Multiple Lines of Evidence Indicate Limited Natural Recruitment of Golden Perch (Macquaria ambigua) in the Highly Regulated Lachlan River
Abstract
1. Introduction
2. Materials and Methods
2.1. Study Species and the Catchment
2.2. Otolith Sampling
2.3. Otolith Processing and Laser Ablation ICP-MS Analysis
2.4. Genetic Data
2.5. Statistical Analysis
2.5.1. Otolith Microchemistry
2.5.2. Single Nucleotide Polymorphism (SNP) and SilicoDArT Data Analysis
3. Results
3.1. Otolith Micro-Chemical Composition
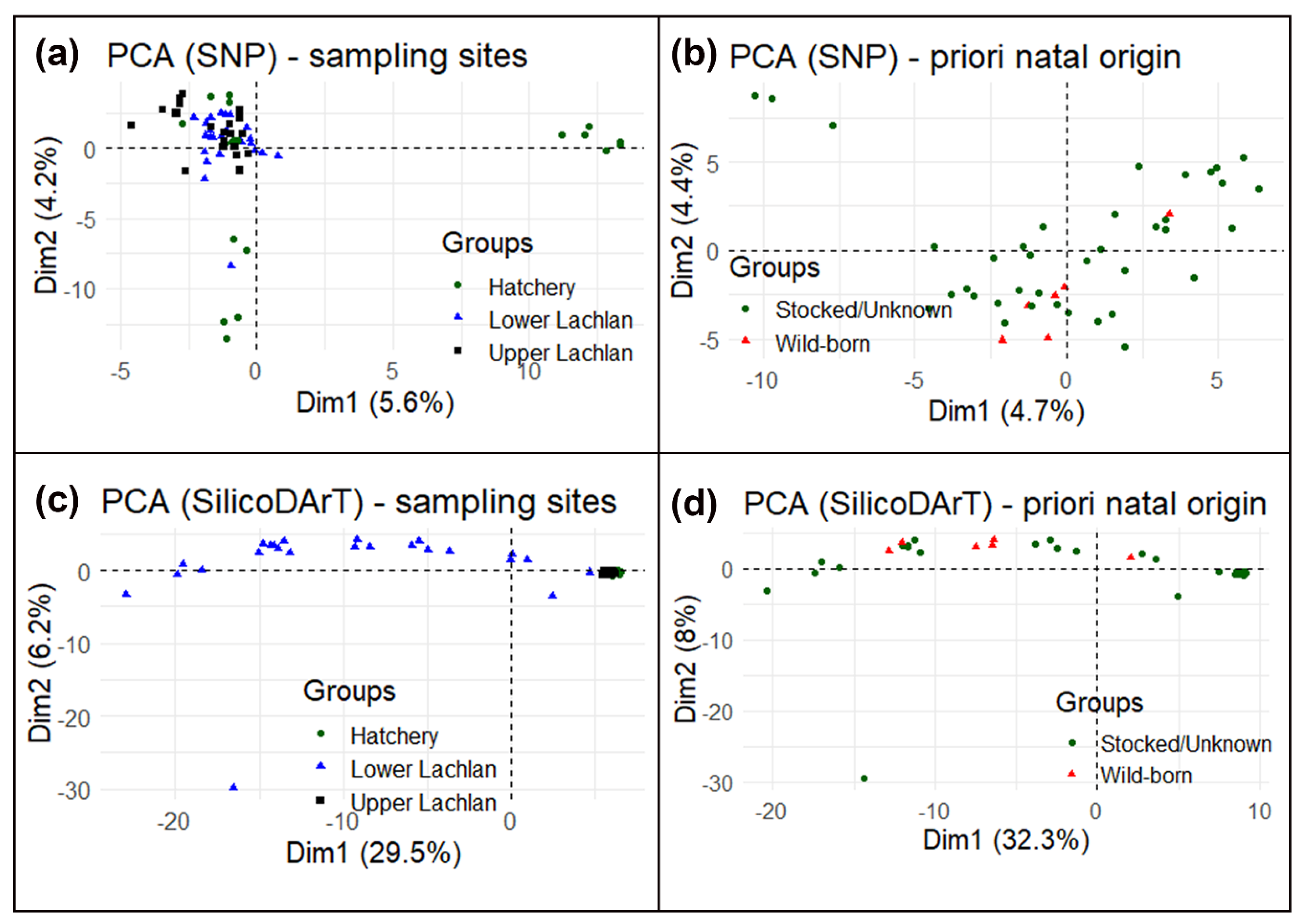
3.2. Genetic Data Analysis
4. Discussion
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Gaillard, J.-M.; Coulson, T.; Festa-Bianchet, M. Recruitment. In Population Dynamics. Vol 4 of Encyclopedia of Ecology; Elsevier: Oxford, UK, 2008; pp. 2982–2986. [Google Scholar]
- Jones, G.P. The importance of recruitment to the dynamics of a coral reef fish population. Ecology 1990, 71, 1691–1698. [Google Scholar] [CrossRef]
- Bradford, M.J.; Irvine, J.R. Land use, fishing, climate change, and the decline of Thompson River, British Columbia, coho salmon. Can. J. Fish. Aquat. Sci. 2000, 57, 13–16. [Google Scholar] [CrossRef]
- Humphries, P.; King, A.J.; Koehn, J.D. Fish, flows and flood plains: Links between freshwater fishes and their environment in the Murray-Darling River system, Australia. Environ. Biol. Fishes 1999, 56, 129–151. [Google Scholar] [CrossRef]
- Pankhurst, N.W.; Munday, P.L. Effects of climate change on fish reproduction and early life history stages. Mar. Freshw. Res. 2011, 62, 1015–1026. [Google Scholar] [CrossRef]
- Tillitt, D.E.; Papoulias, D.M.; Whyte, J.J.; Richter, C.A. Atrazine reduces reproduction in fathead minnow (Pimephales promelas). Aquat. Toxicol. 2010, 99, 149–159. [Google Scholar] [CrossRef]
- Mims, M.C.; Olden, J.D. Fish assemblages respond to altered flow regimes via ecological filtering of life history strategies. Freshw. Biol. 2013, 58, 50–62. [Google Scholar] [CrossRef]
- Zeug, S.; Winemiller, K. Relationships between hydrology, spatial heterogeneity, and fish recruitment dynamics in a temperate floodplain river. River Res. Appl. 2008, 24, 90–102. [Google Scholar] [CrossRef]
- Cowx, I. Stocking strategies. Fish. Manag. Ecol. 1994, 1, 15–30. [Google Scholar] [CrossRef]
- George, A.L.; Kuhajda, B.R.; Williams, J.D.; Cantrell, M.A.; Rakes, P.L.; Shute, J. Guidelines for propagation and translocation for freshwater fish conservation. Fisheries 2009, 34, 529–545. [Google Scholar] [CrossRef]
- Lintermans, M. The rise and fall of a translocated population of the endangered Macquarie perch, Macquaria australasica, in south-eastern Australia. Mar. Freshw. Res. 2013, 64, 838–850. [Google Scholar] [CrossRef]
