Marker- and Microbiome-Based Microbial Source Tracking and Evaluation of Bather Health Risk from Fecal Contamination in Galveston, Texas
Abstract
1. Introduction
2. Materials and Methods
3. Results
3.1. Marker-Based MST
3.2. Microbiome-Based MST
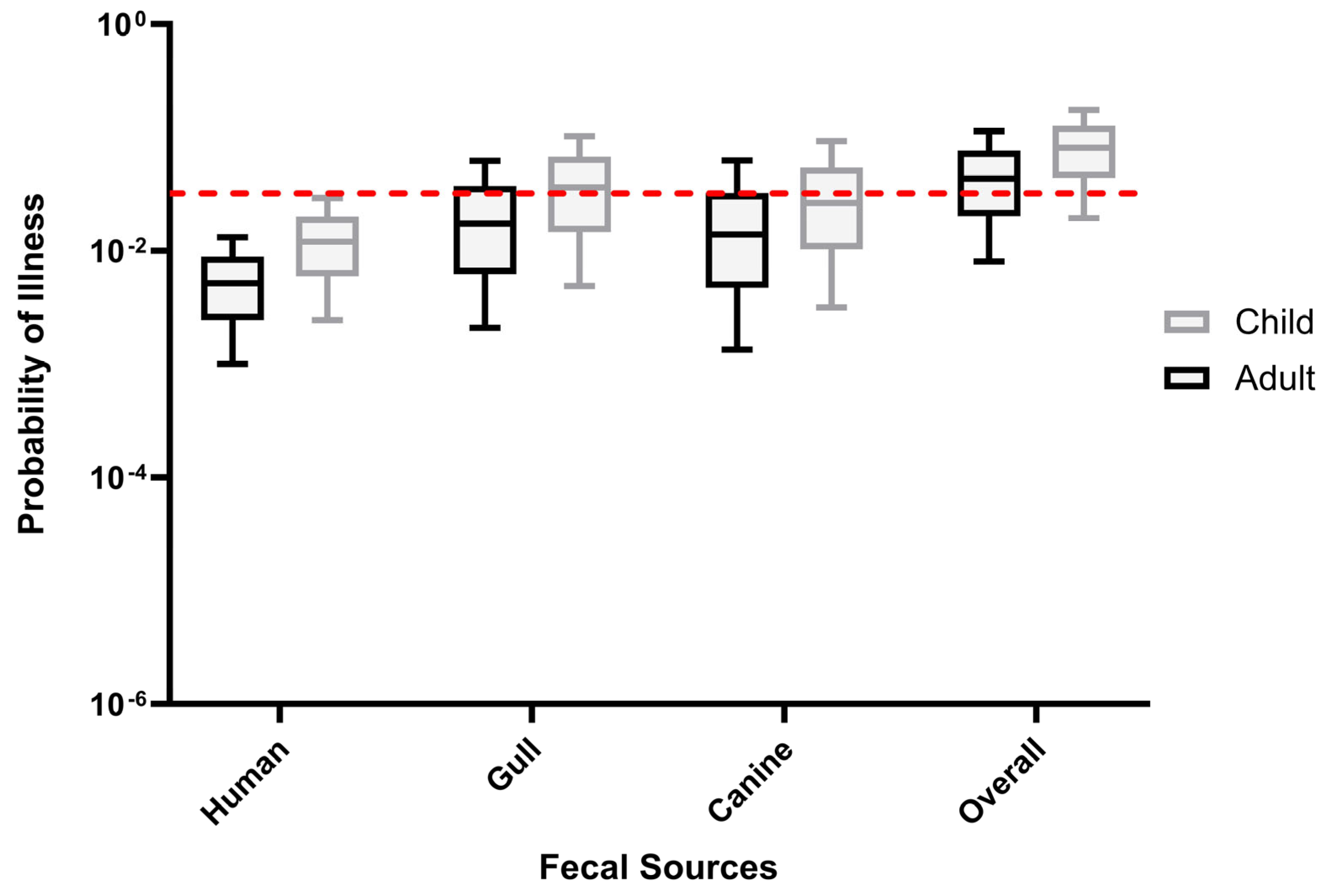
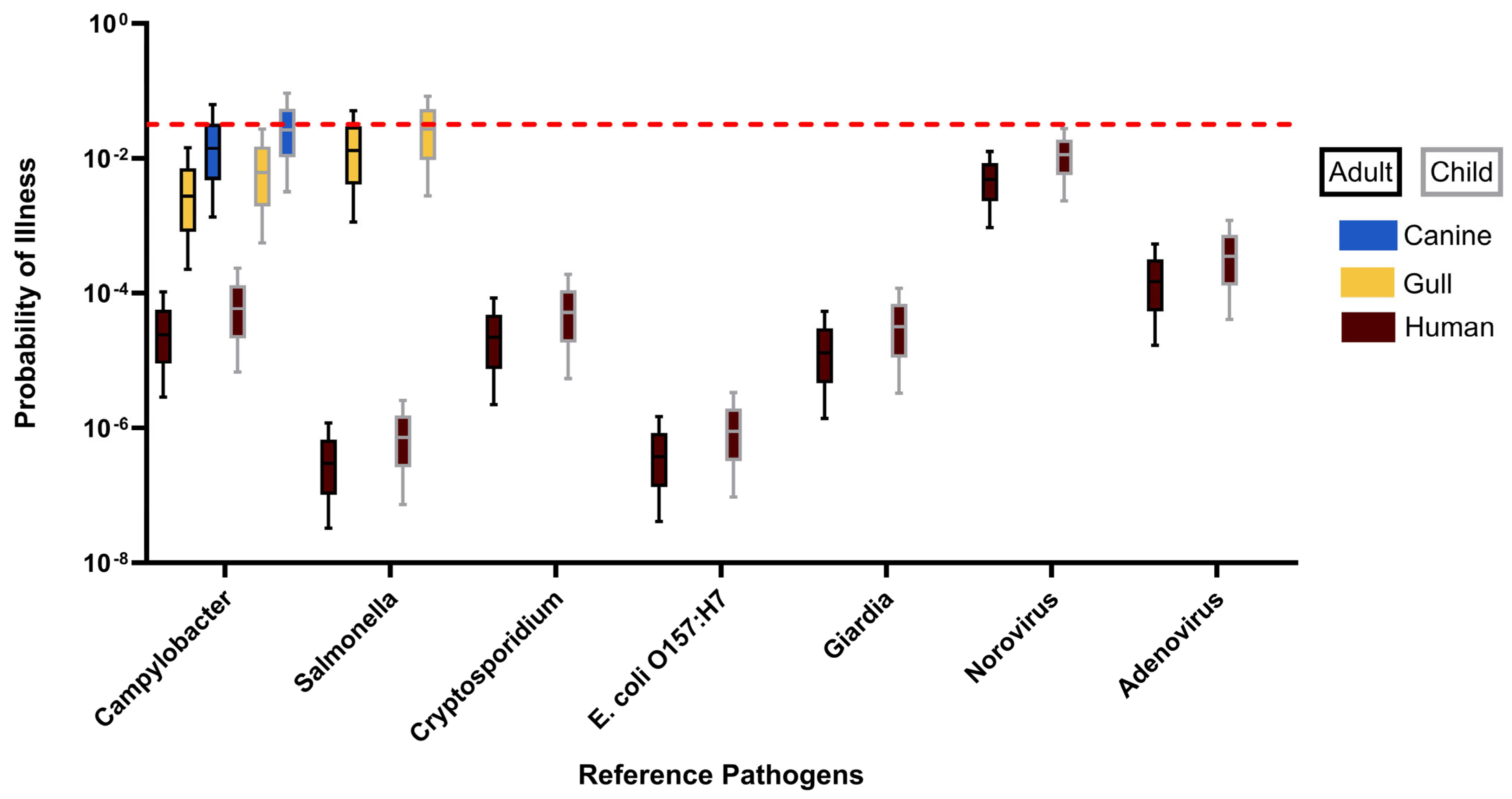
3.3. Human Health Risk Estimates
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| MST | Microbial source tracking |
| QMRA | Quantitative microbial risk assessment |
| BAV | Beach Action Value |
| TBW | Texas Beach Watch |
| TGLO | Texas General Land Office |
| STV | Statistical threshold value |
| OSSF | On Site Sewer Facilities |
| WWTP | Wastewater treatment plant |
| LOQ | Limit of quantification |
| LOD | Limit of detection |
| qPCR | Quantitative polymerase chain reaction |
| TIGSS | Texas A&M Institute for Genome Science and Society |
| AIC | Akaike Information Criterion |
| Quantile-quantile | |
| CDF | Cumulative distribution plots |
| PP | Probability-probability |
References
- DeFlorio-Barker, S.; Wing, C.; Jones, R.M.; Dorevitch, S. Estimate of Incidence and Cost of Recreational Waterborne Illness on United States Surface Waters. Environ. Health 2018, 17, 3. [Google Scholar] [CrossRef]
- Jennings, W.C.; Chern, E.C.; O’Donohue, D.; Kellogg, M.G.; Boehm, A.B. Frequent Detection of a Human Fecal Indicator in the Urban Ocean: Environmental Drivers and Covariation with Enterococci. Environ. Sci. Process. Impacts 2018, 20, 480–492. [Google Scholar] [CrossRef] [PubMed]
- Tomenchok, L.E.; Abdool-Ghany, A.A.; Elmir, S.M.; Gidley, M.L.; Sinigalliano, C.D.; Solo-Gabriele, H.M. Trends in Regional Enterococci Levels at Marine Beaches and Correlations with Environmental, Global Oceanic Changes, Community Populations, and Wastewater Infrastructure. Sci. Total Environ. 2021, 793, 148641. [Google Scholar] [CrossRef] [PubMed]
- Nevers, M.B.; Byappanahalli, M.N.; Nakatsu, C.H.; Kinzelman, J.L.; Phanikumar, M.S.; Shively, D.A.; Spoljaric, A.M. Interaction of Bacterial Communities and Indicators of Water Quality in Shoreline Sand, Sediment, and Water of Lake Michigan. Water Res. 2020, 178, 115671. [Google Scholar] [CrossRef]
- Anderson, K.L.; Whitlock, J.E.; Harwood, V.J. Persistence and Differential Survival of Fecal Indicator Bacteria in Subtropical Waters and Sediments. Appl. Environ. Microbiol. 2005, 71, 3041–3048. [Google Scholar] [CrossRef] [PubMed]
- Ahmed, W.; Hughes, B.; Harwood, V.J. Current Status of Marker Genes of Bacteroides and Related Taxa for Identifying Sewage Pollution in Environmental Waters. Water 2016, 8, 231. [Google Scholar] [CrossRef]
- Harwood, V.J.; Staley, C.; Badgley, B.D.; Borges, K.; Korajkic, A. Microbial Source Tracking Markers for Detection of Fecal Contamination in Environmental Waters: Relationships between Pathogens and Human Health Outcomes. FEMS Microbiol. Rev. 2014, 38, 1–40. [Google Scholar] [CrossRef]
