SARS-CoV-2 Virus-like Particles (VLPs) Specifically Detect Humoral Immune Reactions in an ELISA-Based Platform
Abstract
1. Introduction
2. Materials and Methods
3. Results
3.1. Characterization of SARS-CoV-2 VLPs
3.2. Application of SARS-CoV-2 VLPs as Antigens for Serum Diagnostics
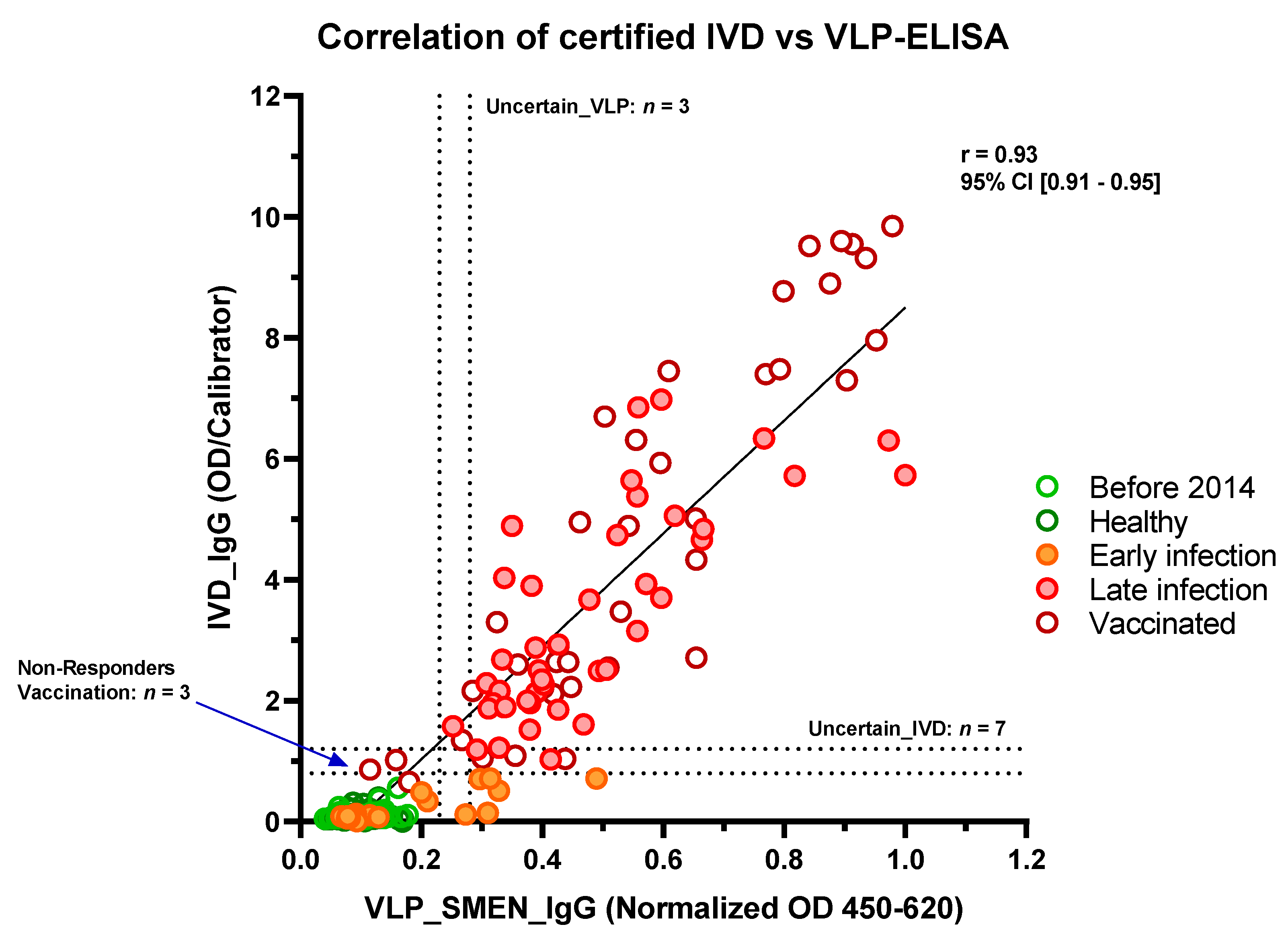
3.3. Comparison of the Performance of SARS-CoV-2 VLP-ELISA with a Certified IVD
4. Discussion
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Wu, F.; Zhao, S.; Yu, B.; Chen, Y.M.; Wang, W.; Song, Z.G.; Hu, Y.; Tao, Z.W.; Tian, J.H.; Pei, Y.Y.; et al. A New Coronavirus Associated with Human Respiratory Disease in China. Nature 2020, 579, 265–269. [Google Scholar] [CrossRef] [PubMed]
- Zhu, N.; Zhang, D.; Wang, W.; Li, X.; Yang, B.; Song, J.; Zhao, X.; Huang, B.; Shi, W.; Lu, R.; et al. A Novel Coronavirus from Patients with Pneumonia in China, 2019. N. Engl. J. Med. 2020, 382, 727–733. [Google Scholar] [CrossRef] [PubMed]
- Guan, W.; Ni, Z.; Hu, Y.; Liang, W.; Ou, C.; He, J.; Liu, L.; Shan, H.; Lei, C.; Hui, D.S.C.; et al. Clinical Characteristics of Coronavirus Disease 2019 in China. N. Engl. J. Med. 2020, 382, 1708–1720. [Google Scholar] [CrossRef] [PubMed]
- Long, Q.X.; Tang, X.J.; Shi, Q.L.; Li, Q.; Deng, H.J.; Yuan, J.; Hu, J.L.; Xu, W.; Zhang, Y.; Lv, F.J.; et al. Clinical and Immunological Assessment of Asymptomatic SARS-CoV-2 Infections. Nat. Med. 2020, 26, 1200–1204. [Google Scholar] [CrossRef] [PubMed]
- Yao, H.; Song, Y.; Chen, Y.; Wu, N.; Xu, J.; Sun, C.; Zhang, J.; Weng, T.; Zhang, Z.; Wu, Z.; et al. Molecular Architecture of the SARS-CoV-2 Virus. Cell 2020, 183, 730–738.e13. [Google Scholar] [CrossRef]
- Kim, D.; Lee, J.Y.; Yang, J.S.; Kim, J.W.; Kim, V.N.; Chang, H. The Architecture of SARS-CoV-2 Transcriptome. Cell 2020, 181, 914–921.e10. [Google Scholar] [CrossRef]
- Xu, R.; Shi, M.; Li, J.; Song, P.; Li, N. Construction of SARS-CoV-2 Virus-Like Particles by Mammalian Expression System. Front. Bioeng. Biotechnol. 2020, 8, 862. [Google Scholar] [CrossRef]
- Swann, H.; Sharma, A.; Preece, B.; Peterson, A.; Eldridge, C.; Belnap, D.M.; Vershinin, M.; Saffarian, S. Minimal System for Assembly of SARS-CoV-2 Virus like Particles. Sci. Rep. 2020, 10, 21877. [Google Scholar] [CrossRef]
