The Spatial Distribution of the Microbial Community in a Contaminated Aquitard below an Industrial Zone
Abstract
1. Introduction
2. Materials and Methods
2.1. Material
2.2. Study Area and Groundwater Sampling
2.3. DNA Extraction, PCR Amplification and Sequencing
2.4. Data Analysis of Sequencing Results
2.5. Microcosm Experiments
2.6. Analytical Methods
3. Results
3.1. Molecular Analysis of the Groundwater Indigenous Bacterial Communities
3.1.1. Sequencing and Taxa Identification
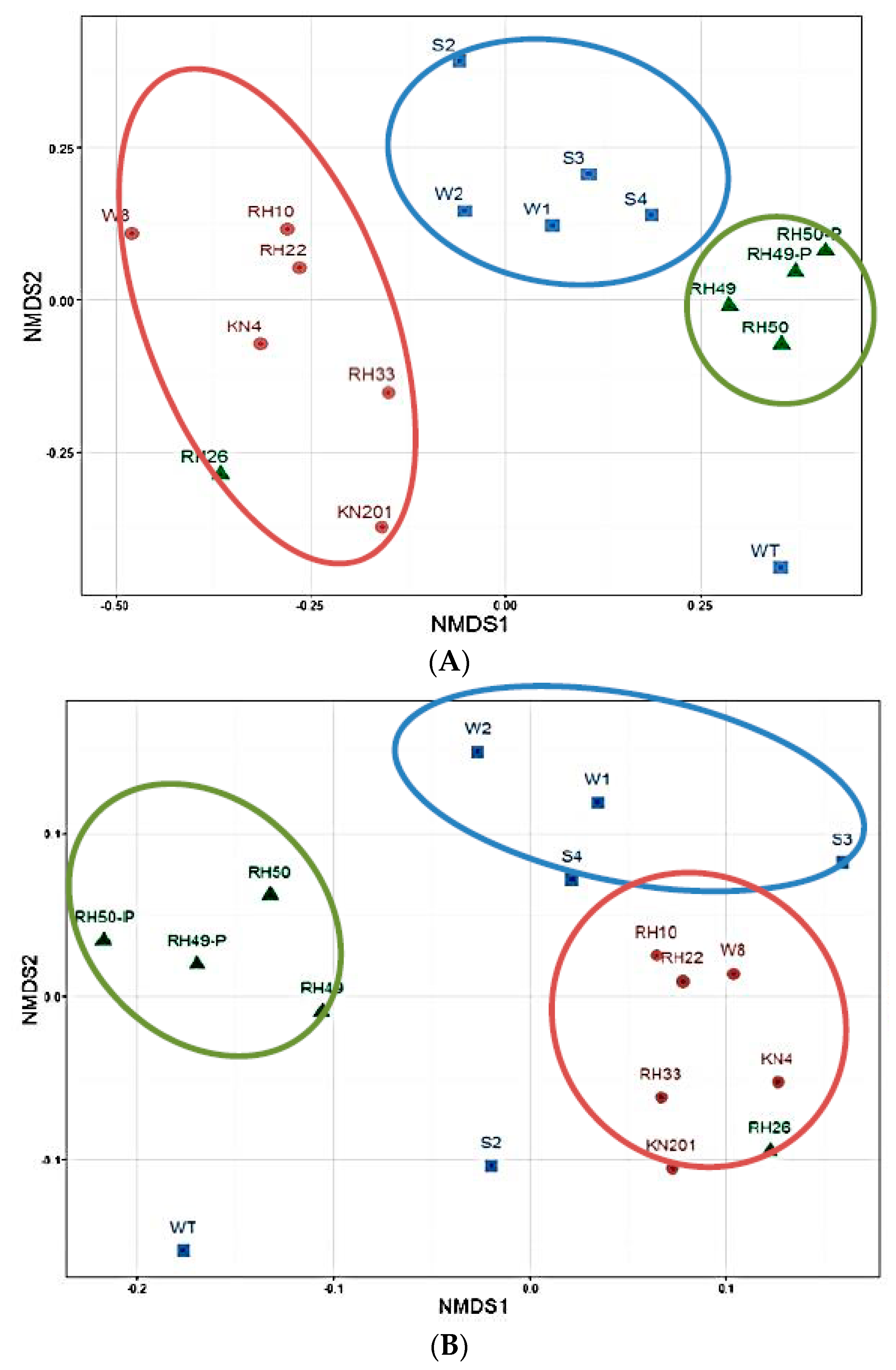
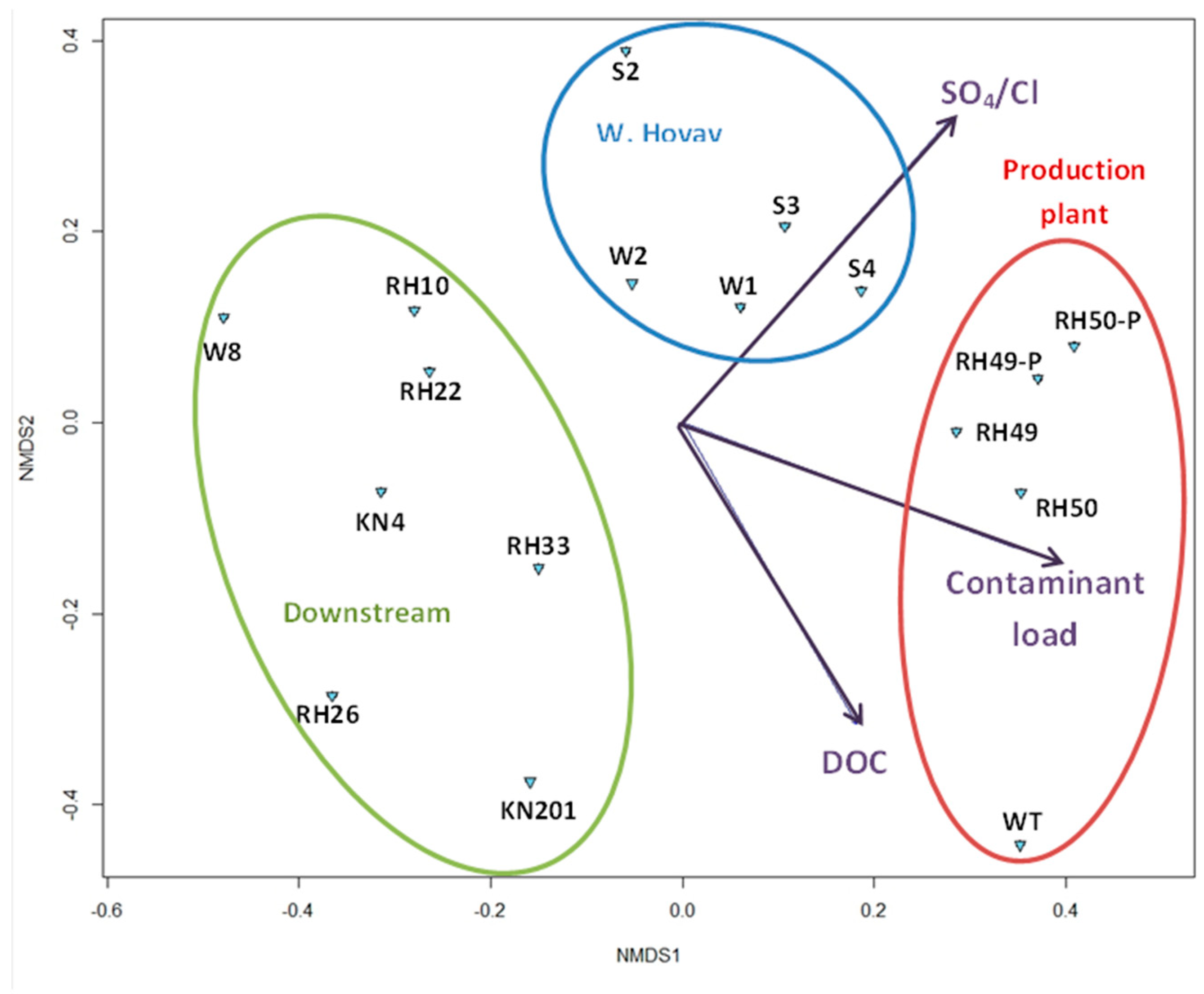
3.1.2. Overall Bacterial Community Structure and Relationship with Environmental Variables
3.1.3. Variations in Microbial Community Diversity
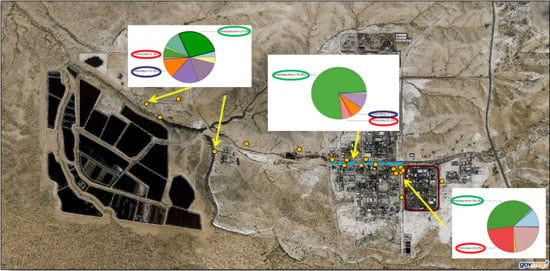
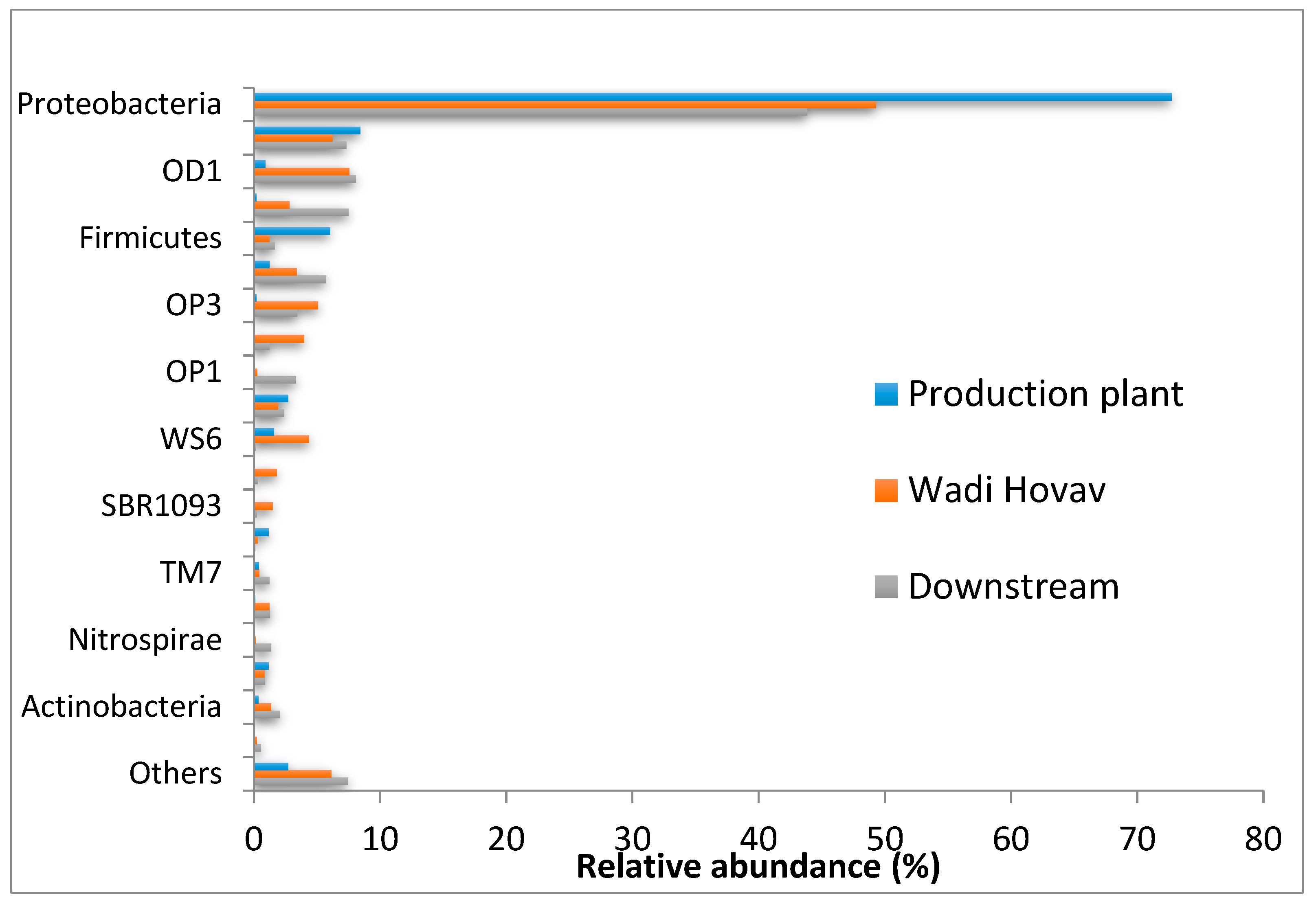
3.1.4. Microbial Community Composition
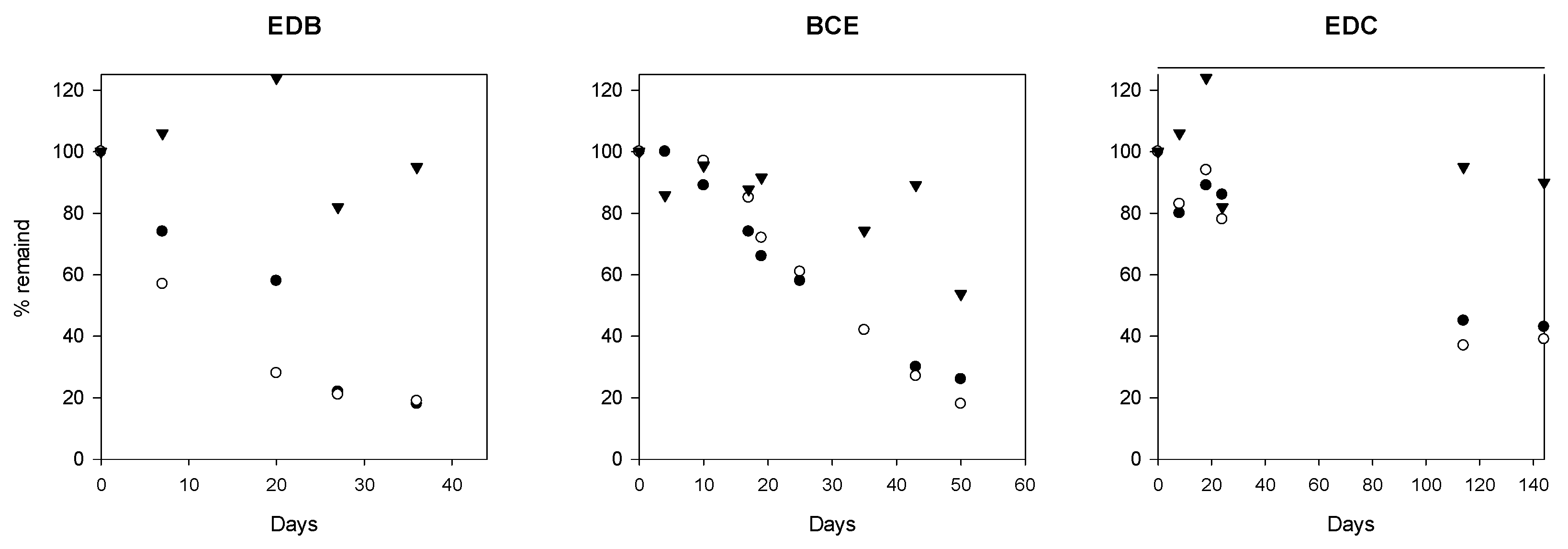
3.2. Dehalogenation Potential of the Groundwater Microbial Community
4. Discussion
4.1. Site Hydrology
4.2. Spatial Distribution of Microbial Community Affected by Environmental Parameters
4.3. Possible Metabolic Features in the Microbial Communities Based on Spatial Location
4.4. Pollution Extent Affects Community Diversity
4.5. Active Dehalogenation Potential
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Griebler, C.; Avramov, M. Groundwater ecosystem services: A review Christian. Freshw. Sci. 2015, 34, 355–367. [Google Scholar] [CrossRef]
- Rodríguez-Delgado, M.; Orona-Navar, C.; García-Morales, R.; Hernandez-Luna, C.; Parra, R.; Mahlknecht, J.; Ornelas-Soto, N. Biotransformation kinetics of pharmaceutical and industrial micropollutants in groundwaters by a laccase cocktail from Pycnoporus sanguineus CS43 fungi. Int. Biodeterior. Biodegrad. 2016, 108, 34–41. [Google Scholar] [CrossRef]