- Shanks, O.C.; White, K.; Kelty, C.A.; Sivaganesan, M.; Blannon, J.; Meckes, M.; Varma, M.; Haugland, R.A. Performance of PCR-Based Assays Targeting Bacteroidales Genetic Markers of Human Fecal Pollution in Sewage and Fecal Samples. Environ. Sci. Technol. 2010, 44, 6281–6288. [Google Scholar] [CrossRef]
- Dick, L.K.; Bernhard, A.E.; Brodeur, T.J.; Santo Domingo, J.W.; Simpson, J.M.; Walters, S.P.; Field, K.G. Host Distributions of Uncultivated Fecal Bacteroidales Bacteria Reveal Genetic Markers for Fecal Source Identification. Appl. Environ. Microbiol. 2005, 71, 3184–3191. [Google Scholar] [CrossRef]
- Sinigalliano, C.D.; Fleisher, J.M.; Gidley, M.L.; Solo-Gabriele, H.M.; Shibata, T.; Plano, L.R.W.; Elmir, S.M.; Wanless, D.; Bartkowiak, J.; Boiteau, R.; et al. Traditional and Molecular Analyses for Fecal Indicator Bacteria in Non-Point Source Subtropical Recreational Marine Waters. Water Res. 2010, 44, 3763–3772. [Google Scholar] [CrossRef]
- Ervin, J.S.; Van De Werfhorst, L.C.; Murray, J.L.S.; Holden, P.A. Microbial Source Tracking in a Coastal California Watershed Reveals Canines as Controllable Sources of Fecal Contamination. Environ. Sci. Technol. 2014, 48, 9043–9052. [Google Scholar] [CrossRef] [PubMed]
- Lee, C.; Marion, J.W.; Lee, J. Development and Application of a Quantitative PCR Assay Targeting Catellicoccus marimammalium for Assessing Gull-Associated Fecal Contamination at Lake Erie Beaches. Sci. Total Environ. 2013, 454–455, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Sinigalliano, C.D.; Ervin, J.S.; Van De Werfhorst, L.C.; Badgley, B.D.; Ballesté, E.; Bartkowiak, J.; Boehm, A.B.; Byappanahalli, M.; Goodwin, K.D.; Gourmelon, M.; et al. Multi-Laboratory Evaluations of the Performance of Catellicoccus marimammalium PCR Assays Developed to Target Gull Fecal Sources. Water Res. 2013, 47, 6883–6896. [Google Scholar] [CrossRef] [PubMed]
- Knights, D.; Kuczynski, J.; Charlson, E.S.; Zaneveld, J.; Mozer, M.C.; Collman, R.G.; Bushman, F.D.; Knight, R.; Kelley, S.T. Bayesian Community-Wide Culture-Independent Microbial Source Tracking. Nat. Methods 2011, 8, 761–763. [Google Scholar] [CrossRef]
- O’Dea, C.; Zhang, Q.; Staley, C.; Masters, N.; Kuballa, A.; Fisher, P.; Veal, C.; Stratton, H.; Sadowsky, M.J.; Ahmed, W.; et al. Compositional and Temporal Stability of Fecal Taxon Libraries for Use with SourceTracker in Sub-Tropical Catchments. Water Res. 2019, 165, 114967. [Google Scholar] [CrossRef]
- Ahmed, W.; Staley, C.; Kaiser, T.; Sadowsky, M.J.; Kozak, S.; Beale, D.; Simpson, S. Decay of Sewage-Associated Bacterial Communities in Fresh and Marine Environmental Waters and Sediment. Appl. Microbiol. Biotechnol. 2018, 102, 7159–7170. [Google Scholar] [CrossRef]