- Lagousi, T.; Routsias, J.; Spoulou, V. Development of an Enzyme-Linked Immunosorbent Assay (ELISA) for Accurate and Prompt Coronavirus Disease 2019 (COVID-19) Diagnosis Using the Rational Selection of Serological Biomarkers. Diagnostics 2021, 11, 1970. [Google Scholar] [CrossRef]
- Kirnbauer, R.; Hubbert, N.L.; Wheeler, C.M.; Becker, T.M.; Lowy, D.R.; Schiller, J.T. A Virus-like Particle Enzyme-Linked Immunosorbent Assay Detects Serum Antibodies in a Majority of Women Infected with Human Papillomavirus Type 16. J. Natl. Cancer Inst. 1994, 86, 494–499. [Google Scholar] [CrossRef]
- Ko, Y.J.; Choi, K.S.; Nah, J.J.; Paton, D.J.; Oem, J.K.; Wilsden, G.; Kang, S.Y.; Jo, N.I.; Lee, J.H.; Kim, J.H.; et al. Noninfectious Virus-Like Particle Antigen for Detection of Swine Vesicular Disease Virus Antibodies in Pigs by Enzyme-Linked Immunosorbent Assay. Clin. Diagn. Lab. Immunol. 2005, 12, 922. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Hernandez, B.Y.; Ton, T.; Shvetsov, Y.B.; Goodman, M.T.; Zhu, X. Human Papillomavirus (HPV) L1 and L1-L2 Virus-like Particle-Based Multiplex Assays for Measurement of HPV Virion Antibodies. Clin. Vaccine Immunol. 2012, 19, 1348–1352. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Wang, Z.; Zhan, Y.; Gong, Q.; Yu, W.; Deng, Z.; Wang, A.; Yang, Y.; Wang, N. Generation of E. Coli-Derived Virus-like Particles of Porcine Circovirus Type 2 and Their Use in an Indirect IgG Enzyme-Linked Immunosorbent Assay. Arch. Virol. 2016, 161, 1485–1491. [Google Scholar] [CrossRef] [PubMed]
- Bai, M.; Wang, R.; Sun, S.; Zhang, Y.; Dong, H.; Guo, H. Development and Validation of a Competitive ELISA Based on Virus-like Particles of Serotype Senecavirus A to Detect Serum Antibodies. AMB Express 2021, 11, 7. [Google Scholar] [CrossRef] [PubMed]
- Arora, K.; Rastogi, R.; Arora, N.M.; Parashar, D.; Paliwal, J.; Naqvi, A.; Srivastava, A.; Singh, S.K.; Kalyanaraman, S.; Potdar, S.; et al. Multi-Antigenic Virus-like Particle of SARS CoV-2 Produced in Saccharomyces Cerevisiae as a Vaccine Candidate. bioRxiv 2020. [Google Scholar] [CrossRef]
- Ward, B.J.; Gobeil, P.; Séguin, A.; Atkins, J.; Boulay, I.; Charbonneau, P.-Y.; Couture, M.; D’Aoust, M.-A.; Dhaliwall, J.; Finkle, C.; et al. Phase 1 Randomized Trial of a Plant-Derived Virus-like Particle Vaccine for COVID-19. Nat. Med. 2021, 27, 1071–1078. [Google Scholar] [CrossRef]