- An, D.; Xi, B.; Wang, Y.; Xu, D.; Tang, J.; Dong, L.; Ren, J.; Pang, C. A sustainability assessment methodology for prioritizing the technologies of groundwater contamination remediation. J. Clean. Prod. 2016, 112, 4647–4656. [Google Scholar] [CrossRef]
- Ezra, S.; Feinstein, S.; Yakirevich, A.; Adar, E.; Bilkis, I. Retardation of organo-bromides in a fractured chalk aquitard. J. Contam. Hydrol. 2006, 86, 195–214. [Google Scholar] [CrossRef]
- Postigo, C.; Barceló, D. Synthetic organic compounds and their transformation products in groundwater: Occurrence, fate and mitigation. Sci. Total Environ. 2015, 503, 32–47. [Google Scholar] [CrossRef]
- Montes-Grajales, D.; Fennix-Agudelo, M.; Miranda-Castro, W. Occurrence of personal care products as emerging chemicals of concern in water resources: A review. Sci. Total Environ. 2017, 595, 601–614. [Google Scholar] [CrossRef]
- Lapworth, D.J.; Baran, N.; Stuart, M.E.; Ward, R.S. Emerging organic contaminants in groundwater: A review of sources, fate and occurrence. Environ. Pollut. 2012, 163, 287–303. [Google Scholar] [CrossRef]
- Van Liedekerke, M.; Prokop, G.; Rabl-Berger, S.; Kibblewhite, M.; Louwagie, G. Progress in the Management of Contaminated Sites in Europe; Joint Research Centre: Ispra, Italy, 2014. [Google Scholar]
- Haggblom, M.M.; Bossert, I.D. Halogenated organic compounds—A global perspective. In Dehalogenation: Microbial Processes and Environmental Applications; Haggblom, M., Bossert, I., Eds.; Kluwer Academic Publishers: New York, NY, USA, Country, 2003. [Google Scholar]
- Birnbaum, L.S.; Staskal, D.F. Brominated flame retardants: Cause for concern? Environ. Health Perspect. 2004, 112, 9–17. [Google Scholar] [CrossRef]
- Alexander, M. Biodegradation and Bioremediation; Academic Press: San Diego, CA, USA, 1994. [Google Scholar]