- Araki, H.; Schmid, C. Is hatchery stocking a help or harm? Evidence, limitations and future directions in ecological and genetic surveys. Aquaculture 2010, 308, S2–S11. [Google Scholar] [CrossRef]
- Brown, C.; Laland, K. Social learning and life skills training for hatchery reared fish. J. Fish Biol. 2001, 59, 471–493. [Google Scholar] [CrossRef]
- Svåsand, T.; Kristiansen, T.S.; Pedersen, T.; Salvanes, A.V.; Engelsen, R.; Naevdal, G.; Nødtvedt, M. The enhancement of cod stocks. Fish Fish. 2000, 1, 173–205. [Google Scholar] [CrossRef]
- Englbrecht, C.C.; Schliewen, U.; Tautz, D. The impact of stocking on the genetic integrity of Arctic charr (Salvelinus) populations from the alpine region. Mol. Ecol. 2002, 11, 1017–1027. [Google Scholar] [CrossRef]
- Borwick, J.; District, A.; Philpot, A.; District, M.; Wilson, K. Review of Lake Simcoe’s Coldwater Fish Stocking Program; Ontario Ministry of Natural Resources: Midhurst (Aurora districts), ON, Canada, 2009.
- Hunt, T.L.; Allen, M.S.; Douglas, J.; Gason, A. Evaluation of a sport fish stocking program in lakes of the southern Murray-Darling Basin, Australia. N. Am. J. Fish. Manag. 2010, 30, 805–811. [Google Scholar] [CrossRef]
- Jennings, M.J.; Kampa, J.M.; Hatzenbeler, G.R.; Emmons, E.E. Evaluation of supplemental walleye stocking in northern Wisconsin lakes. N. Am. J. Fish. Manag. 2005, 25, 1171–1178. [Google Scholar] [CrossRef]
- Cook, G.S. Changes in otolith microchemistry over a protracted spawning season influence assignment of natal origin. Mar. Ecol. Prog. Ser. 2011, 423, 197–209. [Google Scholar] [CrossRef]
- Crook, D.A.; Macdonald, J.I.; McNeil, D.G.; Gilligan, D.M.; Asmus, M.; Maas, R.; Woodhead, J. Recruitment sources and dispersal of an invasive fish in a large river system as revealed by otolith chemistry analysis. Can. J. Fish. Aquat. Sci. 2013, 70, 953–963. [Google Scholar] [CrossRef]
- Elsdon, T.S.; Gillanders, B.M. Alternative life-history patterns of estuarine fish: Barium in otoliths elucidates freshwater residency. Can. J. Fish. Aquat. Sci. 2005, 62, 1143–1152. [Google Scholar] [CrossRef]
- Macdonald, J.; McNeil, D.G.; Crook, D.A. Identification of Carp Recruitment Hotspots in the Lachlan River Using Otolith Chemistry; Report to the Invasive Animals CRC and Lachlan CMa; SaRDI Aquatic Sciences, Invasive Animals Cooperative Research Centre: Canberra, Australia, SARDI Publication No. F2009/000682-1. SARDI Research Report Series No. 434; 2010; Available online: https://www.researchgate.net/publication/247768728_Identification_of_Carp_Recruitment_Hotspots_in_the_Lachlan_River_Using_Otolith_Chemistry (accessed on 28 October 2019).
