Comparison of Community and Function of Dissimilatory Nitrate Reduction to Ammonium (DNRA) Bacteria in Chinese Shallow Lakes with Different Eutrophication Degrees
Abstract
1. Introduction
2. Materials and Methods
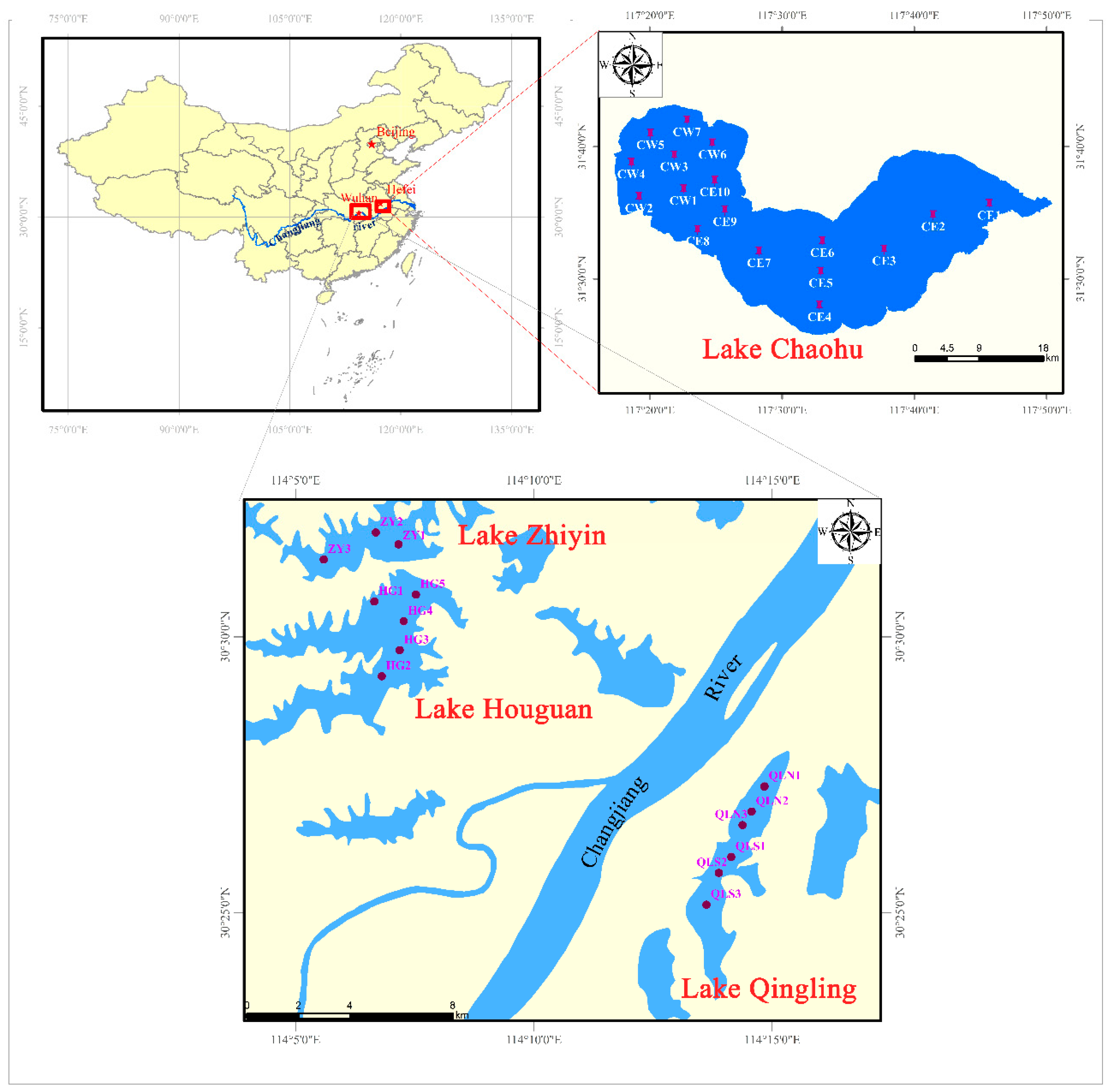
2.1. Study Sites and Sample Collection
2.2. Chemical Analysis
2.3. DNA Extraction and Quantitative PCR
2.4. Cloning, Sequencing, and Phylogenetic Analyses
2.5. Statistical Analysis
3. Results
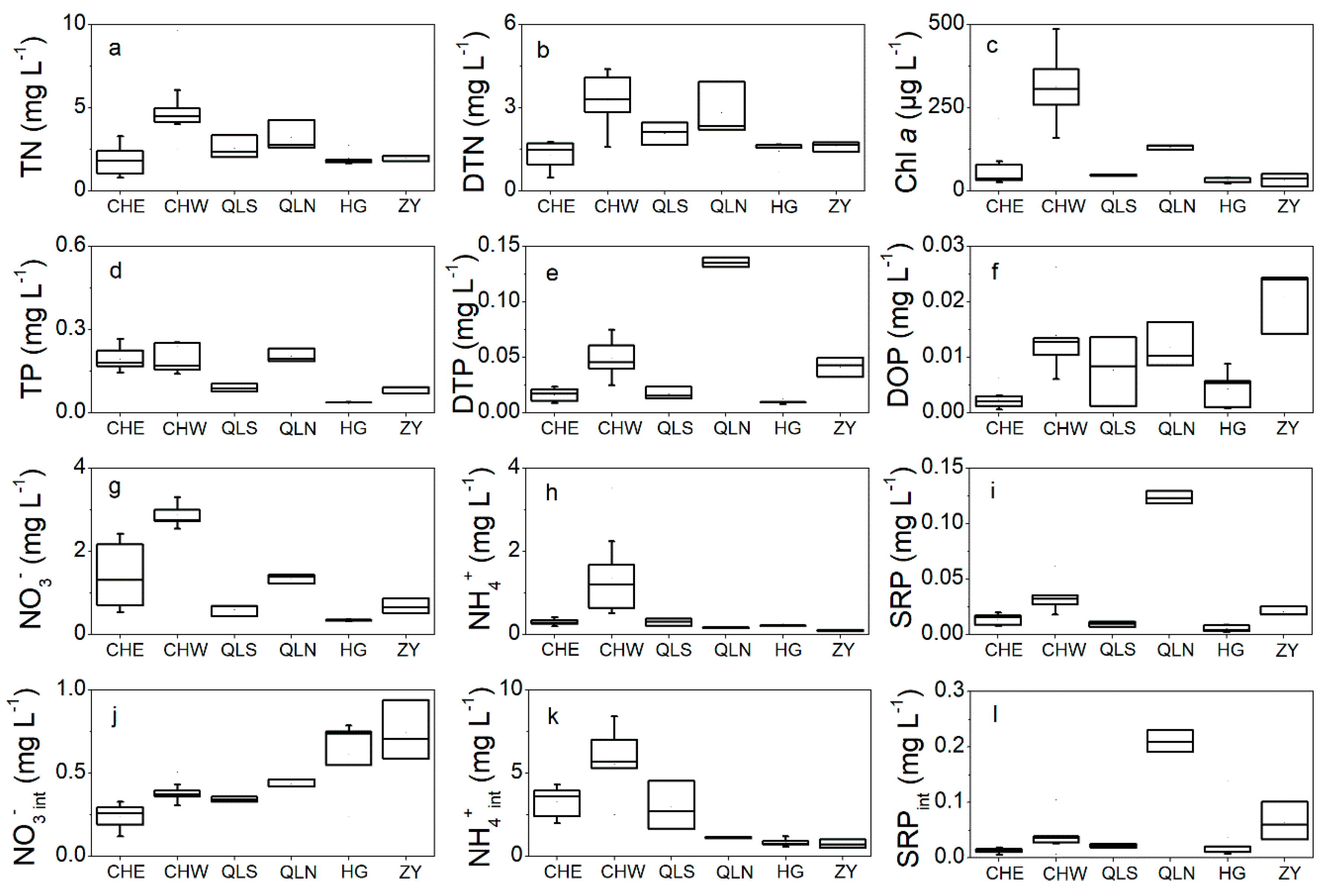
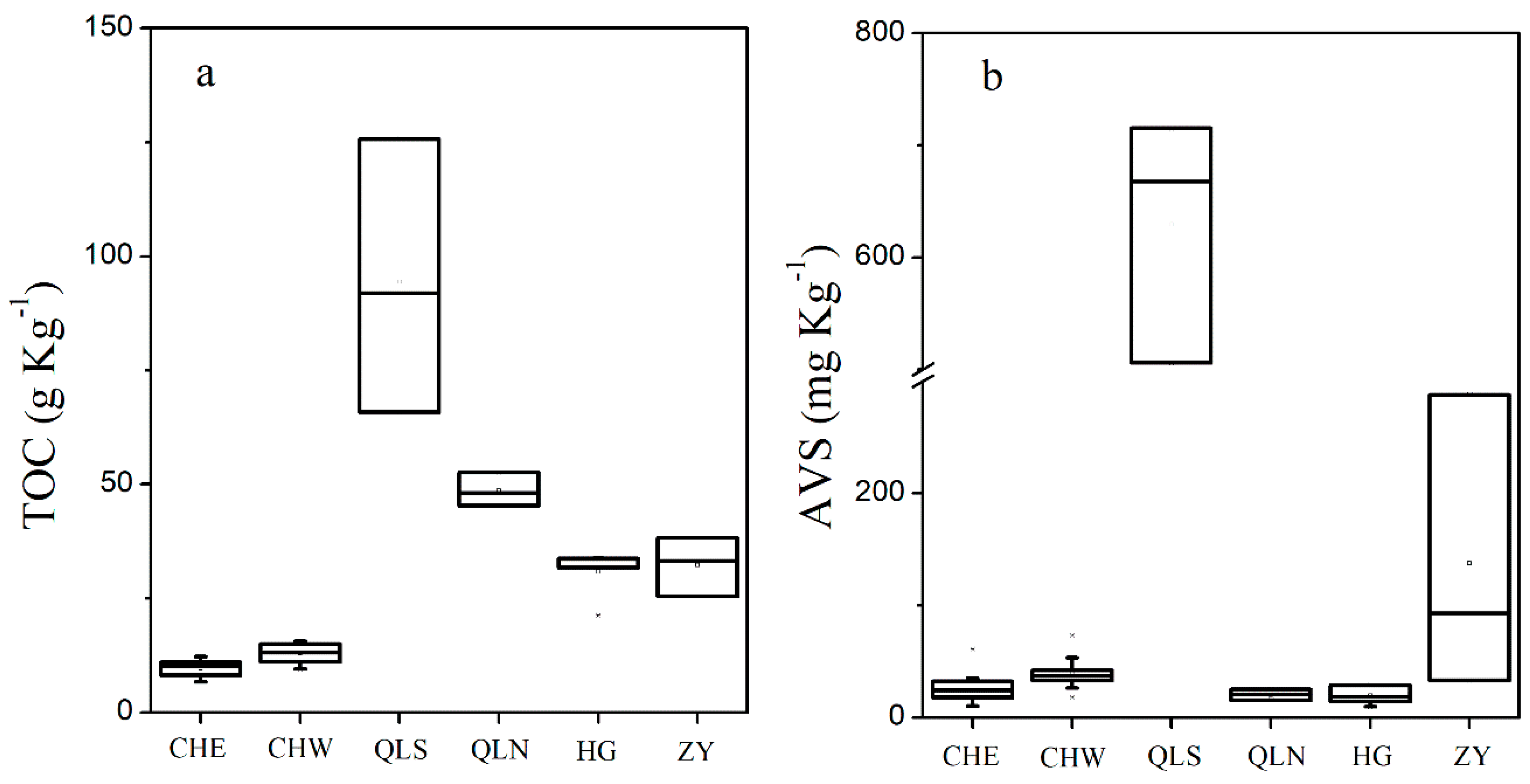
3.1. Nutrients Level
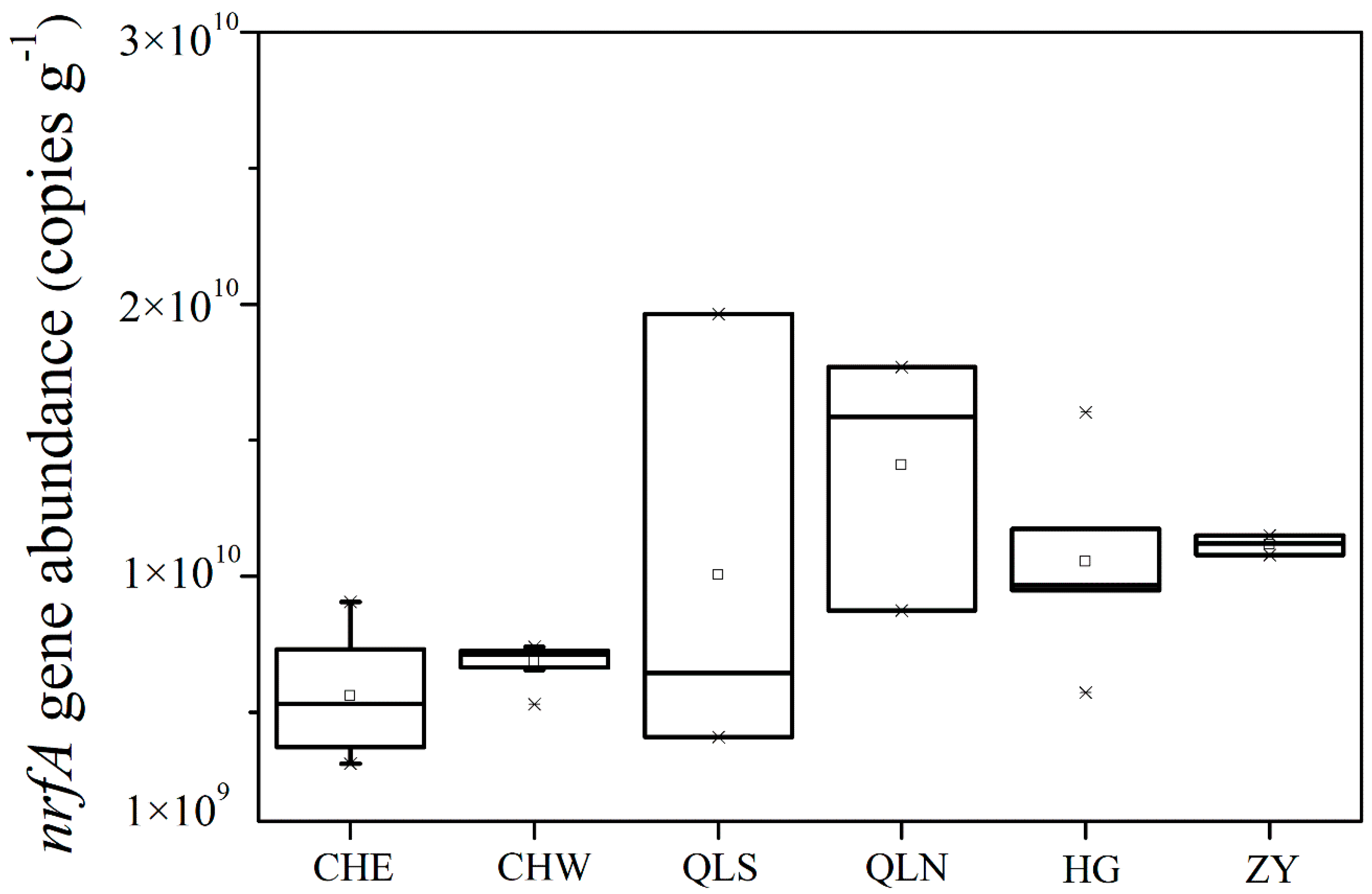
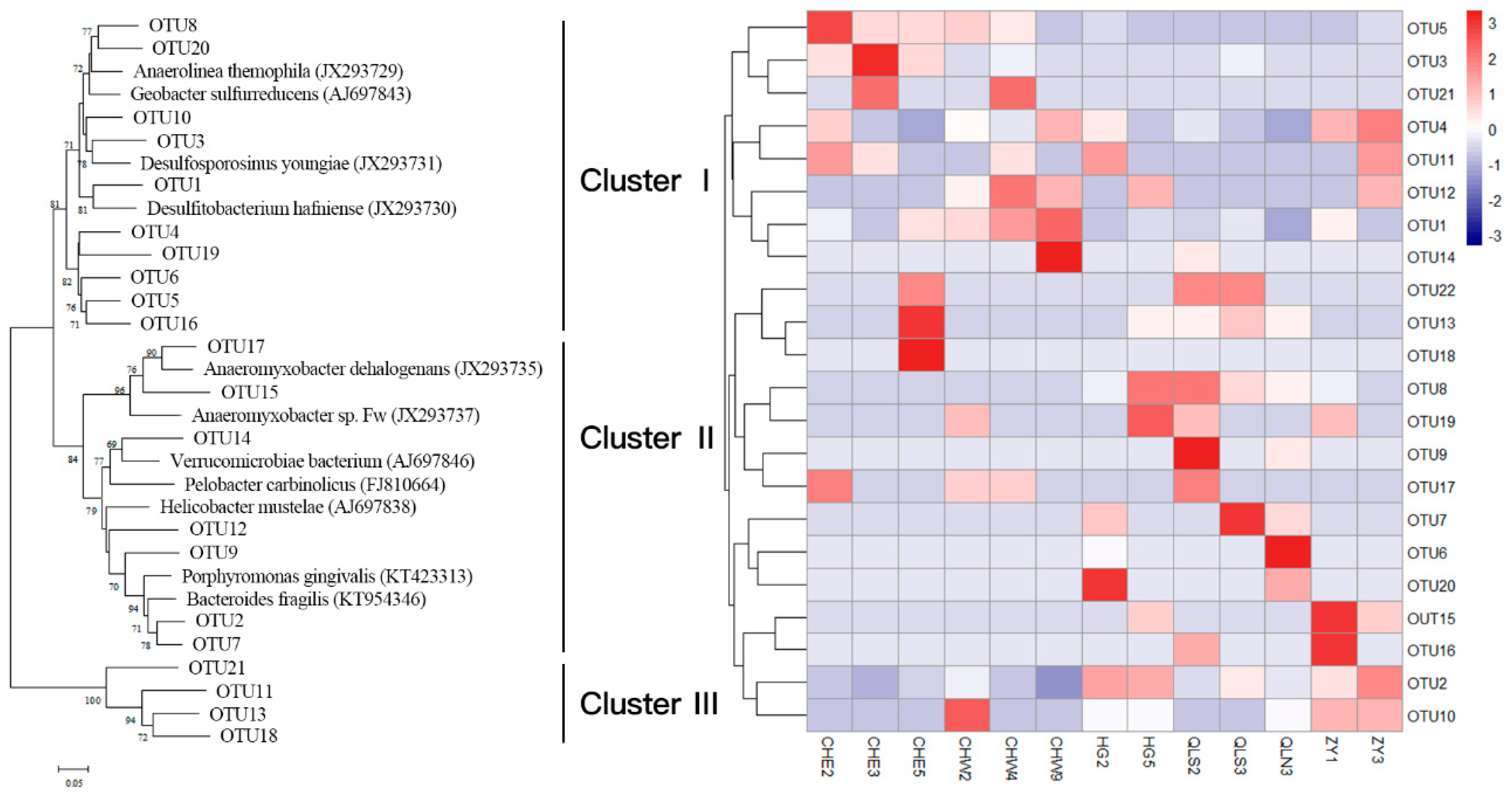
3.2. Abundance and Community Structure of Bacteria Mediating DNRA Process
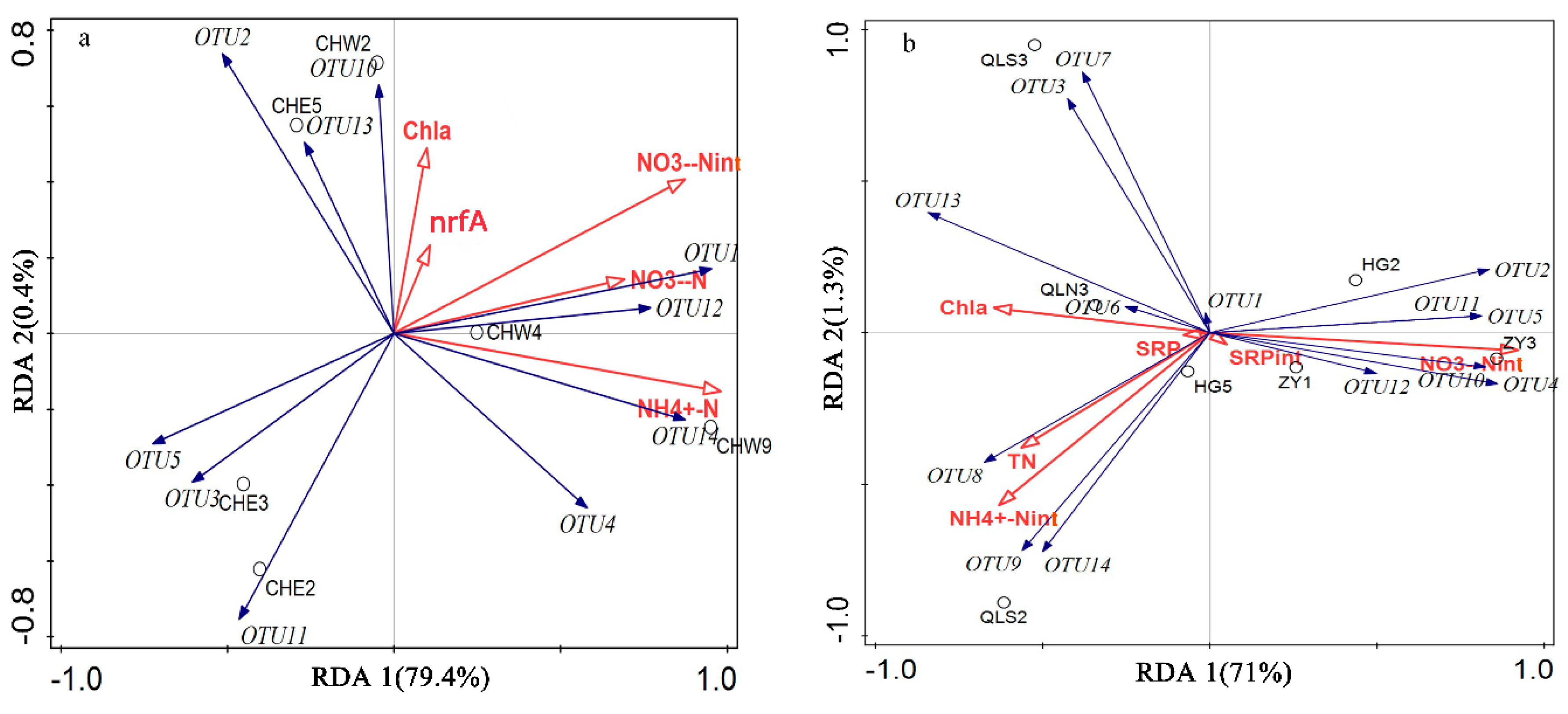
3.3. Relationship Between Abundance, Community Structure of DNRA Bacteria, and Nutrients
4. Discussion
5. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Vollenweider, R.A. Coastal marine eutrophication—Principles and control. Sci. Total. Environ. 1992, 1992, 1–20. [Google Scholar] [CrossRef]
- Carpenter, S.R.; Caraco, N.F.; Correll, D.L.; Howarth, R.W.; Sharpley, A.N.; Smith, V.H. Nonpoint pollution of surface waters with phosphorus and nitrogen. Ecol. Appl. 1998, 8, 559–568. [Google Scholar] [CrossRef]
- Berg, G.M.; Balode, M.; Purina, I.; Bekere, S.; Béchemin, C.; Maestrini, S.Y. Plankton community composition in relation to availability and uptake of oxidized and reduced nitrogen. Aquat. Microb. Ecol. 2003, 30, 263–274. [Google Scholar] [CrossRef]
- Gobler, C.J.; Burkholder, J.M.; Davis, T.W.; Harke, M.J.; Johengen, T.; Stow, C.A.; van de Waal, D.B. The dual role of nitrogen supply in controlling the growth and toxicity of cyanobacterial blooms. Harmful Algae 2016, 54, 87–89. [Google Scholar] [CrossRef] [PubMed]
- Rutting, T.; Boeckx, P.; Muller, C.; Klemedtsson, L. Assessment of the importance of dissimilatory nitrate reduction to ammonium for the terrestrial nitrogen cycle. Biogeosciences 2011, 8, 1779–1791. [Google Scholar] [CrossRef]