- Poursat, B.A.J.; van Spanning, R.J.M.; de Voogt, P.; Parsons, J.R. Implications of microbial adaptation for the assessment of environmental persistence of chemicals. Crit. Rev. Environ. Sci. Technol. 2019, 49, 2220–2255. [Google Scholar] [CrossRef]
- Flynn, T.M.; Sanford, R.A.; Ryu, H.; Bethke, C.M.; Levine, A.D.; Ashbolt, N.J.; Santo Domingo, J.W. Functional microbial diversity explains groundwater chemistry in a pristine aquifer. BMC Microbiol. 2013, 13, 146. [Google Scholar] [CrossRef]
- Dojka, M.; Hugenholtz, P.; Haack, S.; Pace, N. Microbial diversity in a hydrocarbon-and chlorinated-solvent-contaminated aquifer undergoing intrinsic bioremediation. Appl. Environ. Microbiol. 1998, 64, 3869–3877. [Google Scholar] [PubMed]
- Yagi, J.M.; Neuhauser, E.F.; Ripp, J.A.; Mauro, D.M.; Madsen, E.L. Subsurface ecosystem resilience: Long-term attenuation of subsurface contaminants supports a dynamic microbial community. ISME J. 2010, 4, 131–143. [Google Scholar] [CrossRef] [PubMed]
- Meckenstock, R.U.; Elsner, M.; Griebler, C.; Lueders, T.; Stumpp, C.; Aamand, J.; Agathos, S.N.; Albrechtsen, H.J.; Bastiaens, L.; Bjerg, P.L.; et al. Biodegradation: Updating the concepts of control for microbial cleanup in contaminated aquifers. Environ. Sci. Technol. 2015, 49, 7073–7081. [Google Scholar] [CrossRef] [PubMed]
- Tarnawski, S.-E.; Rossi, P.; Brennerova, M.V.; Stavelova, M.; Holliger, C. Validation of an integrative methodology to assess and monitor reductive dechlorination of chlorinated ethenes in contaminated aquifers. Front. Environ. Sci. 2016, 4, 7. [Google Scholar] [CrossRef]