- Arechavala-Lopez, P.; Milošević-González, M.; Sanchez-Jerez, P. Using trace elements in otoliths to discriminate between wild and farmed European sea bass (Dicentrarchus labrax L.) and Gilthead sea bream (Sparus aurata L.). Int. Aquat. Res. 2016, 8, 263–273. [Google Scholar] [CrossRef][Green Version]
- Perrier, C.; Daverat, F.; Evanno, G.; Pécheyran, C.; Bagliniere, J.-L.; Roussel, J.-M. Coupling genetic and otolith trace element analyses to identify river-born fish with hatchery pedigrees in stocked Atlantic salmon (Salmo salar) populations. Can. J. Fish. Aquat. Sci. 2011, 68, 977–987. [Google Scholar] [CrossRef]
- Starrs, D.; Ebner, B.C.; Fulton, C.J. All in the ears: Unlocking the early life history biology and spatial ecology of fishes. Biol. Rev. 2016, 91, 86–105. [Google Scholar] [CrossRef] [PubMed]
- Cochran-Biederman, J.L.; Wyman, K.E.; French, W.E.; Loppnow, G.L. Identifying correlates of success and failure of native freshwater fish reintroductions. Conserv. Biol. 2015, 29, 175–186. [Google Scholar] [CrossRef] [PubMed]
- Murphy, B.R.; Nielsen, L.A.; Turner, B.J. Use of genetic tags to evaluate stocking success for reservoir walleyes. Trans. Am. Fish. Soc. 1983, 112, 457–463. [Google Scholar] [CrossRef]
- Nielsen, E.E.; Hansen, M.M.; Loeschcke, V. Analysis of microsatellite DNA from old scale samples of Atlantic salmon Salmo salar: A comparison of genetic composition over 60 years. Mol. Ecol. 1997, 6, 487–492. [Google Scholar] [CrossRef]
- Nock, C.J.; Ovenden, J.; Butler, G.; Wooden, I.; Moore, A.; Baverstock, P.R. Population structure, effective population size and adverse effects of stocking in the endangered Australian eastern freshwater cod Maccullochella ikei. J. Fish Biol. 2011, 78, 303–321. [Google Scholar] [CrossRef]
- Gillanders, B.M.; Elsdon, T.S.; Munro, A.R. Impacts of native fish stocking on fish within the Murray—Darling Basin. In Murray-Darling Basin Commission Contract Number MD239, Murray-Darling Basin Commission Contract Number MD239; University of Adelaide: Adelaide, Australia, 2006. [Google Scholar]
- Hunt, T.L.; Jones, P. Informing the great fish stocking debate: An Australian case study. Rev. Fish. Sci. Aquac. 2018, 26, 275–308. [Google Scholar] [CrossRef]
- Koehn, J.D.; Lintermans, M. A strategy to rehabilitate fishes of the Murray-Darling Basin, south-eastern Australia. Endanger. Species Res. 2012, 16, 165–181. [Google Scholar] [CrossRef]
- Murray-Darling Basin Commission. Native Fish Strategy for the Murray-Darling Basin 2003–2013; Murray-Darling Basin Commission: Canberra, Australia, 2003.
- Lintermans, M. Fishes of the Murray-Darling Basin: An Introductory Guide; Murray-Darling Basin Commission: Canberra, Australia, 2007.
- Mackay, N. Histological changes in the ovaries of the golden perch, Plectroplites ambiguus, associated with the reproductive cycle. Mar. Freshw. Res. 1973, 24, 95–102. [Google Scholar] [CrossRef]
- Pusey, B.; Kennard, M.J.; Arthington, A.H. Freshwater Fishes of North-Eastern Australia; CSIRO Publishing: Collingwood, Australia, 2004. [Google Scholar]
- Rowland, S.J. Development of techniques for the large-scale rearing of the larvae of the Australian freshwater fish golden perch, Macquaria ambigua (Richardson, 1845). Mar. Freshw. Res. 1996, 47, 233–242. [Google Scholar] [CrossRef]
- Roberts, D.; Duivenvoorden, L.; Stuart, I. Factors influencing recruitment patterns of golden perch (Macquaria ambigua oriens) within a hydrologically variable and regulated Australian tropical river system. Ecol. Freshw. Fish 2008, 17, 577–589. [Google Scholar] [CrossRef]
- Reynolds, L. Migration patterns of five fish species in the Murray-Darling River system. Mar. Freshw. Res. 1983, 34, 857–871. [Google Scholar] [CrossRef]
- Kerezsy, A.; Balcombe, S.R.; Arthington, A.H.; Bunn, S.E. Continuous recruitment underpins fish persistence in the arid rivers of far-western Queensland, Australia. Mar. Freshw. Res. 2011, 62, 1178–1190. [Google Scholar] [CrossRef]
- Driver, P.; Chowdhury, S.; Hameed, T.; Lloyd-Jones, P.; Raisin, G.; Ribbons, C.; Singh, G.; Wettin, P. Natural and Modified Flows of the Lachlan Valley Wetlands; Resource Analysis Unit, Central West Region, NSW Department of Infrastructure, Planning and Natural Resources: Forbes, New South Wales, Australia, 2004.
- Driver, P.; Chowdhury, S.; Hameed, T.; O’Rourke, M.; Shaikh, M. Ecosystem response models for lower Calare (Lachlan River) floodplain wetlands: Managing wetland biota and climate change modelling. In Ecosystem Response Modelling in the Murray-Darling Basin, CSIRO Publishing; CSIRO: Melbourne, Australia, 2010; pp. 183–196. [Google Scholar]
- Higgisson, W.; Higgisson, B.; Powell, M.; Driver, P.; Dyer, F. Impacts of water resource development on hydrological connectivity of different floodplain habitats in a highly variable system. River Res. Appl. 2019, 36, 542–552. [Google Scholar] [CrossRef]
- Dijk, A.I.; Beck, H.E.; Crosbie, R.S.; Jeu, R.A.; Liu, Y.Y.; Podger, G.M.; Timbal, B.; Viney, N.R. The millennium drought in southeast Australia (2001–2009): Natural and human causes and implications for water resources, ecosystems, economy, and society. Water Resour. Res. 2013, 49, 1040–1057. [Google Scholar] [CrossRef]
- Dyer, F.; Broadhurst, B.; Tschierschke, A.; Thiem, J.; Thompson, R.; Driver, P.; Bowen, S. Commonwealth Environmental Water Office Long Term Intervention Monitoring Project: Lower Lachlan River System Selected Area 2017-18 Monitoring and Evaluation Summary Report; Commonwealth of Australia: Canberra, Australia, 2018.
- Rowland, S.J.; Tully, P. Hatchery Quality Assurance Program for Murrary Cod (Maccullochella Peelii Peelii), Golden Perch (Macquaria Ambigua) and Silver Perch (Bidyanus Bidyanus); NSW Department of Primary Industries: Sydney, New South Wales, Australia, 2004.
- Gilligan, D.; Jess, L.; McLean, G.; Asmus, M.; Wooden, I.; Hartwell, D.; McGregor, C.; Stuart, I.; Vey, A.; Jefferies, M. dentifying and implementing targeted carp control options for the Lower Lachlan Catchment. In Fisheries Final Report Series; 2010. Available online: http://www.dpi.nsw.gov.au/__data/assets/pdf_file/0008/344663/AE_2010_Output-1631_Gilligan-et-al_Lachlan-Carp-final-report_REPORT.pdf (accessed on 28 October 2019).
