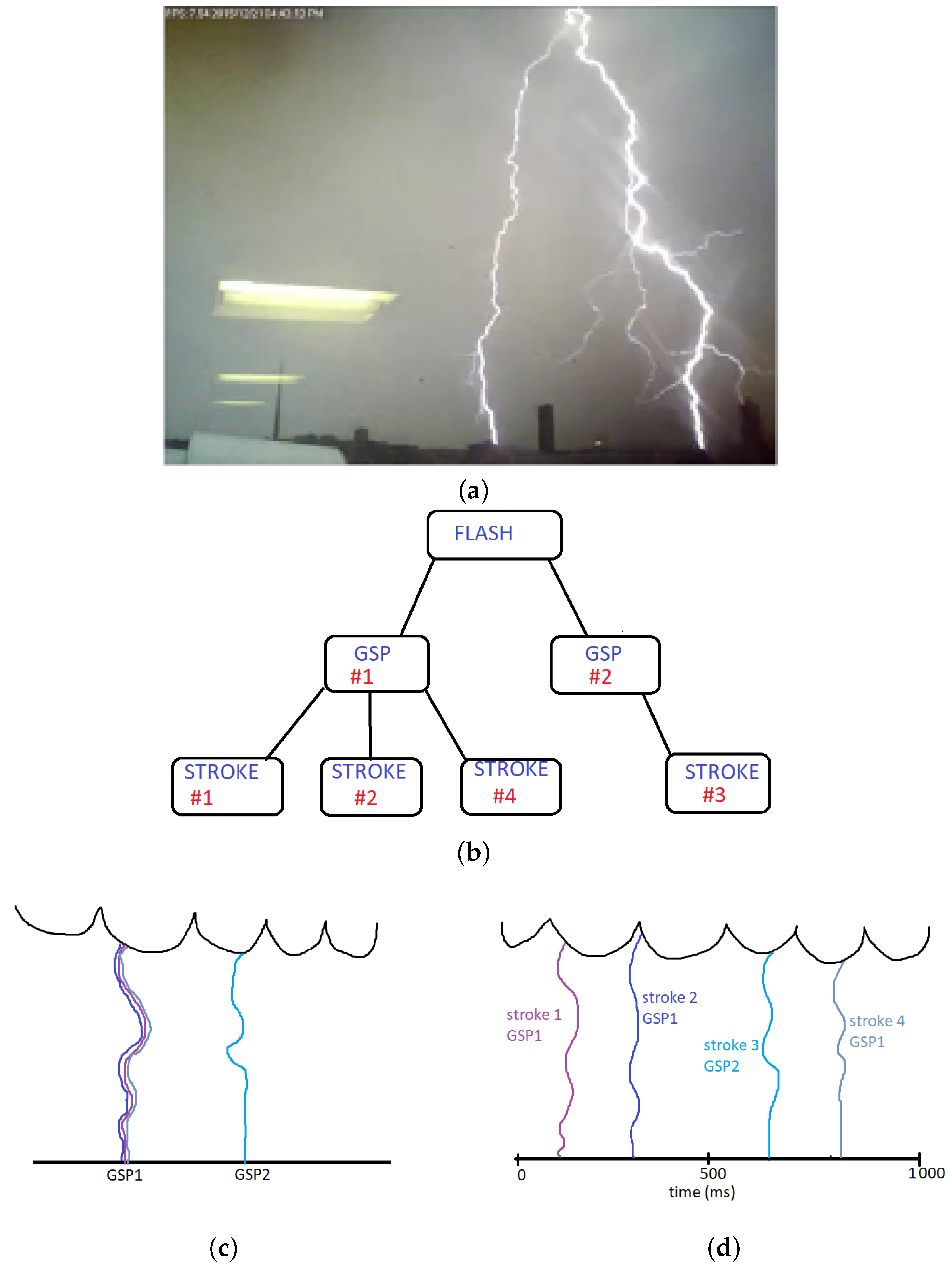
3.1. Bayesian Networks
Bayesian networks provide a compact representation of the joint probability distribution of variables, where each variable’s conditional probability depends on its parent variables [
25]. The joint probability of a Bayesian network is expressed as:
where
are variables, and
are their parent variables [
26,
27].