- Révész, K.M.; Lollar, B.S.; Kirshtein, J.D.; Tiedeman, C.R.; Imbrigiotta, T.E.; Goode, D.J.; Shapiro, A.M.; Voytek, M.A.; Lacombe, P.J.; Busenberg, E. Integration of stable carbon isotope, microbial community, dissolved hydrogen gas, and 2HH2O tracer data to assess bioaugmentation for chlorinated ethene degradation in fractured rocks. J. Contam. Hydrol. 2014, 156, 62–77. [Google Scholar] [CrossRef] [PubMed]
- Bahat, D. Early single-layer and late multi-layer joints in the lower Eocene chalk near Beer Sheva, Israel. Ann. Tect. 1988, 2, 3–11. [Google Scholar]
- Dahan, O.; Nativ, R.; Adar, E.M.; Berkowitz, B.; Weisbrod, N. On fracture structure and preferential flow in unsaturated chalk. Groundwater 2000, 3, 444–451. [Google Scholar] [CrossRef]
- Nativ, R.; Adar, E.; Assaf, L.; Nygaard, E. Characterization of the hydraulic properties of fractures in chalk. Groundwater 2003, 41, 532–543. [Google Scholar] [CrossRef]
- Nativ, R.; Adar, E.; Dahan, O.; Nissim, I. Water salinization in arid regions-observations from the Negev desert, Israel. J. Hydrol. 1997, 196, 271–296. [Google Scholar] [CrossRef]
- Adar, E.; Kaplan, A.; Gilerman, L. Groundwater Quality Monitoring in the Neot Hovav Industrial Complex; Monitoring report for the years 2012–2013, 2013–2014; Neot Hovav Industrial Council: Neot Hovav, Israel, 2015. (In Hebrew) [Google Scholar]
- Maamar, S.B.; Aquilina, L.; Quaiser, A.; Pauwels, H.; Michon-Coudouel, S.; Vergnaud-Ayraud, V.; Labasque, T.; Roques, C.; Abbott, B.W.; Dufresne, A. Groundwater isolation governs chemistry and microbial community structure along hydrologic flowpaths. Front. Microbiol. 2015, 6, 1–13. [Google Scholar]
- Weiss, M.; Rubin, Y.; Adar, E.; Nativ, R. Fracture and bedding plane control on groundwater flow in a chalk aquitard. Hydrogeol. J. 2006, 14, 1081–1093. [Google Scholar] [CrossRef]