- Fallon, S.J.; McCulloch, M.T.; Van Woesik, R.; Sinclair, D.J. Corals at their latitudinal limits: Laser ablation trace element systematics in Porites from Shirigai Bay, Japan. Earth Planet. Sci. Lett. 1999, 172, 221–238. [Google Scholar] [CrossRef]
- Shams, F.; Dyer, F.; Thompson, R.; Duncan, R.P.; Thiem, J.D.; Ezaz, T. Genetic assessment to quantify long term stock enhancement reveals a signature of domestication in two Australian riverine fish species. Conserv. Genet. Under review.
- Kilian, A.; Wenzl, P.; Huttner, E.; Carling, J.; Xia, L.; Blois, H.; Caig, V.; Heller-Uszynska, K.; Jaccoud, D.; Hopper, C. Diversity arrays technology: A generic genome profiling technology on open platforms. Data Prod. Anal. Popul. Genom. Methods Protoc. 2012, 888, 67–89. [Google Scholar] [CrossRef]
- Altshuler, D.; Pollara, V.J.; Cowles, C.R.; Van Etten, W.J.; Baldwin, J.; Linton, L.; Lander, E.S. An SNP map of the human genome generated by reduced representation shotgun sequencing. Nature 2000, 407, 513–516. [Google Scholar] [CrossRef]
- Couch, A.J.; Unmack, P.J.; Dyer, F.J.; Lintermans, M. Who’s your mama? Riverine hybridisation of threatened freshwater Trout Cod and Murray Cod. PeerJ 2016, 4, e2593. [Google Scholar] [CrossRef]
- Melville, J.; Haines, M.L.; Boysen, K.; Hodkinson, L.; Kilian, A.; Date, K.L.S.; Potvin, D.A.; Parris, K.M. Identifying hybridization and admixture using SNPs: Application of the DArTseq platform in phylogeographic research on vertebrates. R. Soc. Open Sci. 2017, 4, 161061. [Google Scholar] [CrossRef] [PubMed]
- Shams, F.; Dyer, F.; Thompson, R.; Duncan, R.P.; Thiem, J.D.; Kilian, A.; Ezaz, T. Application of DArT seq derived SNP tags for comparative genome analysis in fishes; An alternative pipeline using sequence data from a non-traditional model species, Macquaria ambigua. PLoS ONE 2019, 14, e0226365. [Google Scholar] [CrossRef] [PubMed]
- Wells, S.J.; Dale, J. Contrasting gene flow at different spatial scales revealed by genotyping-by-sequencing in Isocladus armatus, a massively colour polymorphic New Zealand marine isopod. PeerJ 2018, 6, e5462. [Google Scholar] [CrossRef] [PubMed]
- R Core Team. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2016; Volume 3.3.1. [Google Scholar]
- Kruskal, J.B. Multidimensional scaling by optimizing goodness of fit to a nonmetric hypothesis. Psychometrika 1964, 29, 1–27. [Google Scholar] [CrossRef]
- Anderson, M.J.; Walsh, D.C. PERMANOVA, ANOSIM, and the Mantel test in the face of heterogeneous dispersions: What null hypothesis are you testing? Ecol. Monogr. 2013, 83, 557–574. [Google Scholar] [CrossRef]
- Oksanen, J.; Blanchet, F.; Friendly, M.; Kindt, R.; Legendre, P.; McGlin, D. Vegan: Community Ecology Package. R Package Version 2.5-3. 2018. Available online: https://CRAN.R-project.org/package=vegan (accessed on 14 June 2019).
- Murtagh, F.; Legendre, P. Ward’s hierarchical agglomerative clustering method: Which algorithms implement Ward’s criterion? J. Classif. 2014, 31, 274–295. [Google Scholar] [CrossRef]
- Ward Jr, J.H. Hierarchical grouping to optimize an objective function. J. Am. Stat. Assoc. 1963, 58, 236–244. [Google Scholar] [CrossRef]
- Gruber, B.; Unmack, P.J.; Berry, O.F.; Georges, A. Dartr: An R package to facilitate analysis of SNP data generated from reduced representation genome sequencing. Mol. Ecol. Resour. 2018, 18, 691–699. [Google Scholar] [CrossRef]
- Kassambara, A.; Mundt, F. Package ‘Factoextra’. Extract and Visualize the Results of Multivariate Data Analyses. R Package Version 1.0.5. 2017. Available online: https://CRAN.R-project.org/package=factoextra (accessed on 14 June 2019).