- Mohsen, M.O.; Balke, I.; Zinkhan, S.; Zeltina, V.; Liu, X.; Chang, X.; Krenger, P.S.; Plattner, K.; Gharailoo, Z.; Vogt, A.C.S.; et al. A Scalable and Highly Immunogenic Virus-like Particle-Based Vaccine against SARS-CoV-2. Allergy 2022, 77, 243–257. [Google Scholar] [CrossRef]
- Yilmaz, I.C.; Ipekoglu, E.M.; Bulbul, A.; Turay, N.; Yildirim, M.; Evcili, I.; Yilmaz, N.S.; Guvencli, N.; Aydin, Y.; Gungor, B.; et al. Development and Preclinical Evaluation of Virus-like Particle Vaccine against COVID-19 Infection. Allergy 2022, 77, 258–270. [Google Scholar] [CrossRef]
- Plescia, C.B.; David, E.A.; Patra, D.; Sengupta, R.; Amiar, S.; Su, Y.; Stahelin, R.V. SARS-CoV-2 Viral Budding and Entry Can Be Modeled Using BSL-2 Level Virus-like Particles. J. Biol. Chem. 2021, 296, 100103. [Google Scholar] [CrossRef]
- Kumar, B.; Hawkins, G.M.; Kicmal, T.; Qing, E.; Timm, E.; Gallagher, T. Assembly and Entry of Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV2): Evaluation Using Virus-Like Particles. Cells 2021, 10, 853. [Google Scholar] [CrossRef]
- Syed, A.M.; Taha, T.Y.; Tabata, T.; Chen, I.P.; Ciling, A.; Khalid, M.M.; Sreekumar, B.; Chen, P.Y.; Hayashi, J.M.; Soczek, K.M.; et al. Rapid Assessment of SARS-CoV-2–Evolved Variants Using Virus-like Particles. Science 2021, 374, 1626–1632. [Google Scholar] [CrossRef]
- Abdullah, S.W.; Jaron, M.; Lehky, M.; Zarà, M.; Zaydowicz, C.N.; Lak, A.; Ballmann, R.; Heine, P.A.; Wenzel, E.V.; Schneider, K.-T.; et al. Baculovirus-Free SARS-CoV-2 Virus-like Particle Production in Insect Cells for Rapid Neutralization Assessment. Viruses 2022, 14, 2087. [Google Scholar] [CrossRef]
- Gossen, M.; Freundlieb, S.; Bender, G.; Müller, G.; Hillen, W.; Bujard, H. Transcriptional Activation by Tetracyclines in Mammalian Cells. Science 1995, 268, 1766–1769. [Google Scholar] [CrossRef] [PubMed]
- Freundlieb, S.; Schirra-Mu Èller Hermann Bujard, C. A Tetracycline Controlled Activation/Repression System with Increased Potential for Gene Transfer into Mammalian Cells. J. Gene Med. 1999, 1, 4–12. [Google Scholar] [CrossRef]
