Rapid Spread of Omicron Sub-Lineage as Evidence by Wastewater Surveillance
Abstract
1. Introduction
2. Materials and Methods
2.1. Study Areas
2.2. Wastewater Environmental RNA Extraction and SARS-CoV-2 RNA Quantification
2.3. Effective Reproduction Number (Reff)
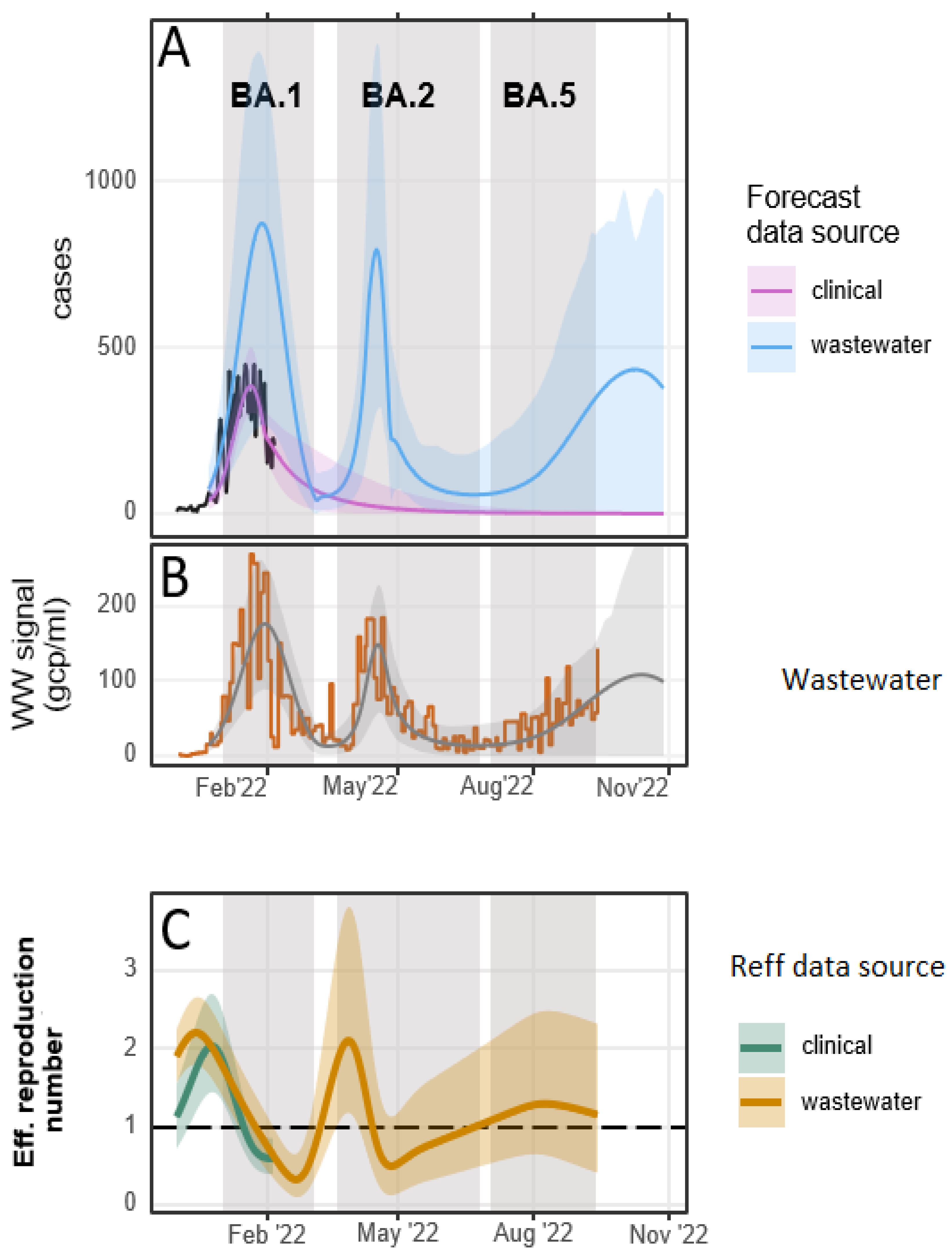
3. Results
4. Discussion
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Rahman, S.; Hossain, M.J.; Nahar, Z.; Shahriar, M.; Bhuiyan, M.A.; Islam, M.R. Emerging SARS-CoV-2 Variants and Subvariants: Challenges and Opportunities in the Context of COVID-19 Pandemic. Environ. Health Insights 2022, 16, 11786302221129396. [Google Scholar] [CrossRef]
- Wolfe, M.; Hughes, B.; Duong, D.; Chan-Herur, V.; Wigginton, K.R.; White, B.J.; Boehm, A.B. Detection of SARS-CoV-2 Variants Mu, Beta, Gamma, Lambda, Delta, Alpha, and Omicron in Wastewater Settled Solids Using Mutation-Specific Assays Is Associated with Regional Detection of Variants in Clinical Samples. Appl. Environ. Microbiol. 2022, 88, e00045-22. [Google Scholar] [CrossRef] [PubMed]
- Chakraborty, C.; Bhattacharya, M.; Sharma, A.R. Present variants of concern and variants of interest of severe acute respiratory syndrome coronavirus 2: Their significant mutations in S-glycoprotein, infectivity, re-infectivity, immune escape and vaccines activity. Rev. Med. Virol. 2022, 32, e2270. [Google Scholar]
- Chemaitelly, H.; Ayoub, H.H.; AlMukdad, S.; Coyle, P.; Tang, P.; Yassine, H.M.; Al-Khatib, H.A.; Smatti, M.K.; Hasan, M.R.; Al-Kanaani, Z. Duration of mRNA vaccine protection against SARS-CoV-2 Omicron BA. 1 and BA. 2 subvariants in Qatar. Nat. Commun. 2022, 13, 3082. [Google Scholar] [CrossRef] [PubMed]
- Lee, W.L.; Armas, F.; Guarneri, F.; Gu, X.; Formenti, N.; Wu, F.; Chandra, F.; Parisio, G.; Chen, H.; Xiao, A. Rapid displacement of SARS-CoV-2 variant Delta by Omicron revealed by allele-specific PCR in wastewater. Water Res. 2022, 221, 118809. [Google Scholar] [CrossRef]
- Lyngse, F.P.; Kirkeby, C.T.; Denwood, M.; Christiansen, L.E.; Mølbak, K.; Møller, C.H.; Skov, R.L.; Krause, T.G.; Rasmussen, M.; Sieber, R.N. Transmission of SARS-CoV-2 Omicron VOC subvariants BA. 1 and BA. 2: Evidence from Danish Households. MedRxiv, 2022; preprint. [Google Scholar]
- Oloye, F.F.; Xie, Y.; Asadi, M.; Cantin, J.; Challis, J.K.; Brinkmann, M.; McPhedran, K.N.; Kristian, K.; Keller, M.; Sadowski, M. Rapid transition between SARS-CoV-2 variants of concern Delta and Omicron detected by monitoring municipal wastewater from three Canadian cities. Sci. Total Environ. 2022, 841, 156741. [Google Scholar]