- Arnon, S.; Ronen, Z.; Yakirevich, A.; Adar, E. Evaluation of soil flushing potential for clean-up of desert soil contaminated by industrial wastewater. Chemosphere 2006, 62, 17–25. [Google Scholar] [CrossRef] [PubMed]
- Nativ, R.; Nissim, I. Characterization of a desert aquitard-hydrologic and hydrochemical considerations. Groundwater 1992, 30, 598–606. [Google Scholar] [CrossRef]
- Takahashi, S.; Tomita, J.; Nishioka, K.; Hisada, T.; Nishijima, M. Development of a prokaryotic universal primer for simultaneous analysis of Bacteria and Archaea using Next-Generation sequencing. PLoS ONE 2014, 9, e105592. [Google Scholar] [CrossRef] [PubMed]
- Quast, C.; Pruesse, E.; Yilmaz, P.; Gerken, J.; Schweer, T.; Yarza, P.; Peplies, J.; Glockner, F.O. The SILVA ribosomal RNA gene database project: Improved data processing and web-based tools. Nucleic Acids Res. 2013, 41, 590–596. [Google Scholar] [CrossRef] [PubMed]
- Staley, C.; Unno, T.; Gould, T.J.; Jarvis, B.; Phillips, J.; Cotner, J.B.; Sadowsky, M.J. Application of Illumina next-generation sequencing to characterize the bacterial community of the Upper Mississippi River. J. Appl. Microbiol. 2013, 115, 1147–1158. [Google Scholar] [CrossRef]
- Team, R. RStudio: Integrated Development Environment for R; RStudio: Boston, MA, USA, 2012; Available online: https://rstudio.com/products/rstudio/ (accessed on 1 July 2016).
- McMurdie, P.J.; Holmes, S. phyloseq: An R package for reproducible interactive analysis and graphics of microbiome census data. PLoS ONE 2013, 8, e61217. [Google Scholar] [CrossRef]
- Oksanen, J.; Blanchet, F.G.; Kindt, R.; Legendre, P.; Minchin, P.; O’Hara, R.; Simpson, G.; Solymos, P.; Stevens, M.; Wagner, H. Vegan: Community Ecology Package, R Package Version 2.0-10; 2013. Available online: https://cran.r-project.org/web/packages/vegan/index.html (accessed on 1 July 2016).