- Urlinger, S.; Baron, U.; Thellmann, M.; Hasan, M.T.; Bujard, H.; Hillen, W. Exploring the Sequence Space for Tetracycline-Dependent Transcriptional Activators: Novel Mutations Yield Expanded Range and Sensitivity. Proc. Natl. Acad. Sci. USA 2000, 97, 7963–7968. [Google Scholar] [CrossRef] [PubMed]
- Reynolds, E.S. The use of lead citrate at high pH as an electron-opaque stain in electron microscopy. J. Cell Biol. 1963, 17, 208–212. [Google Scholar] [CrossRef] [PubMed]
- Schlör, A.; Hirschberg, S.; Amor, G.B.; Meister, T.L.; Arora, P.; Pöhlmann, S.; Hoffmann, M.; Pfaender, S.; Eddin, O.K.; Kamhieh-Milz, J.; et al. SARS-CoV-2 Neutralizing Camelid Heavy-Chain-Only Antibodies as Powerful Tools for Diagnostic and Therapeutic Applications. Front. Immunol. 2022, 13, 930975. [Google Scholar] [CrossRef] [PubMed]
- Schindelin, J.; Arganda-Carreras, I.; Frise, E.; Kaynig, V.; Longair, M.; Pietzsch, T.; Preibisch, S.; Rueden, C.; Saalfeld, S.; Schmid, B.; et al. Fiji: An Open-Source Platform for Biological-Image Analysis. Nat. Methods 2012, 97, 676–682. [Google Scholar] [CrossRef]
- Hager, K.J.; Pérez Marc, G.; Gobeil, P.; Diaz, R.S.; Heizer, G.; Llapur, C.; Makarkov, A.I.; Vasconcellos, E.; Pillet, S.; Riera, F.; et al. Efficacy and Safety of a Recombinant Plant-Based Adjuvanted Covid-19 Vaccine. N. Engl. J. Med. 2022, 386, 2084–2096. [Google Scholar] [CrossRef]
- Moon, K.B.; Jeon, J.H.; Choi, H.; Park, J.S.; Park, S.J.; Lee, H.J.; Park, J.M.; Cho, H.S.; Moon, J.S.; Oh, H.; et al. Construction of SARS-CoV-2 Virus-like Particles in Plant. Sci. Rep. 2022, 121, 1005. [Google Scholar] [CrossRef] [PubMed]
- Naskalska, A.; Dabrowska, A.; Szczepanski, A.; Jasik, K.P.; Gromadzka, B.; Pyrc, K. Functional Severe Acute Respiratory Syndrome Coronavirus 2 Virus-Like Particles from Insect Cells. Front. Microbiol. 2021, 12, 2773. [Google Scholar] [CrossRef] [PubMed]
- Peng, S.; Qi, Y.; Christensen, N.; Hengst, K.; Kennedy, L.; Frazer, I.H.; Tindle, R.W. Capture ElISA and in Vitro Cell Binding Assay for the Detection of Antibodies to Human Papillomavirus Type 6b Virus-like Particles in Patients with Anogenital Warts. Pathology 1999, 31, 418–422. [Google Scholar] [CrossRef] [PubMed]