Figure 2 illustrates a Bayesian network with six variables. The joint probability is expanded using the Bayesian theorem chain rule and Equation (
1):
Learning a Bayesian network involves obtaining a model that describes the joint probability distribution of variables using data samples. This step is crucial for density estimation, inference queries, specific predictions, and knowledge discovery [
29]. Bayesian network models can be trained through structure learning and parameter learning. Parameter learning estimates the parameters of a fixed Bayesian network model from a complete dataset. It can be achieved using maximum likelihood estimation (MLE) or Bayesian statistics [
29]. Inference reduces the global probability distribution to a conditional probability of observed variables, allowing probabilistic queries and data imputation [
29,
30].
Structure learning uncovers variable correlations. It includes score-based structure learning and treats each potential model as a statistical problem, using a scoring function to fit models to data. The model with the highest score is chosen [
29]. Constraint-based structure learning assumes a Bayesian network represents variable independence. Conditional dependence and independence tests are run for all variables to create a suitable model [
29]. Bayesian model averaging combines multiple models to obtain an average [
29]. Bayesian networks utilize various algorithms for structure learning, such as hill climbing and tabu search. Tabu search optimizes graphical models by iteratively searching for the minimum of an objective function [
31]. Bayesian networks rely on probabilistic model selection to identify the best-fitting model using scoring functions like the Gaussian log-likelihood, Akaike information criterion (
AIC), and Bayesian information criterion (
BIC). The Gaussian log-likelihood is defined as [
32]:
where
is the Gaussian function:
The
AIC function estimates the variance between the data used to generate the model and the fitted potential model [
33]:
where
L is the likelihood function and
k is the number of model parameters.
BIC, similar to
AIC, transforms the posterior probability of the model [
34,
35]:
where
k is the number of model parameters,
n is the number of data records, and
L is the likelihood function.
BIC was introduced for independent, identically distributed observations and linear models [
36], assuming the likelihood is from the regular exponential family [
29,
34]. It selects a model by maximizing the posterior probability of a potential model from a dataset [
34,
36,
37]. The posterior probability is described by:
where
is the marginal probability distribution of the data and
is the marginal likelihood of the model [
34,
37].
Maximizing the posterior probability of a potential model and considering it as a continuous function gives:
where the integral is the continuous function of the model’s likelihood given its parameter vectors and prior distribution. From Equation (
4), we have
Expanding the integral using a Taylor expression gives:
with
, the integral becomes [
34,
37]:
Approximating the integral by its symmetry gives:
Substituting Equation (
7) into Equation (
6), we obtain [
34,
37]:
Bayesian networks do not require prior assumptions about the data and are suitable for small datasets. They show which features directly affect the target value and how features are interconnected, making them useful for prediction through inference.
3.2. Data
The data used for this analysis were provided by the South African Lightning Detection Network (SALDN) in Johannesburg, South Africa, for the years 2017 to 2019 [
12,
38]. The dataset consists of 15 features and 1311 entries with no missing data, as described in
Table 2. An additional feature, flash number, was used to categorize the lightning strokes according to IEC 62585 [
1]. Strokes with an inter-stroke delay of 500 milliseconds and a maximum distance of 10 km between them were categorized as a single flash. The ground truth dataset is represented by the feature strike point, which was validated using high-speed cameras capturing events to the millisecond.
Figure 3 shows an image representation of lightning flash 74 from the data over a map of Johannesburg. The red crosses indicate GSP locations of strokes captured by the LLSs.