- Giblin, A.E.; Tobias, C.R.; Song, B.; Weston, N.; Banta, G.T.; Rivera-Monroy, V.H. The importance of dissimilatory nitrate reduction to ammonium (DNRA) in the nitrogen cycle of coastal ecosystems. Oceanography 2013, 26, 124–131. [Google Scholar] [CrossRef]
- Tiedje, J.M. Ecology of denitrification and dissimilatory nitrate reduction to ammonium. Biol. Anaerob. Microorg. 1988, 717, 179–244. [Google Scholar]
- Megonigal, J.P.; Hines, M.E.; Visscher, P.T. Anaerobic metabolism: Linkages to trace gases and aerobic processes. Biogeochemistry 2014, 8, 317–424. [Google Scholar] [CrossRef]
- Brunet, R.C.; Garcia-Gil, L.J. Sulfide-induced dissimilatory nitrate reduction to ammonia in anaerobic freshwater sediments. Fems. Microbiol. Ecol. 1996, 21, 131–138. [Google Scholar] [CrossRef]
- Otte, S.; Kuenen, J.G.; Nielsen, L.P.; Paerl, H.W.; Zopfi, J.; Schulz, H.N. Nitrogen, carbon, and sulfur metabolism in natural Thioploca samples. Appl. Environ. Microb. 1999, 65, 3148–3157. [Google Scholar] [CrossRef]
- Dong, L.F.; Sobey, M.N.; Smith, C.J.; Rusmana, I.; Phillips, W.; Stott, A. Dissimilatory reduction of nitrate to ammonium, not denitrification or anammox, dominates benthic nitrate reduction in tropical estuaries. Limnol. Oceanogr. 2011, 56, 279–291. [Google Scholar] [CrossRef]
- Laverman, A.M.; Canavan, R.W.; Slomp, C.P.; Van Cappellen, P. Potential nitrate removal in a coastal freshwater sediment (Haringvliet Lake, The Netherlands) and response to salinization. Water Res. 2007, 41, 3061–3068. [Google Scholar] [CrossRef] [PubMed]
- Yang, W.H.; Ryals, R.A.; Cusack, D.F.; Silver, W.L. Cross-biome assessment of gross soil nitrogen cycling in California ecosystems. Soil. Biol. Biochem. 2017, 107, 144–155. [Google Scholar] [CrossRef]
- Seitzinger, S.P. Denitrification in fresh-water and coastal marine ecosystems - ecological and geochemical significance. Limnol. Oceanogr. 1988, 33, 702–724. [Google Scholar] [CrossRef]
- van den Berg, E.M.; van Dongen, U.; Abbas, B.; van Loosdrecht, M.C.M. Enrichment of DNRA bacteria in a continuous culture. ISME J. 2015, 9, 2153–2161. [Google Scholar] [CrossRef] [PubMed]