- Mathai, P.P.; Staley, C.; Sadowsky, M.J. Sequence-Enabled Community-Based Microbial Source Tracking in Surface Waters Using Machine Learning Classification: A Review. J. Microbiol. Methods 2020, 177, 106050. [Google Scholar] [CrossRef]
- Haas, C.N.; Rose, J.B.; Gerba, C.P. Quantitative Microbial Risk Assessment; John Wiley & Sons: Hoboken, NJ, USA, 2014; ISBN 978-1-118-91002-3. [Google Scholar]
- Boehm, A.B.; Soller, J.A.; Shanks, O.C. Human-Associated Fecal Quantitative Polymerase Chain Reaction Measurements and Simulated Risk of Gastrointestinal Illness in Recreational Waters Contaminated with Raw Sewage. Environ. Sci. Technol. Lett. 2015, 2, 270–275. [Google Scholar] [CrossRef]
- Brown, K.I.; Graham, K.E.; Boehm, A.B. Risk-Based Threshold of Gull-Associated Fecal Marker Concentrations for Recreational Water. Environ. Sci. Technol. Lett. 2017, 4, 44–48. [Google Scholar] [CrossRef]
- Soller, J.A.; Schoen, M.E.; Varghese, A.; Ichida, A.M.; Boehm, A.B.; Eftim, S.; Ashbolt, N.J.; Ravenscroft, J.E. Human Health Risk Implications of Multiple Sources of Faecal Indicator Bacteria in a Recreational Waterbody. Water Res. 2014, 66, 254–264. [Google Scholar] [CrossRef]
- Gitter, A.; Gidley, M.; Mena, K.D.; Ferguson, A.; Sinigalliano, C.; Bonacolta, A.; Solo-Gabriele, H. Integrating Microbial Source Tracking with Quantitative Microbial Risk Assessment to Evaluate Site Specific Risk Based Thresholds at Two South Florida Beaches. Front. Microbiol. 2023, 14, 1210192. [Google Scholar] [CrossRef]
- Soller, J.A.; Schoen, M.E.; Bartrand, T.; Ravenscroft, J.E.; Ashbolt, N.J. Estimated Human Health Risks from Exposure to Recreational Waters Impacted by Human and Non-Human Sources of Faecal Contamination. Water Res. 2010, 44, 4674–4691. [Google Scholar] [CrossRef] [PubMed]
- Boehm, A.B.; Soller, J.A. Refined Ambient Water Quality Thresholds for Human-Associated Fecal Indicator HF183 for Recreational Waters with and without Co-Occurring Gull Fecal Contamination. Microb. Risk Anal. 2020, 16, 100139. [Google Scholar] [CrossRef]
- Powers, N.C.; Pinchback, J.; Flores, L.; Huang, Y.; Wetz, M.S.; Turner, J.W. Long-Term Water Quality Analysis Reveals Correlation between Bacterial Pollution and Sea Level Rise in the Northwestern Gulf of Mexico. Mar. Pollut. Bull. 2021, 166, 112231. [Google Scholar] [CrossRef] [PubMed]
- USEPA. Method 1696: Characterization of Human Fecal Pollution in Water by HF183/BacR287 TaqMan® Quantitative Polymerase Chain Reaction (qPCR) Assay; USEPA: Washington, DC, USA, 2019. [Google Scholar]
- Dick, L.K.; Simonich, M.; Field, K. Microplate Subtractive Hybridization To Enrich for Bacteroidales Genetic Markers for Fecal Source Identification|Applied and Environmental Microbiology. Available online: https://journals.asm.org/doi/full/10.1128/aem.71.6.3179-3183.2005 (accessed on 21 May 2025).