- Lê, S.; Josse, J.; Husson, F. FactoMineR: An R package for multivariate analysis. J. Stat. Softw. 2008, 25, 1–18. [Google Scholar] [CrossRef]
- Crook, D.A.; O’Mahony, D.J.; Gillanders, B.M.; Munro, A.R.; Sanger, A.C.; Thurstan, S.; Baumgartner, L.J. Contribution of stocked fish to riverine populations of golden perch (Macquaria ambigua) in the Murray-Darling Basin, Australia. Mar. Freshw. Res. 2016, 67, 1401–1409. [Google Scholar] [CrossRef]
- Forbes, J.; Watts, R.J.; Robinson, W.A.; Baumgartner, L.J.; McGuffie, P.; Cameron, L.M.; Crook, D.A. Assessment of stocking effectiveness for Murray cod (Maccullochella peelii) and golden perch (Macquaria ambigua) in rivers and impoundments of south-eastern Australia. Mar. Freshw. Res. 2016, 67, 1410–1419. [Google Scholar] [CrossRef]
- Thiem, J.D.; Wooden, I.J.; Baumgartner, L.J.; Butler, G.L.; Forbes, J.P.; Conallin, J. Recovery from a fish kill in a semi-arid Australian river: Can stocking augment natural recruitment processes? Austral Ecol. 2017, 42, 218–226. [Google Scholar] [CrossRef]
- Amano, Y.; Kuwahara, M.; Takahashi, T.; Shirai, K.; Yamane, K.; Amakawa, H.; Otake, T. Otolith elemental and Sr isotopic composition as a natal tag for Biwa salmon Oncorhynchus masou subsp. in Lake Biwa, Japan. Aquat. Biol. 2013, 19, 85–95. [Google Scholar] [CrossRef][Green Version]
- Artetxe-Arrate, I.; Fraile, I.; Crook, D.A.; Zudaire, I.; Arrizabalaga, H.; Greig, A.; Murua, H. Otolith microchemistry: A useful tool for investigating stock structure of yellowfin tuna (Thunnus albacares) in the Indian Ocean. Mar. Freshw. Res. 2019, 70, 1708–1721. [Google Scholar] [CrossRef]
- Ashford, J.; Arkhipkin, A.; Jones, C. Can the chemistry of otolith nuclei determine population structure of Patagonian toothfish Dissostichus eleginoides? J. Fish Biol. 2006, 69, 708–721. [Google Scholar] [CrossRef]