- Nizzoli, D.; Carraro, E.; Nigro, V.; Viaroli, P. Effect of organic enrichment and thermal regime on denitrification and dissimilatory nitrate reduction to ammonium (DNRA) in hypolimnetic sediments of two lowland lakes. Water Res. 2010, 44, 2715–2724. [Google Scholar] [CrossRef] [PubMed]
- An, S.M.; Gardner, W.S. Dissimilatory nitrate reduction to ammonium (DNRA) as a nitrogen link, versus denitrification as a sink in a shallow estuary (Laguna Madre/Baffin Bay, Texas). Mar. Ecol. Prog. Ser. 2002, 237, 41–50. [Google Scholar] [CrossRef]
- Einsle, O.; Messerschmidt, A.; Stach, P.; Bourenkov, G.P.; Bartunik, H.D.; Huber, R. Structure of cytochrome c nitrite reductase. Nature 1999, 400, 476–480. [Google Scholar] [CrossRef]
- Mohan, S.B.; Schmid, M.; Jetten, M.; Cole, J. Detection and widespread distribution of the nrfA gene encoding nitrite reduction to ammonia, a short circuit in the biological nitrogen cycle that competes with denitrification. FEMS Microbiol. Ecol. 2004, 49, 433–443. [Google Scholar] [CrossRef]
- Giles, M.E.; Morley, N.J.; Baggs, E.M.; Daniell, T.J. Soil nitrate reducing processes–drivers, mechanisms for spatial variation, and significance for nitrous oxide production. Front. Microbiol. 2012, 3, 407. [Google Scholar] [CrossRef]
- Hou, L.; Zheng, Y.; Liu, M.; Li, X.F.; Lin, X.B.; Yin, G.Y.; Gao, J.; Deng, F.Y.; Chen, F.; Jiang, X.F. Anaerobic ammonium oxidation and its contribution to nitrogen removal in China’s coastal wetlands. Sci. Rep. 2015, 5, 15621. [Google Scholar] [CrossRef] [PubMed]
- Smith, C.J.; Dong, L.F.; Wilson, J.; Stott, A.; Osborn, A.M.; Nedwell, D. Seasonal variation in denitrification and dissimilatory nitrate reduction to ammonia process rates and corresponding key functional genes along an estuarine nitrate gradient. Front. Microbiol. 2015, 6, 542. [Google Scholar] [CrossRef] [PubMed]
- Song, B.; Lisa, J.A.; Tobias, C.R. Linking DNRA community structure and activity in a shallow lagoonal estuarine system. Front. Microbiol. 2014, 5, 460. [Google Scholar] [CrossRef] [PubMed]