- Lee, L. NADA: Nondetects and Data Analysis for Environmental Data. R Package Version 1.6-1.1. Available online: https://CRAN.R-Project.Org/package=NADA (accessed on 1 April 2024).
- Claesson, M.J.; Wang, Q.; O’Sullivan, O.; Greene-Diniz, R.; Cole, J.R.; Ross, R.P.; O’Toole, P.W. Comparison of Two Next-Generation Sequencing Technologies for Resolving Highly Complex Microbiota Composition Using Tandem Variable 16S rRNA Gene Regions. Nucleic Acids Res. 2010, 38, e200. [Google Scholar] [CrossRef]
- Bolyen, E.; Rideout, J.R.; Dillon, M.R.; Bokulich, N.A.; Abnet, C.C.; Al-Ghalith, G.A.; Alexander, H.; Alm, E.J.; Arumugam, M.; Asnicar, F.; et al. Reproducible, Interactive, Scalable and Extensible Microbiome Data Science Using QIIME 2. Nat. Biotechnol. 2019, 37, 852–857. [Google Scholar] [CrossRef]
- Callahan, B.J.; McMurdie, P.J.; Rosen, M.J.; Han, A.W.; Johnson, A.J.A.; Holmes, S.P. DADA2: High-Resolution Sample Inference from Illumina Amplicon Data. Nat. Methods 2016, 13, 581–583. [Google Scholar] [CrossRef]
- USEPA. Recreational Water Quality Criteria; USEPA: Washington, DC, USA, 2012. [Google Scholar]
- Delignette-Muller, M.L.; Dutang, C. Fitdistrplus: An R Package for Fitting Distributions. J. Stat. Softw. 2015, 64, 1–34. [Google Scholar] [CrossRef]
- Schoen, M.E.; Ashbolt, N.J.; Jahne, M.A.; Garland, J. Risk-Based Enteric Pathogen Reduction Targets for Non-Potable and Direct Potable Use of Roof Runoff, Stormwater, and Greywater. Microb. Risk Anal. 2017, 5, 32–43. [Google Scholar] [CrossRef]
- Millard, S.; Kowarik, A. EnvStats: Package for Environmental Statistics, Including US EPA Guidance. 2023. Available online: https://cran.r-project.org/web/packages/EnvStats/index.html (accessed on 23 September 2023).
- USEPA. Quantitative Microbial Risk Assessment to Estimate Illness in Freshwater Impacted by Agricultural Animal Sources of Fecal Contamination; USEPA: Washington, DC, USA, 2010. [Google Scholar]
- Brown, K.I.; Graham, K.E.; Soller, J.A.; Boehm, A.B. Estimating the Probability of Illness Due to Swimming in Recreational Water with a Mixture of Human- and Gull-Associated Microbial Source Tracking Markers. Environ. Sci. Process. Impacts 2017, 19, 1528–1541. [Google Scholar] [CrossRef]
- Schoen, M.E.; Ashbolt, N.J. Assessing Pathogen Risk to Swimmers at Non-Sewage Impacted Recreational Beaches. Environ. Sci. Technol. 2010, 44, 2286–2291. [Google Scholar] [CrossRef] [PubMed]
- Campagnolo, E.R.; Philipp, L.M.; Long, J.M.; Hanshaw, N.L. Pet-Associated Campylobacteriosis: A Persisting Public Health Concern. Zoonoses Public Health 2018, 65, 304–311. [Google Scholar] [CrossRef] [PubMed]
- Gras, L.M.; Smid, J.H.; Wagenaar, J.A.; Koene, M.G.J.; Havelaar, A.H.; Friesema, I.H.M.; French, N.P.; Flemming, C.; Galson, J.D.; Graziani, C.; et al. Increased Risk for Campylobacter Jejuni and C. Coli Infection of Pet Origin in Dog Owners and Evidence for Genetic Association between Strains Causing Infection in Humans and Their Pets. Epidemiol. Infect. 2013, 141, 2526–2535. [Google Scholar] [CrossRef] [PubMed]
- Acke, E. Campylobacteriosis in Dogs and Cats: A Review. N. Z. Vet. J. 2018, 66, 221–228. [Google Scholar] [CrossRef]
- DeFlorio-Barker, S.; Arnold, B.F.; Sams, E.A.; Dufour, A.P.; Colford, J.M.; Weisberg, S.B.; Schiff, K.C.; Wade, T.J. Child Environmental Exposures to Water and Sand at the Beach: Findings from Studies of over 68,000 Subjects at 12 Beaches. J. Expo. Sci. Environ. Epidemiol. 2018, 28, 93–100. [Google Scholar] [CrossRef]
- Li, D.; Van De Werfhorst, L.C.; Steets, B.; Ervin, J.; Murray, J.L.S.; Smith, J.; Holden, P.A. Assessing Multiple Fecal Sources to Surf Zone Waters of Two Recreational Beaches by Bacterial Community Analysis. Water Res. 2022, 221, 118781. [Google Scholar] [CrossRef]
- Chaban, B.; Ngeleka, M.; Hill, J.E. Detection and Quantification of 14 Campylobacter Species in Pet Dogs Reveals an Increase in Species Richness in Feces of Diarrheic Animals. BMC Microbiol. 2010, 10, 73. [Google Scholar] [CrossRef]
- Lévesque, B.; Brousseau, P.; Bernier, F.; Dewailly, É.; Joly, J. Study of the Bacterial Content of Ring-Billed Gull Droppings in Relation to Recreational Water Quality. Water Res. 2000, 34, 1089–1096. [Google Scholar] [CrossRef]
- Koivunen, J.; Siitonen, A.; Heinonen-Tanski, H. Elimination of Enteric Bacteria in Biological–Chemical Wastewater Treatment and Tertiary Filtration Units. Water Res. 2003, 37, 690–698. [Google Scholar] [CrossRef]