- Crook, D.; Macdonald, J.; O’Connor, J.; Barry, B. Use of otolith chemistry to examine patterns of diadromy in the threatened Australian grayling Prototroctes maraena. J. Fish Biol. 2006, 69, 1330–1344. [Google Scholar] [CrossRef]
- Ferguson, G.J.; Ward, T.M.; Gillanders, B.M. Otolith shape and elemental composition: Complementary tools for stock discrimination of mulloway (Argyrosomus japonicus) in southern Australia. Fish. Res. 2011, 110, 75–83. [Google Scholar] [CrossRef]
- Hearne, S.; Travers, M.; Evans, R.; Blyth, A.; Trinajstic, K.; McIlwain, J.; Newman, S. Population Connectivity of Two Reef Fish Species in Northwestern Australia Using Otolith Geochemistry: A Pilot Study; Report of Project 1.1.3—Project 1.1.3.4c Prepared for the Kimberley Marine Research Program; Western Australian Marine Science Institution: Perth, Western Australia, Australia, 2017. [Google Scholar]
- Coghlan, S.M., Jr.; Lyerly, M.S.; Bly, T.R.; Williams, J.S.; Bowman, D.; Hannigan, R. Otolith chemistry discriminates among hatchery-reared and tributary-spawned salmonines in a tailwater system. N. Am. J. Fish. Manag. 2007, 27, 531–541. [Google Scholar] [CrossRef]
- Olley, R.; Young, R.; Closs, G.P.; Kristensen, E.A.; Bickel, T.O.; Deans, N.; Davey, L.; Eggins, S. Recruitment sources of brown trout identified by otolith trace element signatures. N. Z. J. Mar. Freshw. Res. 2011, 45, 395–411. [Google Scholar] [CrossRef]
- Ebner, B.C.; Scholz, O.; Gawne, B. Golden perch Macquaria ambigua are flexible spawners in the Darling River, Australia. N. Z. J. Mar. Freshw. Res. 2009, 43, 571–578. [Google Scholar] [CrossRef]
- Attard, C.R.; Brauer, C.J.; Sandoval-Castillo, J.; Faulks, L.K.; Unmack, P.J.; Gilligan, D.M.; Beheregaray, L.B. Ecological disturbance influences adaptive divergence despite high gene flow in golden perch (Macquaria ambigua): Implications for management and resilience to climate change. Mol. Ecol. 2018, 27, 196–215. [Google Scholar] [CrossRef] [PubMed]