- Arndt, D.; Xia, J.; Liu, Y.; Zhou, Y.; Guo, A.C.; Cruz, J.A.; Sinelnikov, I.; Budwill, K.; Nesbø, C.L.; Wishart, D.S. METAGENassist: A comprehensive web server for comparative metagenomics. Nucleic Acids Res. 2012, 40, 88–95. [Google Scholar] [CrossRef]
- Balaban, N.; Bernstein, A.; Gelman, F.; Ronen, Z. Microbial degradation of the brominated flame retardant TBNPA by groundwater bacteria: Laboratory and field study. Chemosphere 2016, 156, 367–373. [Google Scholar] [CrossRef]
- Adar, E. Groundwater Quality Monitoring in the Neot Hovav Industrial Complex; Neot Hovav Industrial Council: Neot Hovav, Israel, 2014. (In Hebrew) [Google Scholar]
- Weber, K.P.; Gehder, M.; Legge, R.L. Assessment of changes in the microbial community of constructed wetland mesocosms in response to acid mine drainage exposure. Water Res. 2008, 42, 180–188. [Google Scholar] [CrossRef]
- Singh, B.K.; Quince, C.; Macdonald, C.A.; Khachane, A.; Thomas, N.; Al-Soud, W.A.; Soren, J.S.; He, Z.; White, D.; Sinclair, A.; et al. Loss of microbial diversity in soils is coincident with reductions in some specialized functions. Environ. Microbiol. 2014, 16, 2408–2420. [Google Scholar] [CrossRef] [PubMed]
- Fan, M.; Lin, Y.; Huo, H.; Liu, Y.; Zhao, L.; Wang, E.; Chen, W.; Wei, G. Microbial communities in riparian soils of a settling pond for mine drainage treatment. Water Res. 2016, 96, 198–207. [Google Scholar] [CrossRef] [PubMed]
- Yankelzon, I.; Ronen, Z.; Gelman, F. Multi-elemental (δ13C, δ81Br and δ37Cl) isotope effects in biotic and abiotic degradation of 1-bromo-2-chloro-ethane. In Proceedings of the Isotopes 2019, The Cross-Disciplinary Conference on Stable Isotope Sciences, Raitenhaslach, Germany, 7–12 July 2019; pp. 114–115. [Google Scholar]
- Adar, E.; Nativ, R. Isotopes as tracers in a contaminated fractured chalk aquitard. J. Contam. Hydrol. 2003, 65, 19–39. [Google Scholar] [CrossRef]
- Nativ, R.; Adar, E.M.; Becker, A. Designing a monitoring network for contaminated ground water in fractured chalk. Groundwater 1999, 37, 38–47. [Google Scholar] [CrossRef]
- Nativ, R.; Adar, E. Evaluation of the Extent of Groundwater Contamination in the Ramat Hovav Industrial Park Area; Concluding Report 1995–1997; Neot Hovav Industrial Council: Neot Hovav, Israel, 1997. [Google Scholar]
- Brock, T.D.; Gustafson, J. Ferric Iron Reduction by Sulfur- and Iron-Oxidizing Bacteria. Appl. Environ. Microbiol. 1976, 32, 567–571. [Google Scholar]
- Kodama, Y.; Watanabe, K. Isolation and characterization of a sulfur-oxidizing chemolithotroph growing on crude oil under anaerobic conditions isolation and characterization of a sulfur-oxidizing chemolithotroph growing on crude oil under anaerobic conditions. Society 2003, 69, 107–112. [Google Scholar]
- Odum, E.P.; Finn, J.T.; Franz, E.H. Pertubation theory and the subsidy-stress gradient. Bioscience 1979, 29, 349–352. [Google Scholar] [CrossRef]
- He, Z.; Zhang, P.; Wu, L.; Rocha, A.M.; Tu, Q.; Shi, Z.; Wu, B.; Qin, Y.; Wang, J.; Yan, Q.; et al. Microbial functional gene diversity predicts groundwater contamination and ecosystem functioning. MBio 2018, 9, e02435-17. [Google Scholar] [CrossRef]
- Ning, Z.; Zhang, M.; He, Z.; Cai, P.; Guo, C.; Wang, P. Spatial pattern of bacterial community diversity formed in different groundwater field corresponding to electron donors and acceptors distributions at a petroleum-contaminated site. Water 2018, 10, 842. [Google Scholar] [CrossRef]