- Kim, H.; Bae, H.S.; Reddy, K.R.; Ogram, A. Distributions, abundances and activities of microbes associated with the nitrogen cycle in riparian and stream sediments of a river tributary. Water Res. 2016, 106, 51–61. [Google Scholar] [CrossRef] [PubMed]
- Hu, C.Y. The density and diversity of zooplanktons in the succession of five lakes (in Chinese). Acta Hydrobiol. 1999, 23, 217–226. [Google Scholar] [CrossRef]
- Ghadouani, A.; Smith, R.E.H. Phytoplankton distribution in Lake Erie as assessed by a new in situ spectrofluorometric technique. J. Great Lakes Res. 2005, 31, 154–167. [Google Scholar] [CrossRef]
- Murphy, J.; Riley, J.P. A modified single solution method for determination of phosphate in natural waters. Anal. Chim. Acta 1962, 26, 31–36. [Google Scholar] [CrossRef]
- Beattie, D.M.; Golterman, H.L.; Vijverberg, J. Introduction to limnology of Friesian lakes. Hydrobiol 1978, 58, 49–64. [Google Scholar] [CrossRef]
- Greenberd, A.E.; Clesceri, L.S.; Eaton, A.D. Standard Methods for the Examination of Water and Wastewater, 22nd ed.; American Public Health Association: Washington, DC, USA, 2012. [Google Scholar] [CrossRef]
- Guan, Y.C.; Yang, H.; Zhao, L.; Li, X.R.; Li, J. Determination of sulfide in soil and sediment by methylene blue spectrophotometry. J. Anhui Agri. Sci. 2016, 44, 50–52. [Google Scholar]
- Welsh, A.; Chee-Sanford, J.C.; Connor, L.M.; Loffler, F.E.; Sanford, R.A. Refined nrfA phylogeny improves PCR-based nrfA gene detection. Appl. Environ. Microb. 2014, 80, 2110–2119. [Google Scholar] [CrossRef]
- Yang, S.Z.; Liebner, S.; Alawi, M.; Ebenhoh, O.; Wagner, D. Taxonomic database and cut-off value for processing mcrA gene 454 pyrosequencing data by MOTHUR. J. Microbiol. Meth. 2014, 103, 3–5. [Google Scholar] [CrossRef] [PubMed]
- Xu, J.J.; Liu, Z.; Bechtel, A.; Sachsenhofer, R.F.; Jia, J.; Meng, Q. Organic matter accumulation in the Upper Cretaceous Qingshankou and Nenjiang Formations, Songliao Basin (NE China): Implications from high-resolution geochemical analysis. Mar. Petrol. Geol. 2019, 102, 187–201. [Google Scholar] [CrossRef]