- Du, P.; Brendle, S.; Milici, J.; Camacho, F.; Zurlo, J.; Christensen, N.; Meyers, C. Comparisons of VLP-Based ELISA, Neutralization Assays with Native HPV, and Neutralization Assays with PsV in Detecting HPV Antibody Responses in HIV-Infected Women. J. AIDS Clin. Res. 2015, 6, 433. [Google Scholar] [CrossRef] [PubMed]


| Before 2014 | Healthy | Early Infection | Late Infection | Vaccinated | |
|---|---|---|---|---|---|
| No. samples | 42 | 89 | 17 | 44 | 38 |
| 1 E.I. pos | 0 | 0 | 0 | 42 | 32 |
| 1 E.I. neg | 42 | 89 | 17 | 0 | 1 |
| 1 E.I. UC | 0 | 0 | 0 | 2 | 5 |
| % correct | 100.0 | 100.0 | 0.0 | 95.5 | 84.2 |
| VLP pos | 0 | 0 | 6 | 44 | 35 |
| VLP neg | 42 | 89 | 9 | 0 | 3 |
| VLP UC | 0 | 0 | 2 | 1 | 0 |
| % correct | 100.0 | 100.0 | 35.3 | 97.7 | 92.1 |
| S pos | 2 | 2 | 6 | 44 | 38 |
| S neg | 39 | 87 | 10 | 0 | 0 |
| S UC | 1 | 0 | 1 | 0 | 0 |
| % correct | 92.9 | 97.8 | 35.3 | 100.0 | 100.0 |
| N pos | 0 | 3 | 5 | 40 | 9 |
| N neg | 42 | 83 | 12 | 3 | 28 |
| N UC | 0 | 3 | 0 | 1 | 1 |
| % correct | 100.0 | 93.3 | 29.4 | 90.9 | 23.7 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hirschberg, S.; Bauer, H.; Kamhieh-Milz, J.; Ringel, F.; Harms, C.; Eddin, O.K.; Pruß, A.; Hanack, K.; Schulze-Forster, K. SARS-CoV-2 Virus-like Particles (VLPs) Specifically Detect Humoral Immune Reactions in an ELISA-Based Platform. Antibodies 2022, 11, 76. https://doi.org/10.3390/antib11040076
Hirschberg S, Bauer H, Kamhieh-Milz J, Ringel F, Harms C, Eddin OK, Pruß A, Hanack K, Schulze-Forster K. SARS-CoV-2 Virus-like Particles (VLPs) Specifically Detect Humoral Immune Reactions in an ELISA-Based Platform. Antibodies. 2022; 11(4):76. https://doi.org/10.3390/antib11040076
Chicago/Turabian StyleHirschberg, Stefan, Hannes Bauer, Julian Kamhieh-Milz, Frauke Ringel, Christoph Harms, Omar Kamal Eddin, Axel Pruß, Katja Hanack, and Kai Schulze-Forster. 2022. "SARS-CoV-2 Virus-like Particles (VLPs) Specifically Detect Humoral Immune Reactions in an ELISA-Based Platform" Antibodies 11, no. 4: 76. https://doi.org/10.3390/antib11040076
APA StyleHirschberg, S., Bauer, H., Kamhieh-Milz, J., Ringel, F., Harms, C., Eddin, O. K., Pruß, A., Hanack, K., & Schulze-Forster, K. (2022). SARS-CoV-2 Virus-like Particles (VLPs) Specifically Detect Humoral Immune Reactions in an ELISA-Based Platform. Antibodies, 11(4), 76. https://doi.org/10.3390/antib11040076