- Lemarchand, K.; Lebaron, P. Occurrence of Salmonella Spp. and Cryptosporidium Spp. in a French Coastal Watershed: Relationship with Fecal Indicators. FEMS Microbiol. Lett. 2003, 218, 203–209. [Google Scholar] [CrossRef]
- Stampi, S.; Varoli, O.; Zanetti, F.; De Luca, G. Arcobacter cryaerophilus and Thermophilic Campylobacters in a Sewage Treatment Plant in Italy: Two Secondary Treatments Compared. Epidemiol. Infect. 1993, 110, 633–639. [Google Scholar] [CrossRef]
- García-Aljaro, C.; Bonjoch, X.; Blanch, A.R. Combined Use of an Immunomagnetic Separation Method and Immunoblotting for the Enumeration and Isolation of Escherichia Coli O157 in Wastewaters. J. Appl. Microbiol. 2005, 98, 589–597. [Google Scholar] [CrossRef] [PubMed]
- Harwood, V.J.; Levine, A.D.; Scott, T.M.; Chivukula, V.; Lukasik, J.; Farrah, S.R.; Rose, J.B. Validity of the Indicator Organism Paradigm for Pathogen Reduction in Reclaimed Water and Public Health Protection. Appl. Environ. Microbiol. 2005, 71, 3163–3170. [Google Scholar] [CrossRef] [PubMed]
- Crockett, C.S. The Role of Wastewater Treatment in Protecting Water Supplies Against Emerging Pathogens. Water Environ. Res. 2007, 79, 221–232. [Google Scholar] [CrossRef] [PubMed]
- Nasser, A.M. Removal of Cryptosporidium by Wastewater Treatment Processes: A Review. J. Water Health 2015, 14, 1–13. [Google Scholar] [CrossRef]
- Yang, J.; Schneider, O.D.; Jjemba, P.K.; Lechevallier, M.W. Microbial Risk Modeling for Main Breaks. J. Am. Water Work. Assoc. 2015, 107, E97–E108. [Google Scholar] [CrossRef]
- Kitajima, M.; Haramoto, E.; Iker, B.C.; Gerba, C.P. Occurrence of Cryptosporidium, Giardia, and Cyclospora in Influent and Effluent Water at Wastewater Treatment Plants in Arizona. Sci. Total Environ. 2014, 484, 129–136. [Google Scholar] [CrossRef]
- Eftim, S.E.; Hong, T.; Soller, J.; Boehm, A.; Warren, I.; Ichida, A.; Nappier, S.P. Occurrence of Norovirus in Raw Sewage—A Systematic Literature Review and Meta-Analysis. Water Res. 2017, 111, 366–374. [Google Scholar] [CrossRef]
- Hurst, C.J.; McClellan, K.A.; Benton, W.H. Comparison of Cytopathogenicity, Immunofluorescence and In Situ DNA Hybridization as Methods for the Detection of Adenoviruses. Water Res. 1988, 22, 1547–1552. [Google Scholar] [CrossRef]
- Hewitt, J.; Leonard, M.; Greening, G.E.; Lewis, G.D. Influence of Wastewater Treatment Process and the Population Size on Human Virus Profiles in Wastewater. Water Res. 2011, 45, 6267–6276. [Google Scholar] [CrossRef]
- Soller, J.A.; Schoen, M.; Steele, J.A.; Griffith, J.F.; Schiff, K.C. Incidence of Gastrointestinal Illness Following Wet Weather Recreational Exposures: Harmonization of Quantitative Microbial Risk Assessment with an Epidemiologic Investigation of Surfers. Water Res. 2017, 121, 280–289. [Google Scholar] [CrossRef]
- Fenlon, D.R. A Comparison of Salmonella Serotypes Found in the Faeces of Gulls Feeding at a Sewage Works with Serotypes Present in the Sewage. Epidemiol. Infect. 1983, 91, 47–52. [Google Scholar] [CrossRef]
- Gitter, A.; Mena, K.D.; Wagner, K.L.; Boellstorff, D.E.; Borel, K.E.; Gregory, L.F.; Gentry, T.J.; Karthikeyan, R. Human Health Risks Associated with Recreational Waters: Preliminary Approach of Integrating Quantitative Microbial Risk Assessment with Microbial Source Tracking. Water 2020, 12, 327. [Google Scholar] [CrossRef]
- Regli, S.; Rose, J.B.; Haas, C.N.; Gerba, C.P. Modeling the Risk From Giardia and Viruses in Drinking Water. J. Am. Water Work. Assoc. 1991, 83, 76–84. [Google Scholar] [CrossRef]
- Teunis, P.F.M.; Nagelkerke, N.J.D.; Haas, C.N. Dose Response Models For Infectious Gastroenteritis. Risk Anal. 1999, 19, 1251–1260. [Google Scholar] [CrossRef] [PubMed]
- Medema, G.J.; Teunis, P.F.M.; Havelaar, A.H.; Haas, C.N. Assessment of the Dose-Response Relationship of Campylobacter jejuni. Int. J. Food Microbiol. 1996, 30, 101–111. [Google Scholar] [CrossRef] [PubMed]
- Teunis, P.F.M.; Ogden, I.D.; Strachan, N.J.C. Hierarchical Dose Response of E. Coli O157:H7 from Human Outbreaks Incorporating Heterogeneity in Exposure. Epidemiol. Infect. 2008, 136, 761–770. [Google Scholar] [CrossRef]
- USEPA National Primary Drinking Water Regulations: Long Term 2 Enhanced Surface Water Treatment Rule 2006. Available online: https://www.regulations.gov/document/EPA-HQ-OW-2002-0039-0720 (accessed on 18 July 2025).