- Wang, W.D.; Liu, W.Y.; Wu, D.; Wang, X.X.; Zhu, G.B. Differentiation of nitrogen and microbial community in the littoral and limnetic sediments of a large shallow eutrophic lake (Chaohu Lake, China). J. Soil Sediment 2019, 19, 1005–1016. [Google Scholar] [CrossRef]
- Bu, C.N.; Wang, Y.; Ge, C.H.; Ahmad, H.A.; Gao, B.Y.; Ni, S.Q. Dissimilatory nitrate reduction to ammonium in the Yellow River Estuary: Rates, abundance, and community diversity. Sci. Rep. UK 2017, 7, 6830. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Song, C.L.; Zhou, Z.J.; Cao, X.Y.; Zhou, Y.Y. Coupling between Nitrification and Denitrification as well as Its Effect on Phosphorus Release in Sediments of Chinese Shallow Lakes. Water 2019, 11, 1809. [Google Scholar] [CrossRef]
- Simon, J. Enzymology and bioenergetics of respiratory nitrite ammonification. FEMS Microbiol. Rev. 2002, 26, 285–309. [Google Scholar] [CrossRef]
- Kartal, B.; Kuypers, M.M.M.; Lavik, G.; Schalk, J.; Op den Camp, H.J.M.; Jetten, M.S.M. Anammox bacteria disguised as denitrifiers: Nitrate reduction to dinitrogen gas via nitrite and ammonium. Environ. Microbiol. 2007, 9, 635–642. [Google Scholar] [CrossRef]
- Plummer, P.; Tobias, C.; Cady, D. Nitrogen reduction pathways in estuarine sediments: Influences of organic carbon and sulfide. J. Geophys Res. Biogeosci. 2015, 120, 1958–1972. [Google Scholar] [CrossRef]
- Robertson, W.D.; Russell, B.M.; Cherry, J.A. Attenuation of nitrate in aquitard sediments of southern Ontario. J. Hydrol. 1996, 180, 267–281. [Google Scholar] [CrossRef]
- Tesoriero, A.J.; Liebscher, H.; Cox, S.E. Mechanism and rate of denitrification in an agricultural watershed: Electron and mass balance along groundwater flow paths. Water Resour. Res. 2000, 36, 1545–1559. [Google Scholar] [CrossRef]
- Heffernan, J.B.; Albertin, A.R.; Fork, M.L.; Katz, B.G.; Cohen, M.J. Denitrification and inference of nitrogen sources in the karstic Floridan Aquifer. Biogeosciences 2012, 9, 1671–1690. [Google Scholar] [CrossRef]
- Eisenmann, E.; Beuerie, J.; Sulger, K.; Kroneck, P.M.H.; Schumacher, W. Lithotrophic growth of Sulforosprillum delianum with sulfide as electron donor coupled to respiratory reduction of nitrate to ammonia. Arch. Microbiol. 1995, 164, 180–185. [Google Scholar] [CrossRef]