- Beheregaray, L.B.; Pfeiffer, L.V.; Attard, C.R.; Sandoval-Castillo, J.; Domingos, F.M.; Faulks, L.K.; Gilligan, D.M.; Unmack, P.J. Genome-wide data delimits multiple climate-determined species ranges in a widespread Australian fish, the golden perch (Macquaria ambigua). Mol. Phylogenet. Evol. 2017, 111, 65–75. [Google Scholar] [CrossRef] [PubMed]
- Sturrock, A.M.; Hunter, E.; Milton, J.A.; Johnson, R.C.; Waring, C.P.; Trueman, C.N.; Eimf, E. Quantifying physiological influences on otolith microchemistry. Methods Ecol. Evol. 2015, 6, 806–816. [Google Scholar] [CrossRef]
- Sturrock, A.M.; Trueman, C.N.; Milton, J.A.; Waring, C.P.; Cooper, M.J.; Hunter, E. Physiological influences can outweigh environmental signals in otolith microchemistry research. Mar. Ecol. Prog. Ser. 2014, 500, 245–264. [Google Scholar] [CrossRef]
- Hughes, J.M.; Schmidt, D.J.; Macdonald, J.I.; Huey, J.A.; Crook, D.A. Low interbasin connectivity in a facultatively diadromous fish: Evidence from genetics and otolith chemistry. Mol. Ecol. 2014, 23, 1000–1013. [Google Scholar] [CrossRef] [PubMed]
- Reis-Santos, P.; Tanner, S.E.; Aboim, M.A.; Vasconcelos, R.P.; Laroche, J.; Charrier, G.; Pérez, M.; Presa, P.; Gillanders, B.M.; Cabral, H.N. Reconciling differences in natural tags to infer demographic and genetic connectivity in marine fish populations. Sci. Rep. 2018, 8, 1–12. [Google Scholar] [CrossRef]
- Schmidt, D.J.; Crook, D.A.; Macdonald, J.I.; Huey, J.A.; Zampatti, B.P.; Chilcott, S.; Raadik, T.A.; Hughes, J.M. Migration history and stock structure of two putatively diadromous teleost fishes, as determined by genetic and otolith chemistry analyses. Freshw. Sci. 2014, 33, 193–206. [Google Scholar] [CrossRef]
