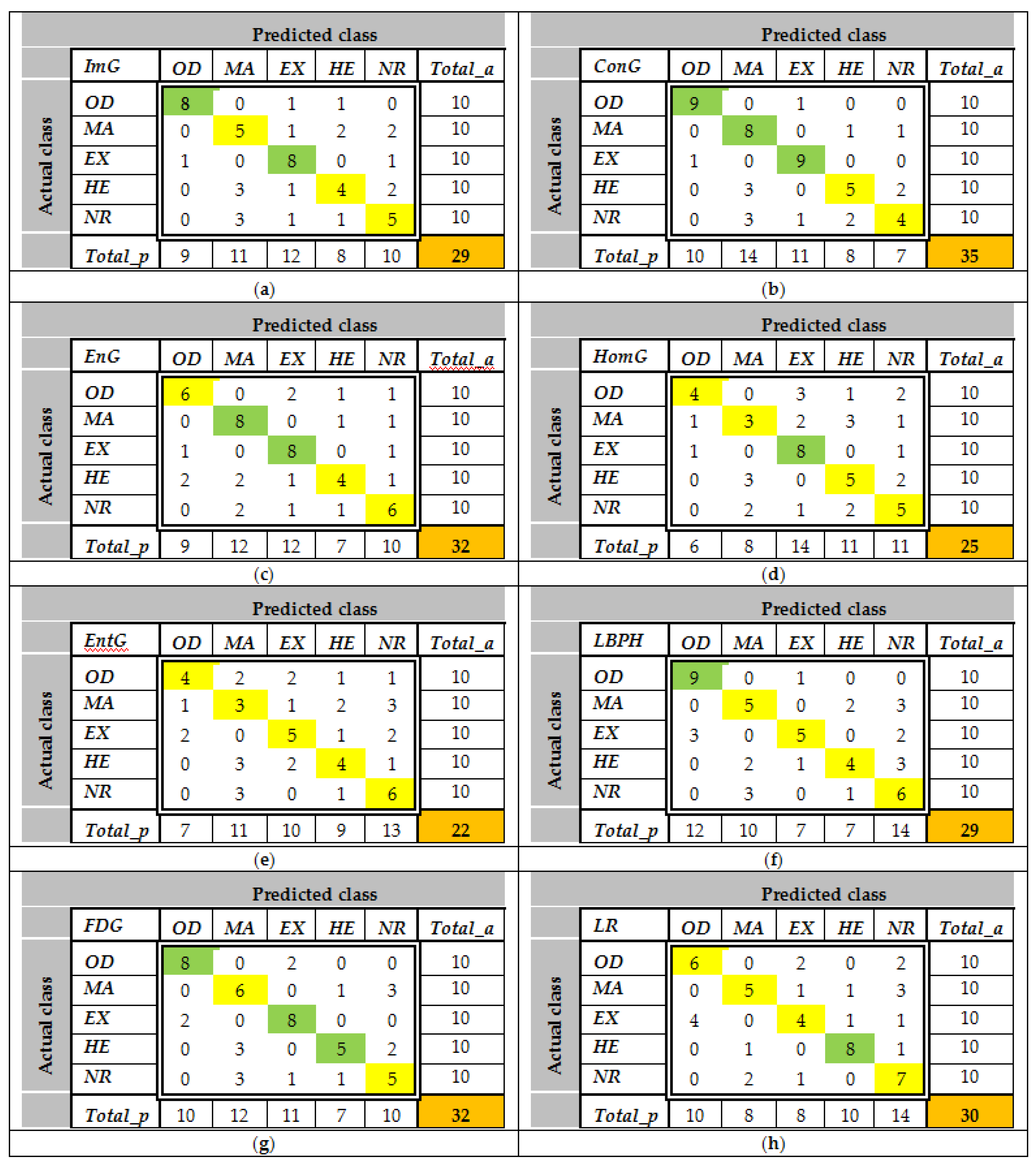
The confusion matrices are calculated using the set
LS2 for all features and all classes. Some examples of
CMs are presented in
Figure 9. As can be observed, the values greater than 8 (greater than 0.75
Cia—see feature selection from the
Section 2.3.2) are highlighted with green. For example, according to
Figure 9a,
ImG is a feature selected for
OD and
EX detection. Similarly,
Figure 9b determines the classifiers which use
ConG,
Figure 9c determines the classifiers which use
EnG,
Figure 9d determines the classifiers which use
HomG,
Figure 9f determines the classifiers which use
LBPH,
Figure 9g determines the classifiers which use
FDG, and
Figure 9h determines the classifiers which use
LR. It can be observed in
Figure 9e that
EntG is not a proper feature for any class.
Table 5 presents the traces of all confusion matrices associated with features (the rows represent such
CM traces). The fulfillment of the selection criterion for the feature-class pair (from the confusion matrices) and also the associated weights for classifiers are highlighted in gray. The last column indicates the class which uses the corresponding feature in their classifier. The features and associated weights for each RoI can be extracted from
Table 5. The classifiers from Equation (12)—
D1,
D2,
D3, and
D4—were implemented taking into account the confidence intervals and band associated with the weights and the corresponding RoIs (
OD,
MA,
EX, and
HE) (
Table 6). For the operational phase testing, both MESSIDOR and STARE databases were used (
Figure 10). Some results concerning the box classification are given in
Figure 11. The boxes classified as
EX or
HE have undergone a segmentation process (
Figure 11, boxes labeled with SB). The results of size evaluation of
EXs and
HEs are presented in
Table 7 and
Table 8, respectively. Then, by summing the partial areas of
EXs (
HEs) from boxes belonging to the same image, a global evaluation in pixels and percent can be obtained (
Table 9).