- Rose, J.B.; Gerba, C.P. Use of Risk Assessment for Development of Microbial Standards. Water Sci. Technol. 1991, 24, 29–34. [Google Scholar] [CrossRef]
- Eisenberg, J.N.; Seto, E.Y.W.; Olivieri, A.W.; Spear, R.C. Quantifying Water Pathogen Risk in an Epidemiological Framework. Risk Anal. 1996, 16, 549–563. [Google Scholar] [CrossRef]
- Messner, M.J.; Berger, P.; Nappier, S.P. Fractional Poisson—A Simple Dose-Response Model for Human Norovirus. Risk Anal. 2014, 34, 1820–1829. [Google Scholar] [CrossRef]
- Van Abel, N.; Schoen, M.E.; Kissel, J.C.; Meschke, J.S. Comparison of Risk Predicted by Multiple Norovirus Dose–Response Models and Implications for Quantitative Microbial Risk Assessment. Risk Anal. 2017, 37, 245–264. [Google Scholar] [CrossRef]
- Couch, R.B.; Knight, V.; Douglas, R.G.; Black, S.H.; Hamory, B.H. The Minimal Infectious Dose of Adenovirus Type 4; the Case for Natural Transmission by Viral Aerosol. Trans. Am. Clin. Climatol. Assoc. 1969, 80, 205–211. [Google Scholar]
- Crabtree, K.D.; Gerba, C.P.; Rose, J.B.; Haas, C.N. Waterborne Adenovirus: A Risk Assessment. Water Sci. Technol. 1997, 35, 1–6. [Google Scholar] [CrossRef]
- Brown, K.I.; Boehm, A.B. Comparative Decay of Catellicoccus marimmalium and Enterococci in Beach Sand and Seawater. Water Res. 2015, 83, 377–384. [Google Scholar] [CrossRef]
- Dhar, D.A.; Vadde, K.K.; Narimani, R.; Murgulet, D.; Felix, J.D.; Matta, A.; Pinchback, J.; Kapoor, V. Occurrence of Fecal Indicator Bacteria and Microbial Source Tracking Markers along the Texas Gulf Coast Adjoining Two Barrier Islands. Sci. Total Environ. 2025, 975, 179323. [Google Scholar] [CrossRef] [PubMed]
- Sefcik, K.; Speshock, J.; Brady, S.; Meik, J.M.; Brady, J.A. Tracking the Culprits: Microbial Source Tracking Uncovers Elevated Fecal Indicators along the Texas Coast. Adv. Microbiol. 2025, 15, 217–231. [Google Scholar] [CrossRef]
- Li, D.; Van De Werfhorst, L.C.; Steets, B.; Ervin, J.; Murray, J.L.S.; Devarajan, N.; Holden, P.A. Bather Shedding as a Source of Human Fecal Markers to a Recreational Beach. Front. Microbiol. 2021, 12, 673190. [Google Scholar] [CrossRef] [PubMed]
- Gerba, C.P. Assessment of Enteric Pathogen Shedding by Bathers during Recreational Activity and Its Impact on Water Quality. Quant. Microbiol. 2000, 2, 55–68. [Google Scholar] [CrossRef]
- Elmir, S.M.; Shibata, T.; Solo-Gabriele, H.M.; Sinigalliano, C.D.; Gidley, M.L.; Miller, G.; Plano, L.R.W.; Kish, J.; Withum, K.; Fleming, L.E. Quantitative Evaluation of Enterococci and Bacteroidales Released by Adults and Toddlers in Marine Water. Water Res. 2009, 43, 4610–4616. [Google Scholar] [CrossRef]
- Henry, R.; Schang, C.; Coutts, S.; Kolotelo, P.; Prosser, T.; Crosbie, N.; Grant, T.; Cottam, D.; O’Brien, P.; Deletic, A.; et al. Into the Deep: Evaluation of SourceTracker for Assessment of Faecal Contamination of Coastal Waters. Water Res. 2016, 93, 242–253. [Google Scholar] [CrossRef]
- Zimmer-Faust, A.G.; Steele, J.A.; Xiong, X.; Staley, C.; Griffith, M.; Sadowsky, M.J.; Diaz, M.; Griffith, J.F. A Combined Digital PCR and Next Generation DNA-Sequencing Based Approach for Tracking Nearshore Pollutant Dynamics Along the Southwest United States/Mexico Border. Front. Microbiol. 2021, 12, 674214. [Google Scholar] [CrossRef]
- Lobos, A.E.; Brandt, A.M.; Gallard-Góngora, J.F.; Korde, R.; Brodrick, E.; Harwood, V.J. Persistence of Sewage-Associated Genetic Markers in Advanced and Conventional Treated Recycled Water: Implications for Microbial Source Tracking in Surface Waters. mBio 2024, 15, e00655-24. [Google Scholar] [CrossRef]