- Xie, Y.; Challis, J.K.; Oloye, F.F.; Asadi, M.; Cantin, J.; Brinkmann, M.; McPhedran, K.N.; Hogan, N.; Sadowski, M.; Jones, P.D. RNA in Municipal Wastewater Reveals Magnitudes of COVID-19 Outbreaks across Four Waves Driven by SARS-CoV-2 Variants of Concern. ACS EST Water 2022, 2, 1852–1862. [Google Scholar] [CrossRef]
- Karthikeyan, S.; Levy, J.I.; De Hoff, P.; Humphrey, G.; Birmingham, A.; Jepsen, K.; Farmer, S.; Tubb, H.M.; Valles, T.; Tribelhorn, C.E. Wastewater sequencing reveals early cryptic SARS-CoV-2 variant transmission. Nature 2022, 609, 101–108. [Google Scholar] [CrossRef]
- Peccia, J.; Zulli, A.; Brackney, D.E.; Grubaugh, N.D.; Kaplan, E.H.; Casanovas-Massana, A.; Ko, A.I.; Malik, A.A.; Wang, D.; Wang, M. Measurement of SARS-CoV-2 RNA in wastewater tracks community infection dynamics. Nat. Biotechnol. 2020, 38, 1164–1167. [Google Scholar] [CrossRef]
- Dietz, K. The estimation of the basic reproduction number for infectious diseases. Stat. Methods Med. Res. 1993, 2, 23–41. [Google Scholar] [CrossRef]
- Huisman, J.S.; Scire, J.; Angst, D.C.; Li, J.; Neher, R.A.; Maathuis, M.H.; Bonhoeffer, S.; Stadler, T. Estimation and worldwide monitoring of the effective reproductive number of SARS-CoV-2. eLife 2022, 11, e71345. [Google Scholar]
- Galani, A.; Aalizadeh, R.; Kostakis, M.; Markou, A.; Alygizakis, N.; Lytras, T.; Adamopoulos, P.G.; Peccia, J.; Thompson, D.C.; Kontou, A. SARS-CoV-2 wastewater surveillance data can predict hospitalizations and ICU admissions. Sci. Total Environ. 2022, 804, 150151. [Google Scholar] [PubMed]
- Wong, T.E.; Thurston, G.M.; Barlow, N.; Cahill, N.D.; Carichino, L.; Maki, K.; Ross, D.; Schneider, J. Evaluating the sensitivity of SARS-CoV-2 infection rates on college campuses to wastewater surveillance. Infect. Dis. Model. 2021, 6, 1144–1158. [Google Scholar] [CrossRef] [PubMed]
- Amman, F.; Markt, R.; Endler, L.; Hupfauf, S.; Agerer, B.; Schedl, A.; Richter, L.; Zechmeister, M.; Bicher, M.; Heiler, G. Viral variant-resolved wastewater surveillance of SARS-CoV-2 at national scale. Nat. Biotechnol. 2022, 40, 1814–1822. [Google Scholar] [CrossRef] [PubMed]
- Nourbakhsh, S.; Fazil, A.; Li, M.; Mangat, C.S.; Peterson, S.W.; Daigle, J.; Langner, S.; Shurgold, J.; D’Aoust, P.; Delatolla, R. A wastewater-based epidemic model for SARS-CoV-2 with application to three Canadian cities. Epidemics 2022, 39, 100560. [Google Scholar] [CrossRef]
- Csilléry, K.; François, O.; Blum, M.G. abc: An R package for approximate Bayesian computation (ABC). Methods Ecol. Evol. 2012, 3, 475–479. [Google Scholar]