- Atashgahi, S.; Lu, S.; Smidt, H. Overview of known organohalide respiring bacteria—Phylogenetic diversity and environmental distribution. In Organohalide Respiring Bacteria; Adrian, L., Loffler, F., Eds.; Springer: Berlin/Heidelberg, Germany, 2016. [Google Scholar]
- Nijenhuis, I.; Nikolausz, M.; Köth, A.; Felföldi, T.; Weiss, H.; Drangmeister, J.; Grossmann, J.; Kästner, M.; Richnow, H.H. Assessment of the natural attenuation of chlorinated ethenes in an anaerobic contaminated aquifer in the Bitterfeld/Wolfen area using stable isotope techniques, microcosm studies and molecular biomarkers. Chemosphere 2007, 67, 300–311. [Google Scholar] [CrossRef]
- Lovley, D.R.; Ueki, T.; Zhang, T.; Malvankar, N.S.; Shrestha, P.M.; Flanagan, K.A.; Aklujkar, M.; Butler, J.E.; Giloteaux, L.; Rotaru, A.-E.; et al. Geobacter: The Microbe electric’s physiology, ecology, and practical applications. In Advances in Microbial Physiology; Academic Press: Cambridge, MA, USA, 2011; Volume 59, ISBN 9780123876614. [Google Scholar]
- Iasur-Kruh, L.; Ronen, Z.; Arbeli, Z.; Nejidat, A. Characterization of an enrichment culture debrominating tetrabromobisphenol A and optimization of its activity under anaerobic conditions. J. Appl. Microbiol. 2010, 109, 707–715. [Google Scholar] [CrossRef] [PubMed]
- Ben-Dov, E.; Brenner, A.; Kushmaro, A. Quantification of sulfate-reducing bacteria in industrial wastewater, by real-time polymerase chain reaction (PCR) using dsrA and apsA genes. Microb. Ecol. 2007, 54, 439–451. [Google Scholar] [CrossRef] [PubMed]
- Yang, C.; Kublik, A.; Weidauer, C.; Seiwert, B.; Adrian, L. reductive dehalogenation of oligocyclic phenolic bromoaromatics by Dehalococcoides mccartyi Strain CBDB1. Environ. Sci. Technol. 2015, 49, 8497–8505. [Google Scholar] [CrossRef] [PubMed]
- Futagami, T.; Morono, Y.; Terada, T.; Kaksonen, A.H.; Inagaki, F. Dehalogenation activities and distribution of reductive dehalogenase homologous genes in marine subsurface sediments. Appl. Environ. Microbiol. 2009, 75, 6905–6909. [Google Scholar] [CrossRef]
- Kuppusamy, S.; Palanisami, T.; Megharaj, M.; Venkateswarlu, K.; Naisu, R. Ex-situ remediation technologies for environmental pollutants: A critical perspective. In Reviews of Environmental Contamination and Toxicology; Voogt, P., Ed.; Springer: Cham, Switzerland, 2015. [Google Scholar]
- Rhee, S.K.; Fennell, D.E.; Häggblom, M.M.; Kerkhof, L.J. Detection by PCR of reductive dehalogenase motifs in a sulfidogenic 2-bromophenol-degrading consortium enriched from estuarine sediment. FEMS Microbiol. Ecol. 2003, 43, 317–324. [Google Scholar] [CrossRef]
- Villemur, R.; Lanthier, M.; Beaudet, R.; Lépine, F. The Desulfitobacterium genus. FEMS Microbiol. Rev. 2006, 30, 706–733. [Google Scholar] [CrossRef]
- Ronen, Z.; Abeliovich, A. Anaerobic-aerobic process for microbial degradation of tetrabromobisphenol A. Appl. Environ. Microbiol. 2000, 66, 2372–2377. [Google Scholar] [CrossRef]
- Luo, J.; Kurt, Z.; Hou, D.; Spain, J.C. Modeling aerobic biodegradation in the capillary fringe. Environ. Sci. Technol. 2015, 49, 1501–1510. [Google Scholar] [CrossRef]
- Maphosa, F.; Van Passel, M.W.J.; De Vos, W.M.; Smidt, H. Metagenome analysis reveals yet unexplored reductive dechlorinating potential of Dehalobacter sp. E1 growing in co-culture with Sedimentibacter sp. Environ. Microbiol. Rep. 2012, 4, 604–616. [Google Scholar]
- Muyzer, G.; Stams, A.J.M. The ecology and biotechnology of sulphate-reducing bacteria. Nat. Rev. Microbiol. 2008, 6, 441–454. [Google Scholar] [CrossRef]
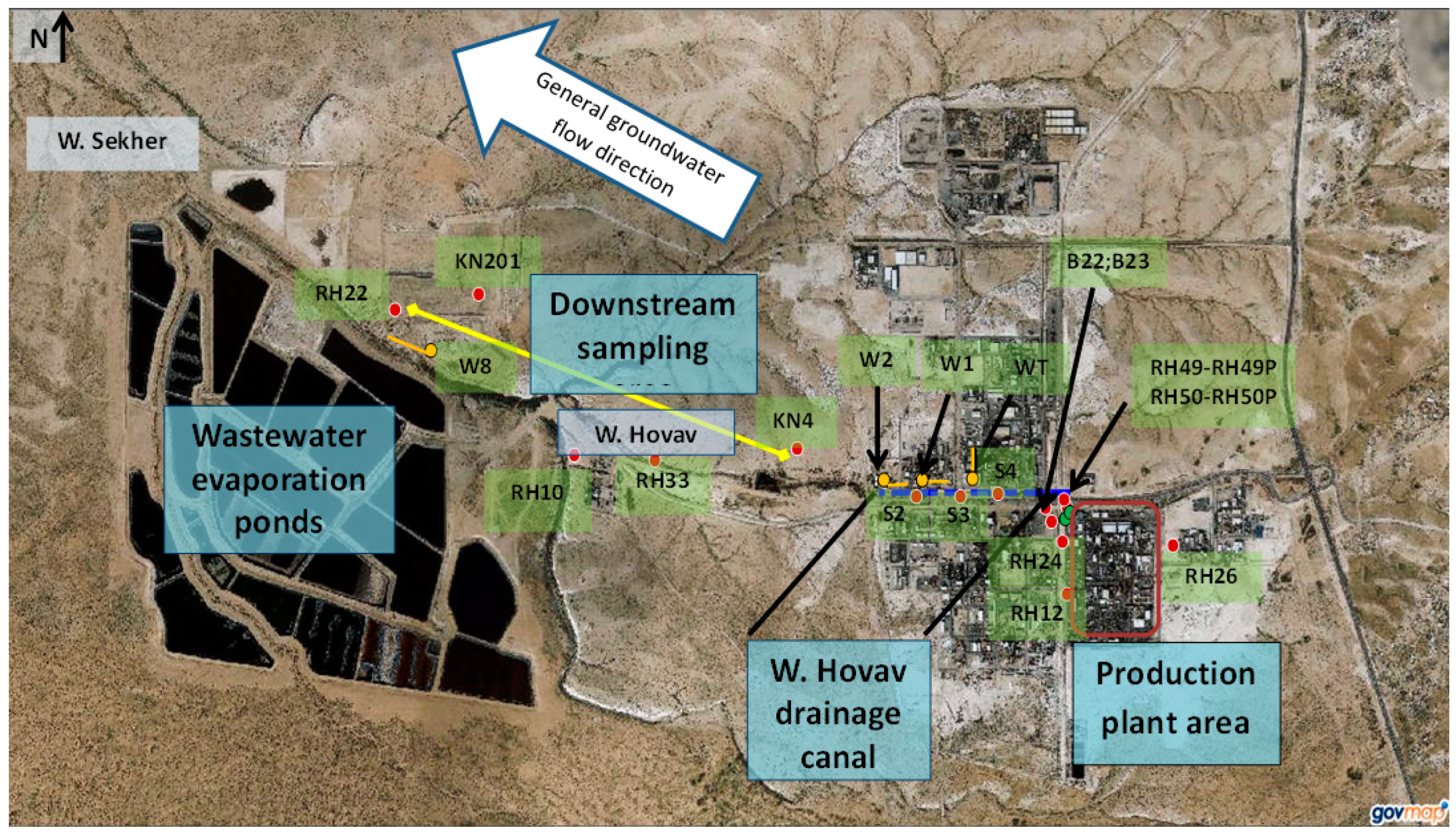
| Borehole | Location | Comments |
|---|---|---|