- Svedäng, H.; André, C.; Jonsson, P.; Elfman, M.; Limburg, K.E. Migratory behaviour and otolith chemistry suggest fine-scale sub-population structure within a genetically homogenous Atlantic Cod population. Environ. Biol. Fishes 2010, 89, 383–397. [Google Scholar] [CrossRef]
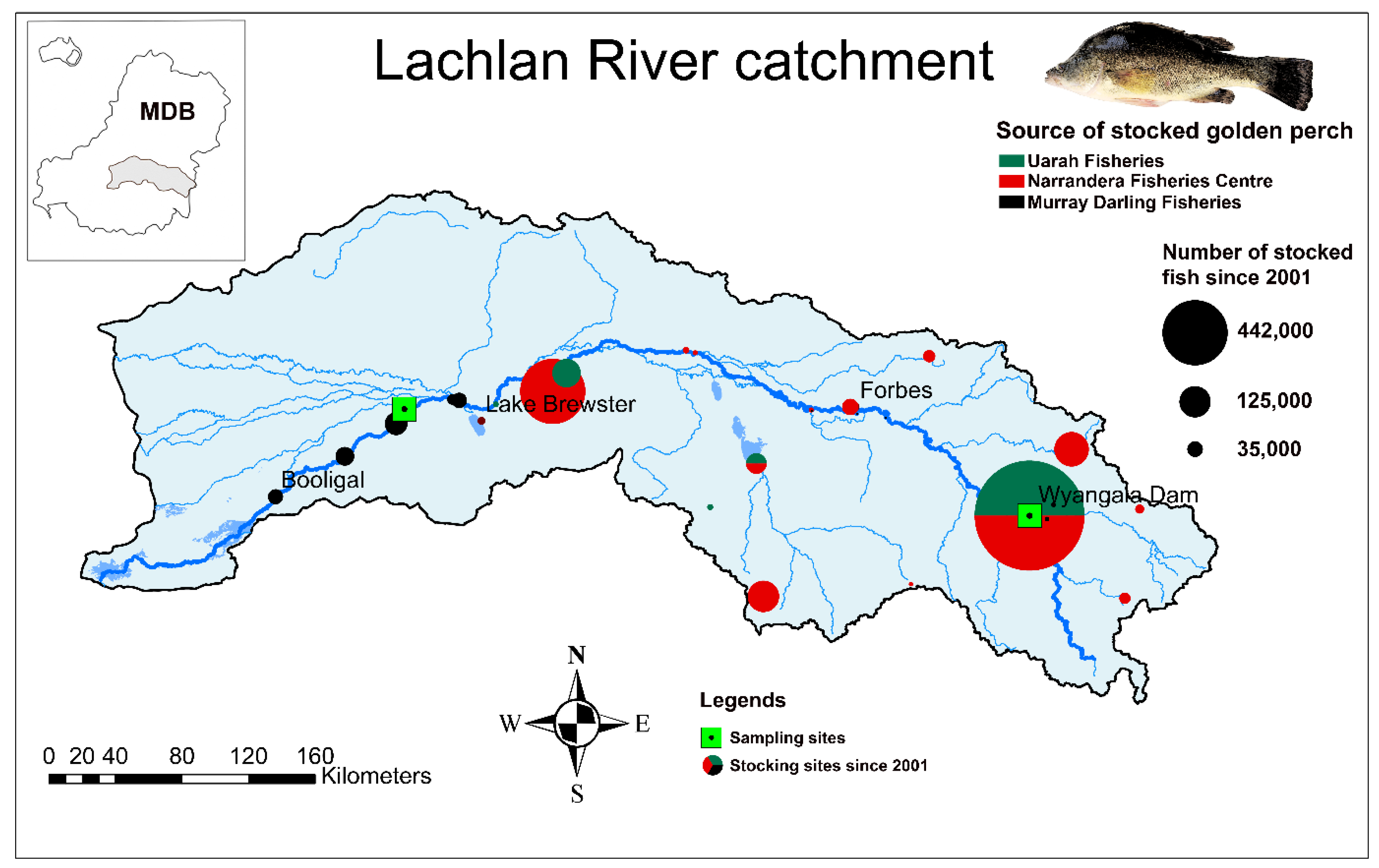
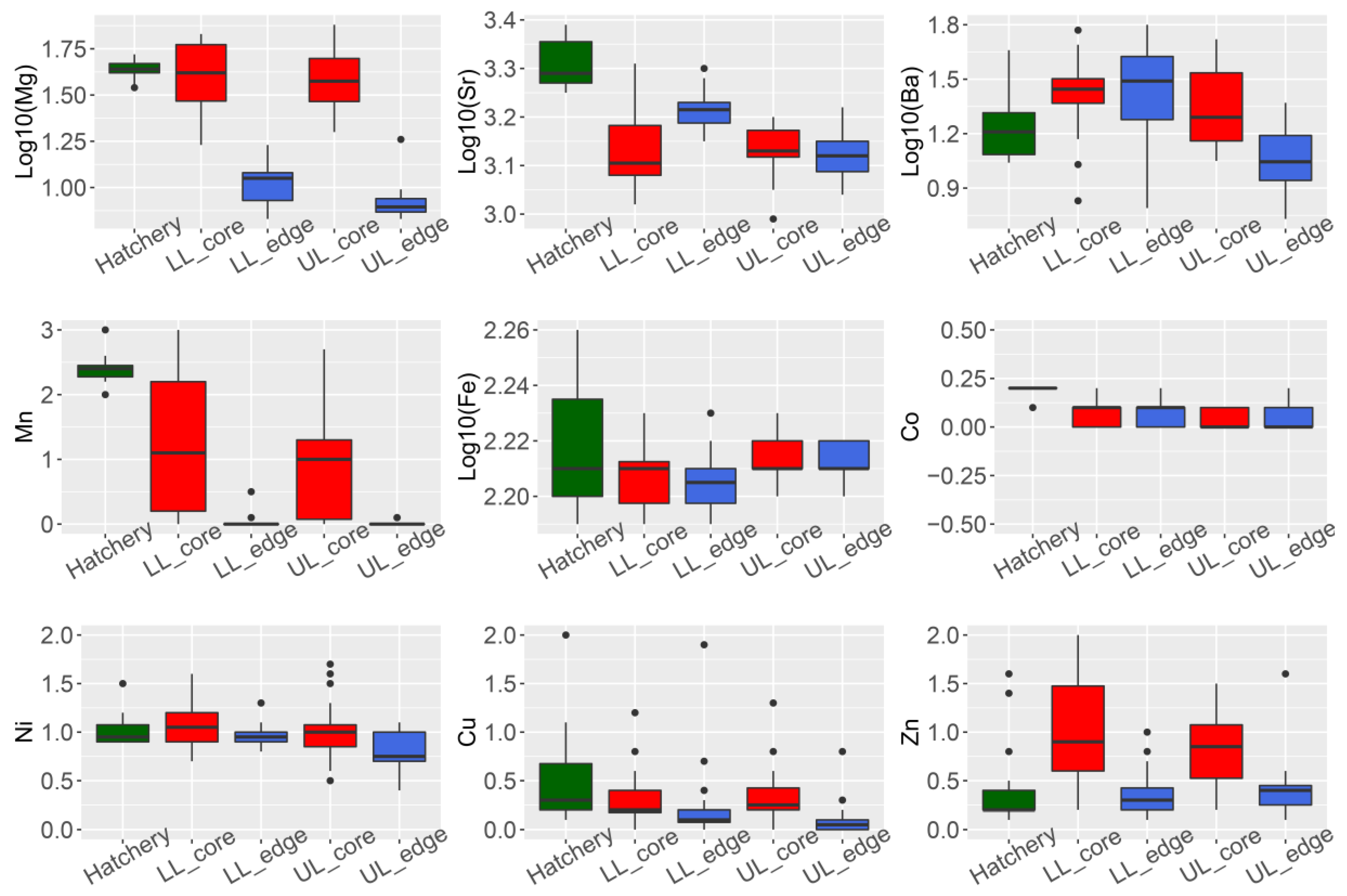
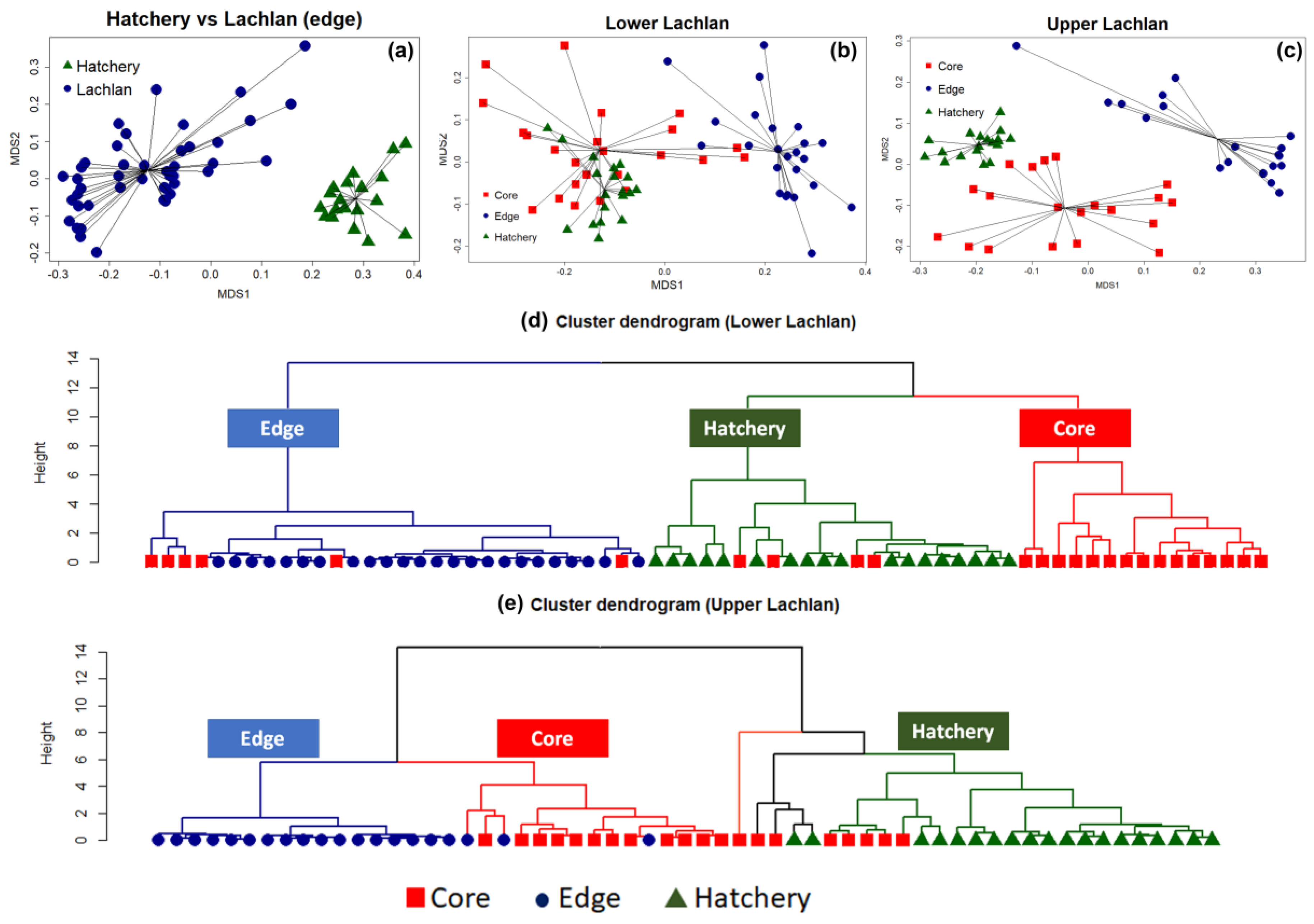
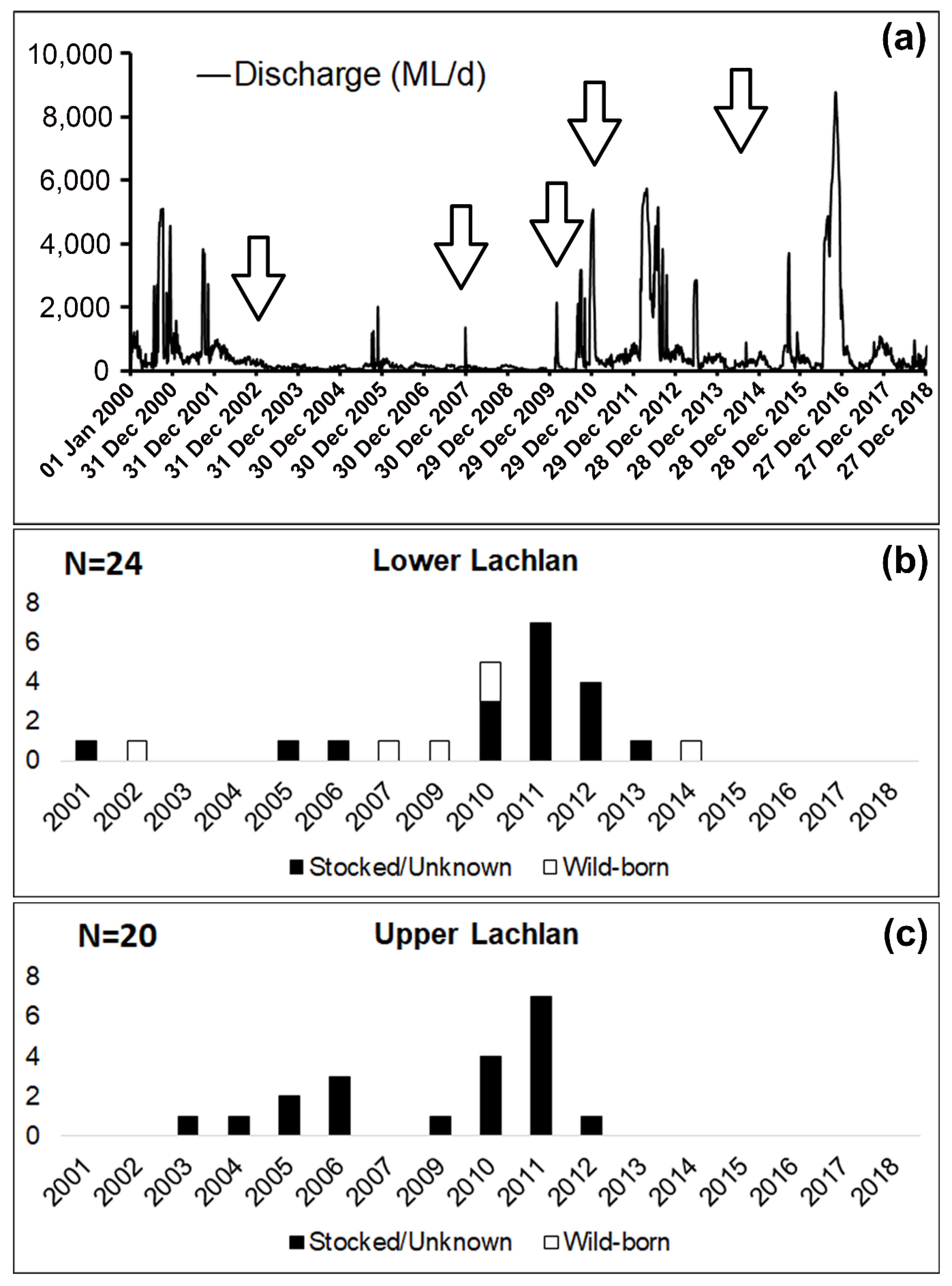
| Locality | Sampling Year | N (Sample Size) | Spawning Year Range |
|---|---|---|---|
| Lower Lachlan | 2015 | 24 | 2001–2014 |
| Upper Lachlan | 2017 | 20 | 2003–2012 |
| Hatchery (YOY) | 2017 | 19 | 2016 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Shams, F.; Dyer, F.; Thompson, R.; Duncan, R.P.; Thiem, J.D.; Enge, T.G.; Ezaz, T. Multiple Lines of Evidence Indicate Limited Natural Recruitment of Golden Perch (Macquaria ambigua) in the Highly Regulated Lachlan River. Water 2020, 12, 1636. https://doi.org/10.3390/w12061636
Shams F, Dyer F, Thompson R, Duncan RP, Thiem JD, Enge TG, Ezaz T. Multiple Lines of Evidence Indicate Limited Natural Recruitment of Golden Perch (Macquaria ambigua) in the Highly Regulated Lachlan River. Water. 2020; 12(6):1636. https://doi.org/10.3390/w12061636
Chicago/Turabian StyleShams, Foyez, Fiona Dyer, Ross Thompson, Richard P. Duncan, Jason D. Thiem, T. Gabriel Enge, and Tariq Ezaz. 2020. "Multiple Lines of Evidence Indicate Limited Natural Recruitment of Golden Perch (Macquaria ambigua) in the Highly Regulated Lachlan River" Water 12, no. 6: 1636. https://doi.org/10.3390/w12061636
APA StyleShams, F., Dyer, F., Thompson, R., Duncan, R. P., Thiem, J. D., Enge, T. G., & Ezaz, T. (2020). Multiple Lines of Evidence Indicate Limited Natural Recruitment of Golden Perch (Macquaria ambigua) in the Highly Regulated Lachlan River. Water, 12(6), 1636. https://doi.org/10.3390/w12061636