- Locas, A.; Demers, J.; Payment, P. Evaluation of Photoreactivation of Escherichia Coli and Enterococci after UV Disinfection of Municipal Wastewater. Can. J. Microbiol. 2008, 54, 971–975. [Google Scholar] [CrossRef] [PubMed]
- Dragoni-Rosado, J.J.; Tavárez-Hernández, G.; González-Mederos, A.; Pérez-Matos, A.E. Evaluation of Photoreactivation and Dark Repair of Total and Fecal Coliforms and Enterococci in Wastewater Treated with Ultraviolet Light. Am. J. Water Resour. 2022, 10, 54–58. [Google Scholar] [CrossRef]
- McKee, B.A.; Molina, M.; Cyterski, M.; Couch, A. Microbial Source Tracking (MST) in Chattahoochee River National Recreation Area: Seasonal and Precipitation Trends in MST Marker Concentrations, and Associations with E. coli Levels, Pathogenic Marker Presence, and Land Use. Water Res. 2020, 171, 115435. [Google Scholar] [CrossRef] [PubMed]
- Skiendzielewski, K.; Burch, T.; Stokdyk, J.; McGinnis, S.; McLoughlin, S.; Firnstahl, A.; Spencer, S.; Borchardt, M.; Murphy, H.M. Two Risk Assessments: Evaluating the Use of Indicator HF183 Bacteroides versus Pathogen Measurements for Modelling Recreational Illness Risks in an Urban Watershed. Water Res. 2024, 259, 121852. [Google Scholar] [CrossRef]
- Burch, T.R.; Stokdyk, J.P.; Firnstahl, A.D.; Opelt, S.A.; Cook, R.M.; Heffron, J.A.; Brown, A.; Hruby, C.; Borchardt, M.A. Quantitative Microbial Risk Assessment with Microbial Source Tracking for Mixed Fecal Sources Contaminating Recreational River Waters, Iowa, USA. ACS EST Water 2024, 4, 2789–2802. [Google Scholar] [CrossRef]
- Powers, N.C.; Wallgren, H.R.; Marbach, S.; Turner, J.W. Relationship between Rainfall, Fecal Pollution, Antimicrobial Resistance, and Microbial Diversity in an Urbanized Subtropical Bay. Appl. Environ. Microbiol. 2020, 86, e01229-20. [Google Scholar] [CrossRef]
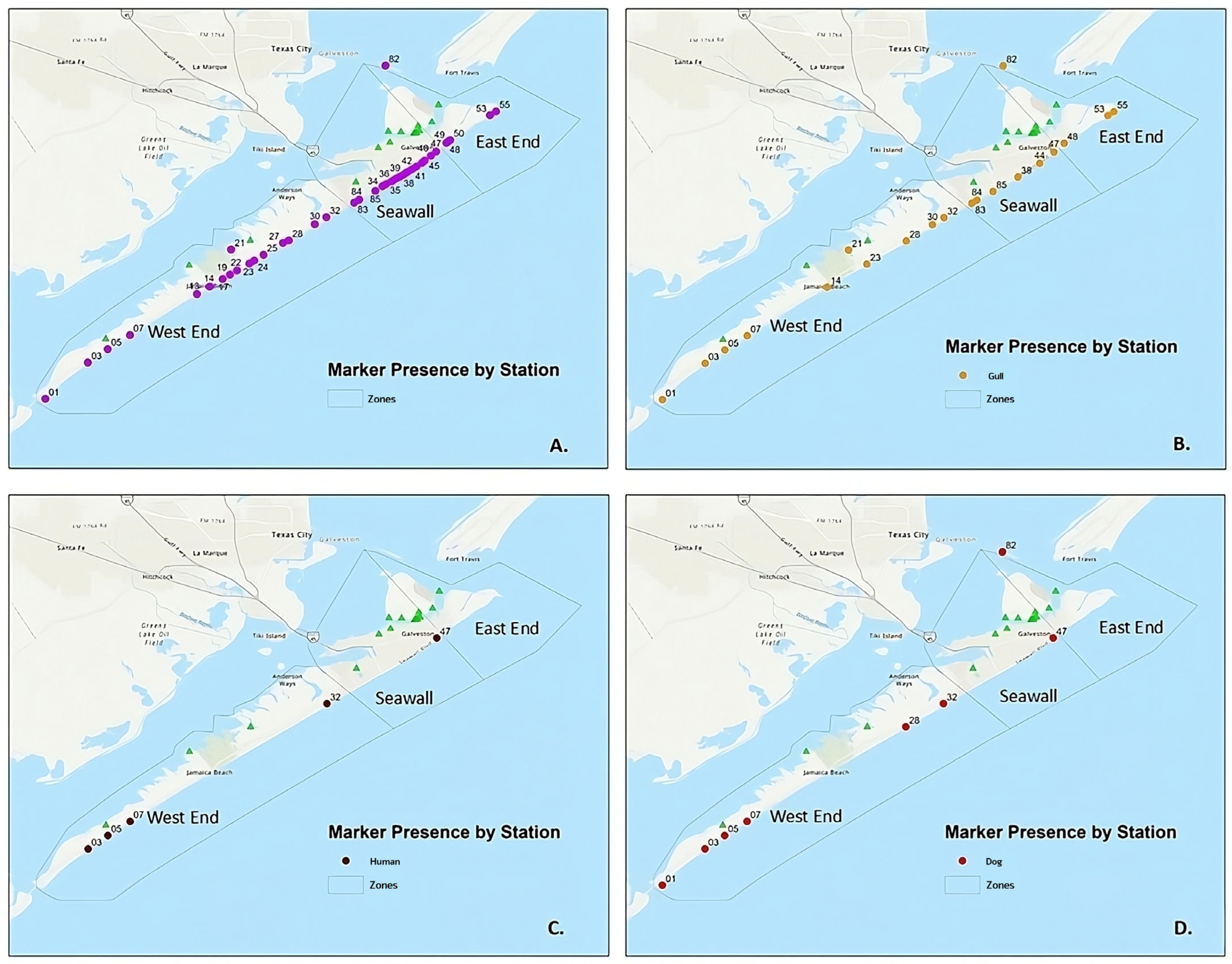

| Number | Station | Date Collected | Enterococci Result (MPN/100 mL) |
|---|---|---|---|
| 1 | GAL001 | 31 August 2022 | 496 |
| 2 | GAL003 | 31 August 2022 | 512 |
| 3 | GAL013 | 17 August 2022 | 161 |
| 4 | GAL014 | 24 August 2022 | 295 |
| 5 | GAL014 | 29 August 2022 | 288 |
| 6 | GAL021 | 24 August 2022 | 350 |
| 7 | GAL023 | 24 August 2022 | 193 |
| 8 | GAL023 | 29 August 2022 | 311 |
| 9 | GAL025 | 25 August 2022 | 9210 |
| 10 | GAL026 | 24 August 2022 | 1020 |
| 11 | GAL026 | 29 August 2022 | 331 |
| 12 | GAL027 | 4 July 2022 | 122 |
| 13 | GAL027 | 24 August 2022 | 1470 |
| 14 | GAL027 | 29 August 2022 | 206 |
| 15 | GAL028 | 24 August 2022 | 1240 |
| 16 | GAL028 | 29 August 2022 | 243 |
| 17 | GAL030 | 24 August 2022 | 2590 |
| 18 | GAL030 | 29 August 2022 | 399 |
| 19 | GAL032 | 24 August 2022 | 14,100 |
| 20 | GAL032 | 26 August 2022 | 459 |
| 21 | GAL032 | 29 August 2022 | 345 |
| 22 * | GAL032 | 14 September 2022 | 30 |
| 23 | GAL035 | 12 July 2022 | 148 |
| 24 | GAL037 | 26 July 2022 | 108 |
| 25 | GAL037 | 23 August 2022 | 309 |
| 26 | GAL044 | 23 August 2022 | 122 |
| 27 | GAL044 | 25 August 2022 | 107 |
| 28 | GAL044 | 29 August 2022 | 351 |
| 29 | GAL045 | 23 August 2022 | 132 |
| 30 | GAL045 | 29 August 2022 | 404 |
| 31 | GAL046 | 23 August 2022 | 135 |
| 32 | GAL046 | 25 August 2022 | 109 |
| 33 | GAL046 | 29 August 2022 | 332 |
| 34 | GAL047 | 23 August 2022 | 108 |
| 35 | GAL049 | 23 August 2022 | 132 |
| 36 | GAL053 | 31 August 2022 | 106 |
| 37 | GAL055 | 23 August 2022 | 108 |
| 38 | GAL055 | 31 August 2022 | 138 |