- Burgin, A.J.; Hamilton, S.K. Have we overemphasized the role of denitrification in aquatic ecosystems? A review of nitrate removal pathways. Front. Ecol. Environ. 2007, 5, 89–96. [Google Scholar] [CrossRef]
- Palacin-Lizarbe, C.; Camarero, L.; Hallin, S.; Jones, C.M.; Caliz, J.; Casamayor, E.O.; Catalan, J. The DNRA-Denitrification Dichotomy Differentiates Nitrogen Transformation Pathways in Mountain Lake Benthic Habitats. Front. Microbiol. 2019, 10. [Google Scholar] [CrossRef] [PubMed]
- Li, X.F.; Sardans, J.; Hou, L.J.; Gao, D.Z.; Liu, M.; Penuelas, J. Dissimilatory Nitrate/Nitrite Reduction Processes in River Sediments Across Climatic Gradient: Influences of Biogeochemical Controls and Climatic Temperature Regime. J. Geophys. Res. Biogeosci. 2019, 124, 2305–2320. [Google Scholar] [CrossRef]
- Zhang, M.; Zhang, Y.C.; Yang, Z.; Wei, L.J.; Yang, W.B.; Chen, C. Spatial and seasonal shifts in bloom-forming cyanobacteria in Lake Chaohu: Patterns and driving factors. Phycol. Res. 2016, 64, 44–55. [Google Scholar] [CrossRef]
- Blomqvist, P.; Pettersson, A.; Hyenstrand, P. Ammonium-nitrogen: A key regulatory factor causing dominance of non-nitrogen-fixing cyanobacteria in aquatic systems. Arch. Hydrobiol. 1994, 132, 141–164. [Google Scholar] [CrossRef]
| □ | nrfA | TN | DTN | TP | DTP | DOP | SRP | Chl a | NH4+ | NO3− | SRPint | NH4+int | NO3−int | AVS | TOC | T | DO | pH |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| nrfA | 1 | |||||||||||||||||
| TN | 0.16 | 1 | ||||||||||||||||
| DTN | 0.20 | 0.92 ** | 1 | |||||||||||||||
| TP | −0.14 | 0.47 | 0.30 | 1 | ||||||||||||||
| DTP | 0.53 * | 0.51 * | 0.48 * | −0.16 | 1 | |||||||||||||
| DOP | 0.31 | 0.56 * | 0.51 * | −0.02 | 0.89 ** | 1 | ||||||||||||
| SRP | 0.57 * | 0.47 | 0.46 | −0.20 | 0.97 ** | 0.79 ** | 1 | |||||||||||
| Chl a | 0.66 ** | 0.66 ** | 0.67 ** | −0.04 | 0.79 ** | 0.73 ** | 0.77 ** | 1 | ||||||||||
| NH4+ | 0.29 | 0.62 ** | 0.58 * | −0.00 | 0.79 ** | 0.77 ** | 0.77 ** | 0.77 ** | 1 | |||||||||