- Minter, A.; Retkute, R. Approximate Bayesian Computation for infectious disease modelling. Epidemics 2019, 29, 100368. [Google Scholar] [CrossRef] [PubMed]
- Beaumont, M.A. Approximate bayesian computation. Annu. Rev. Stat. Its Appl. 2019, 6, 379–403. [Google Scholar] [CrossRef]
- Huisman, J.S.; Scire, J.; Caduff, L.; Fernandez-Cassi, X.; Ganesanandamoorthy, P.; Kull, A.; Scheidegger, A.; Stachler, E.; Boehm, A.B.; Hughes, B. Wastewater-based estimation of the effective reproductive number of SARS-CoV-2. Environ. Health Perspect. 2022, 130, 057011. [Google Scholar] [CrossRef]
- Johnson, R.; Mangwana, N.; Sharma, J.R.; Muller, C.J.; Malemela, K.; Mashau, F.; Dias, S.; Ramharack, P.; Kinnear, C.; Glanzmann, B. Delineating the Spread and Prevalence of SARS-CoV-2 Omicron Sublineages (BA. 1–BA. 5) and Deltacron Using Wastewater in the Western Cape, South Africa. J. Infect. Dis. 2022, 226, 1418–1427. [Google Scholar] [CrossRef]
- Jahn, K.; Dreifuss, D.; Topolsky, I.; Kull, A.; Ganesanandamoorthy, P.; Fernandez-Cassi, X.; Bänziger, C.; Devaux, A.J.; Stachler, E.; Caduff, L.; et al. Early detection and surveillance of SARS-CoV-2 genomic variants in wastewater using COJAC. Nat. Microbiol. 2022, 7, 1151–1160. [Google Scholar] [CrossRef] [PubMed]
- Tegally, H.; Moir, M.; Everatt, J.; Giovanetti, M.; Scheepers, C.; Wilkinson, E.; Subramoney, K.; Makatini, Z.; Moyo, S.; Amoako, D.G.; et al. Emergence of SARS-CoV-2 omicron lineages BA. 4 and BA. 5 in South Africa. Nat. Med. 2022, 28, 1785–1790. [Google Scholar] [CrossRef]
- Davies, M.A.; Morden, E.; Rousseau, P.; Arendse, J.; Bam, J.L.; Boloko, L.; Cloete, K.; Cohen, C.; Chetty, N.; Dane, P.; et al. Outcomes of laboratory-confirmed SARS-CoV-2 infection during resurgence driven by Omicron lineages BA. 4 and BA. 5 compared with previous waves in the Western Cape Province, South Africa. Int. J. Infect. Dis. 2023, 127, 63–68. [Google Scholar] [CrossRef] [PubMed]
- Jiang, G.; Wu, J.; Weidhaas, J.; Li, X.; Chen, Y.; Mueller, J.; Li, J.; Kumar, M.; Zhou, X.; Arora, S. Artificial neural network-based estimation of COVID-19 case numbers and effective reproduction rate using wastewater-based epidemiology. Water Res. 2022, 218, 118451. [Google Scholar]
- Russell, S.L.; Klaver, B.R.; Harrigan, S.P.; Kamelian, K.; Tyson, J.; Hoang, L.; Taylor, M.; Sander, B.; Mishra, S.; Prystajecky, N.; et al. Clinical severity of Omicron subvariants BA. 1, BA. 2, and BA. 5 in a population-based cohort study in British Columbia, Canada. J. Med. Virol. 2023, 95, e28423. [Google Scholar]