| Parameter | Units | Concentration | Source |
|---|---|---|---|
| HF183 measured in the environment | Copies/100 mL | Distribution: (0, 383.43, 0.12) a | Environmental data |
| Geometric mean: 131.50 | |||
| Maximum: 892.36 | |||
| Minimum: 46.72 | |||
| LeeSeaGull in environment | Copies/100 mL | Distribution: (0, 39,761.29, 0.10) a | Environmental data |
| Geometric mean: 12,248.21 | |||
| Maximum: 47,713.34 | |||
| Minimum: 2125.37 | |||
| DogBact in environment | Copies/100 mL | Distribution: (3558.99, 34,528.73) b | Environmental data |
| Geometric mean: 1828.64 | |||
| Maximum: 3692.74 | |||
| Minimum: 1390.84 | |||
| HF183 in human sewage | Copies/mL | (5.21, 0.57) c | [8] |
| LeeSeaGull in gull waste | Copies/g | (0, 8.7, 8.3) d | [20] |
| DogBact marker in dog waste | Copies/g | (5, 9) e | [11] |
| Campylobacter in dog feces | Organisms/g | (3, 8) e | [44] |
| Campylobacter in gull feces | CFU/g | (3.3, 6) e | [45] |
| Salmonella in gull feces | CFU/g | (2.3, 9.0) e | [45] |
| Salmonella in sewage | CFU/L | (0.5, 5) e | [46,47] |
| Campylobacter in sewage | MPN/L | (2.9, 4.6) e | [48] |
| E. coli O157:H7 in sewage | CFU/L | (−1, 3.3) e,f | [49] |
| Cryptosporidium in sewage | oocysts/L | (−0.52, 3.7) e | [34,50,51,52,53] |
| Giardia in sewage | cysts/L | (0.51, 4.2) e | [50,54] |
| Norovirus in sewage | copy/L | (4.7, 1.5) c | [55] |
| Adenovirus in sewage | IU/L | (1.75, 3.84) e | [56,57,58] |
| Volume water ingested (swimming) | Adults (mL) | 9, 64 g,j | [42] |
| Children (mL) | 36, 150 g,k | [42] | |
| Fraction of pathogenic species | Gull | 0.01–0.4 h | [38,59] |
| Sewage | 1 i | Assumed | |
| Dog | 0.02–0.1 h | [40] |
| Pathogen | Probability of Infection | Morbidity Ratio | Reference |
|---|---|---|---|
| Salmonella spp. | 0.17–0.4 a | [18,62] | |
| Campylobacter | 0.1–0.6 a | [63] | |
| E. coli 0157:H7 | 0.2–0.6 a | [64] | |
| Cryptosporidium | 0.3–0.7 a | [65] | |
| Giardia | 0.2–0.7 a | [66,67] | |
| Norovirus | c | 0.3–0.8 a | [68,69] |
| Adenovirus | 0.5 b | [70,71] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Corbeil, K.A.; Gitter, A.; Ruvalcaba, V.; Powers, N.C.; Hossain, M.S.; Bonaiti, G.; Flores, L.; Pinchback, J.; Jantrania, A.; Gentry, T. Marker- and Microbiome-Based Microbial Source Tracking and Evaluation of Bather Health Risk from Fecal Contamination in Galveston, Texas. Water 2025, 17, 2310. https://doi.org/10.3390/w17152310
Corbeil KA, Gitter A, Ruvalcaba V, Powers NC, Hossain MS, Bonaiti G, Flores L, Pinchback J, Jantrania A, Gentry T. Marker- and Microbiome-Based Microbial Source Tracking and Evaluation of Bather Health Risk from Fecal Contamination in Galveston, Texas. Water. 2025; 17(15):2310. https://doi.org/10.3390/w17152310
Chicago/Turabian StyleCorbeil, Karalee A., Anna Gitter, Valeria Ruvalcaba, Nicole C. Powers, Md Shakhawat Hossain, Gabriele Bonaiti, Lucy Flores, Jason Pinchback, Anish Jantrania, and Terry Gentry. 2025. "Marker- and Microbiome-Based Microbial Source Tracking and Evaluation of Bather Health Risk from Fecal Contamination in Galveston, Texas" Water 17, no. 15: 2310. https://doi.org/10.3390/w17152310
APA StyleCorbeil, K. A., Gitter, A., Ruvalcaba, V., Powers, N. C., Hossain, M. S., Bonaiti, G., Flores, L., Pinchback, J., Jantrania, A., & Gentry, T. (2025). Marker- and Microbiome-Based Microbial Source Tracking and Evaluation of Bather Health Risk from Fecal Contamination in Galveston, Texas. Water, 17(15), 2310. https://doi.org/10.3390/w17152310