| NO3− | 0.63 ** | 0.65 ** | 0.71 ** | −0.08 | 0.64 ** | 0.55 * | 0.63 ** | 0.81 ** | 0.51 * | 1 | ||||||||
| SRPint | 0.42 | 0.59 * | 0.65 ** | 0.08 | 0.62 ** | 0.49 * | 0.67 ** | 0.65 ** | 0.81 ** | 0.53 * | 1 | |||||||
| NH4+int | 0.63 ** | 0.32 | 0.35 | −0.03 | 0.58 * | 0.64 ** | 0.53 * | 0.64 ** | 0.45 | 0.47 | 0.24 | 1 | ||||||
| NO3−int | 0.65 ** | 0.46 | 0.43 | −0.04 | 0.83 ** | 0.82 ** | 0.79 ** | 0.79 ** | 0.70 ** | 0.64 ** | 0.65 ** | 0.62 ** | 1 | |||||
| AVS | 0.53 * | 0.27 | 0.12 | −0.09 | 0.50 * | 0.42 | 0.49 * | 0.65 ** | 0.52 * | 0.29 | 0.18 | 0.38 | 0.36 | 1 | ||||
| TOC | 0.57 * | 0.47 | 0.41 | 0.03 | 0.48 | 0.48 | 0.43 | 0.56 * | 0.62 ** | 0.34 | 0.37 | 0.43 | 0.26 | 0.59 * | 1 | |||
| T | 0.20 | 0.53 * | 0.69 ** | 0.09 | 0.31 | 0.44 | 0.26 | 0.52 * | 0.44 | 0.41 | 0.49 * | 0.42 | 0.42 | −0.04 | 0.35 | 1 | ||
| DO | 0.13 | 0.26 | 0.38 | 0.29 | 0.11 | 0.17 | 0.12 | 0.30 | 0.45 | 0.11 | 0.60 * | 0.33 | 0.37 | −0.01 | 0.13 | 0.51 * | 1 | |
| pH | 0.17 | 0.14 | 0.27 | 0.33 | 0.06 | 0.12 | 0.06 | 0.18 | 0.30 | 0.01 | 0.41 | 0.43 | 0.31 | −0.13 | 0.10 | 0.52 * | 0.91 ** | 1 |
| □ | nrfA | TN | DTN | TP | DTP | DOP | SRP | Chl a | NH4+ | NO3− | SRPint | NH4+int | NO3−int | AVS | TOC | T | DO | pH |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| nrfA | 1 | |||||||||||||||||
| TN | 0.04 | 1 | ||||||||||||||||
| DTN | 0.05 | 0.83 ** | 1 | |||||||||||||||
| TP | 0.22 | 0.61 * | 0.67 ** | 1 | ||||||||||||||
| DTP | 0.29 | 0.61 * | 0.63 * | 0.89 ** | 1 | |||||||||||||
| DOP | 0.37 | 0.35 | 0.19 | 0.44 | 0.61 * | 1 | ||||||||||||
| SRP | 0.19 | 0.58 * | 0.68 ** | 0.89 ** | 0.95 ** | 0.35 | 1 | |||||||||||
| Chl a | 0.20 | 0.71 ** | 0.72 ** | 0.69 ** | 0.68 ** | 0.09 | 0.76 ** | 1 | ||||||||||
| NH4+ | −0.34 | 0.12 | 0.03 | −0.24 | −0.52 | −0.62 * | −0.42 | 0.05 | 1 | |||||||||
| NO3− | 0.28 | 0.65 * | 0.65 * | 0.93 ** | 0.92 ** | 0.58 * | 0.87 ** | 0.69 ** | −0.34 | 1 | ||||||||
| SRPint | 0.19 | 0.59 * | 0.69 ** | 0.88 ** | 0.94 ** | 0.34 | 0.99 ** | 0.76 ** | −0.41 | 0.86 ** | 1 | |||||||
| NH4+int | −0.35 | 0.54 * | 0.38 | 0.31 | 0.01 | 0.02 | −0.01 | 0.22 | 0.62 * | 0.24 | −0.01 | 1 | ||||||