| RH49, RH49P, RH50, and RH50P | West to the production plant | Slanted borehole drilled through vertical fractures that lie beneath the BOC’s production plant |
| RH26 | 30 m’ east to the production plant | |
| W1, W2, S2, S3, and S4 | Along Wadi Hovav | Drains shallow groundwater within saturated fractures intersected by the Wadi Hovav Trench (an east-west trench) |
| WT | Drains a north-south trench intersecting with Wadi Hovav | |
| KN4 | Downstream of the production plant | Located at the downstream edge of the industrial area, on the northern bank of Wadi Hovav |
| RH33 | South to Wadi Hovav | |
| RH10 | Upstream to the Wadi Hovav-Wadi Sekher confluence | |
| KN201 | North to Wadi Sekher | |
| W8 | In Wadi Sekher, north to the wastewater evaporation ponds. Drains a trench that drains polluted groundwater of Wadi Secker, including leaks from the wastewater ponds | |
| RH22 | Located west to W8, on the northern bank of Wadi Sekher |
| Genera | Name | Abundance (%) |
|---|---|---|
| phylum | Proteobacteria | 54.2 |
| Bacteriodetes | 7.3 | |
| OD1 | 5.8 | |
| Chloroflexi | 3.6 | |
| OP3 | 3.0 | |
| Firmicutes | 2.8 | |
| unclassified | 0.3 | |
| Class | Gammaproteobacteria | 19.3 |
| Epsilonproteobacteria | 10.4 | |
| Deltaproteobacteriav | 8.0 | |
| Alphaproteobacteria | 11.5 | |
| unclassified | 2.4 | |
| Family | Helicobacteraceae | 7.0 |
| Pseudomonadaceae | 4.6 | |
| Desulfobacteraceae | 3.8 | |
| Campylobacteraceae | 3.5 | |
| Piscirickettsiaceae | 2.8 | |
| Flavobacteriaceae | 2.7 | |
| Rhodobacteraceae | 2.5 | |
| Rhodospirillaceae | 2.5 | |
| unclassified | 36.3 |
| Contaminant Load | DOC | ||||
|---|---|---|---|---|---|
| Location | Boreholes in group | mean | Range (mg/L) | mean | Range (mg/L) |
| Production plant | RH49P; RH50P; WT | 203.6 | 103.7–297.4 | 233.9 | 25–821 |
| Wadi Hovav crossing the industrial area | W1; W2; S2; S3; S4 | 8.2 | 1.5–13.9 | 41.2 | 29–49 |
| Downstream to the industrial area | RH26; KN4; RH10; RH22; RH33; W8; KN201 | 0.2 | 0.0007–0.4 | 26.9 | 4.9–48 |
| Location | Boreholes in Group | Observed | Chao1 | Shannon | ||||
|---|---|---|---|---|---|---|---|---|
| N | Mean | Range | Mean | Range | Mean | Range | ||
| production plant | RH49; RH49P; RH50; RH50P; WT | 5 | 4247 | 3317–4678 | 5505 | 4016–6228 | 6.33 | 6.13–6.51 |
| Wadi Hovav crossing the industrial area | W1; W2; S2; S3; S4 | 5 | 4781 | 2769–6281 | 6952 | 3838–8877 | 6.17 | 5.38–6.71 |
| Downstream to the industrial area | RH26; KN4; RH10; RH22; RH33; W8; KN201 | 7 | 5115 | 3392–7241 | 7430 | 4621–9962 | 6.27 | 5.60–7.12 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Balaban, N.; Yankelzon, I.; Adar, E.; Gelman, F.; Ronen, Z.; Bernstein, A. The Spatial Distribution of the Microbial Community in a Contaminated Aquitard below an Industrial Zone. Water 2019, 11, 2128. https://doi.org/10.3390/w11102128
Balaban N, Yankelzon I, Adar E, Gelman F, Ronen Z, Bernstein A. The Spatial Distribution of the Microbial Community in a Contaminated Aquitard below an Industrial Zone. Water. 2019; 11(10):2128. https://doi.org/10.3390/w11102128
Chicago/Turabian StyleBalaban, Noa, Irina Yankelzon, Eilon Adar, Faina Gelman, Zeev Ronen, and Anat Bernstein. 2019. "The Spatial Distribution of the Microbial Community in a Contaminated Aquitard below an Industrial Zone" Water 11, no. 10: 2128. https://doi.org/10.3390/w11102128
APA StyleBalaban, N., Yankelzon, I., Adar, E., Gelman, F., Ronen, Z., & Bernstein, A. (2019). The Spatial Distribution of the Microbial Community in a Contaminated Aquitard below an Industrial Zone. Water, 11(10), 2128. https://doi.org/10.3390/w11102128