- Kopsidas, I.; Karagiannidou, S.; Kostaki, E.G.; Kousi, D.; Douka, E.; Sfikakis, P.P.; Moustakidis, S.; Kokkotis, C.; Tsaopoulos, D.; Tseti, I. Global Distribution, Dispersal Patterns, and Trend of Several Omicron Subvariants of SARS-CoV-2 across the Globe. Trop. Med. Infect. Dis. 2022, 7, 373. [Google Scholar] [CrossRef]
- Mengist, H.M.; Kombe, A.J.K.; Mekonnen, D.; Abebaw, A.; Getachew, M.; Jin, T. Mutations of SARS-CoV-2 Spike Protein: Implications on Immune Evasion and Vaccine-Induced Immunity, Seminars in Immunology; Elsevier: Amsterdam, The Netherlands, 2021; p. 101533. [Google Scholar]
- Kumar, S.; Karuppanan, K.; Subramaniam, G. Omicron (BA. 1) and Sub-Variants (BA. 1.1, BA. 2 and BA. 3) of SARS-CoV-2 Spike Infectivity and Pathogenicity: A Comparative Sequence and Structural-based Computational Assessment. J. Med. Virol. 2022, 94, 4780–4791. [Google Scholar]
- Roy, U. Molecular Investigations of Selected Spike Protein Mutations in SARS-CoV-2: Delta and Omicron Variants and Omicron BA. 2 Sub Variant. bioRxiv 2022. [Google Scholar] [CrossRef]
- Chen, J.; Wei, G.-W. Omicron ba. 2 (b. 1.1. 529.2): High potential for becoming the next dominant variant. J. Phys. Chem. Lett. 2022, 13, 3840–3849. [Google Scholar]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Oloye, F.F.; Asadi, M.; Yusuf, W.; Champredon, D.; Pu, X.; Femi-Oloye, O.P.; De Lange, C.; El-Baroudy, S.; Osunla, C.A.; Xie, Y.; et al. Rapid Spread of Omicron Sub-Lineage as Evidence by Wastewater Surveillance. Water 2024, 16, 318. https://doi.org/10.3390/w16020318
Oloye FF, Asadi M, Yusuf W, Champredon D, Pu X, Femi-Oloye OP, De Lange C, El-Baroudy S, Osunla CA, Xie Y, et al. Rapid Spread of Omicron Sub-Lineage as Evidence by Wastewater Surveillance. Water. 2024; 16(2):318. https://doi.org/10.3390/w16020318
Chicago/Turabian StyleOloye, Femi F., Mohsen Asadi, Warsame Yusuf, David Champredon, Xia Pu, Oluwabunmi P. Femi-Oloye, Chantel De Lange, Seba El-Baroudy, Charles Ayodeji Osunla, Yuwei Xie, and et al. 2024. "Rapid Spread of Omicron Sub-Lineage as Evidence by Wastewater Surveillance" Water 16, no. 2: 318. https://doi.org/10.3390/w16020318
APA StyleOloye, F. F., Asadi, M., Yusuf, W., Champredon, D., Pu, X., Femi-Oloye, O. P., De Lange, C., El-Baroudy, S., Osunla, C. A., Xie, Y., Cantin, J., McPhedran, K. N., Brinkmann, M., Servos, M. R., Jones, P. D., & Giesy, J. P. (2024). Rapid Spread of Omicron Sub-Lineage as Evidence by Wastewater Surveillance. Water, 16(2), 318. https://doi.org/10.3390/w16020318