| NO3−int | 0.11 | −0.32 | −0.45 | −0.21 | −0.08 | 0.26 | −0.20 | −0.41 | −0.40 | −0.13 | −0.21 | −0.38 | 1 | |||||
| AVS | −0.27 | 0.09 | 0.07 | 0.16 | −0.07 | 0.19 | −0.12 | −0.11 | 0.12 | 0.00 | −0.14 | 0.62 * | −0.25 | 1 | ||||
| TOC | −0.35 | 0.57 * | 0.48 | 0.45 | 0.14 | −0.02 | 0.19 | 0.34 | 0.51 | 0.35 | 0.19 | 0.93 ** | −0.54 * | 0.63 * | 1 | |||
| T | −0.05 | −0.47 | −0.25 | −0.23 | −0.01 | −0.15 | 0.06 | −0.10 | −0.53 * | −0.18 | 0.06 | −0.75 ** | 0.49 | −0.46 | −0.69 ** | 1 | ||
| DO | −0.40 | −0.01 | −0.04 | −0.21 | −0.21 | −0.26 | −0.12 | 0.19 | 0.06 | −0.23 | −0.11 | 0.03 | 0.00 | 0.23 | 0.06 | 0.38 | 1 | |
| pH | −0.23 | −0.37 | −0.49 | −0.40 | −0.26 | −0.24 | −0.23 | −0.08 | −0.13 | −0.41 | −0.22 | −0.42 | 0.44 | −0.18 | −0.45 | 0.65 * | 0.74 ** | 1 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, X.; Song, C.; Zhou, Z.; Xiao, J.; Wang, S.; Yang, L.; Cao, X.; Zhou, Y. Comparison of Community and Function of Dissimilatory Nitrate Reduction to Ammonium (DNRA) Bacteria in Chinese Shallow Lakes with Different Eutrophication Degrees. Water 2020, 12, 174. https://doi.org/10.3390/w12010174
Li X, Song C, Zhou Z, Xiao J, Wang S, Yang L, Cao X, Zhou Y. Comparison of Community and Function of Dissimilatory Nitrate Reduction to Ammonium (DNRA) Bacteria in Chinese Shallow Lakes with Different Eutrophication Degrees. Water. 2020; 12(1):174. https://doi.org/10.3390/w12010174
Chicago/Turabian StyleLi, Xiaowen, Chunlei Song, Zijun Zhou, Jian Xiao, Siyang Wang, Liu Yang, Xiuyun Cao, and Yiyong Zhou. 2020. "Comparison of Community and Function of Dissimilatory Nitrate Reduction to Ammonium (DNRA) Bacteria in Chinese Shallow Lakes with Different Eutrophication Degrees" Water 12, no. 1: 174. https://doi.org/10.3390/w12010174
APA StyleLi, X., Song, C., Zhou, Z., Xiao, J., Wang, S., Yang, L., Cao, X., & Zhou, Y. (2020). Comparison of Community and Function of Dissimilatory Nitrate Reduction to Ammonium (DNRA) Bacteria in Chinese Shallow Lakes with Different Eutrophication Degrees. Water, 12(1), 174. https://doi.org/10.3390/w12010174