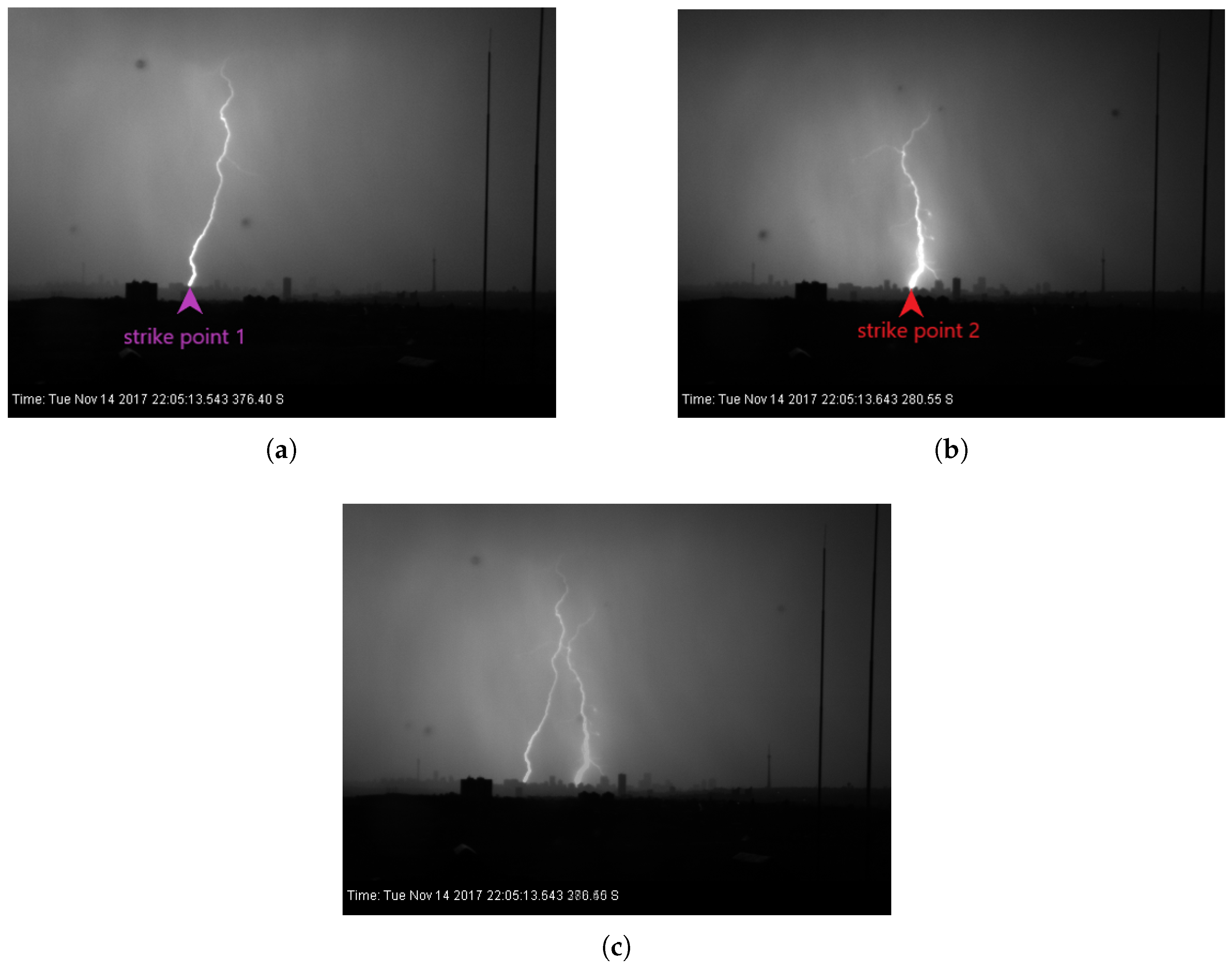
Figure 4 displays images of the two strike points captured by a high-speed camera. The time stamps indicate that the strokes occurred within a tenth of a second apart, confirming they are part of the same flash.
The semi-major and semi-minor distances were converted from kilometers to degrees for consistency. Time was converted from hours:minutes:seconds to seconds to be read as a float. The dataset has 463 flashes, with 213 single-stroke flashes excluded as they do not have a PSP variable.
3.3. Methods
The aim of this analysis was to determine the GSP of a stroke given the GSP of the previous stroke for flashes that have more than one stroke. To make such predictions, another feature was added: the previous strike point (PSP). First, the data were categorized according to the flashes, then single-stroke flashes were removed as they were not significant for the purpose of the project. The stroke labels in each flash were observed and were used to determine the value of the PSP. If a stroke label is the same as the previous one, PEC, then the PSP label for that entry is 1; if it is not the same, NGC, then the PSP label is 0. The data have more strike points classified as NGCs than PECs: this is in alignment with the observation made in the research of global GSP characteristics in negative downward flashes [
12], which then creates a class imbalance. It should also be noted that the initial stroke of each flash was removed since it does not have a PSP value.
BN-learn is an R package that is capable of Bayesian network modeling analysis by means of structure learning, parameter learning, and inference [
41]. The package contains algorithms for data pre-processing, inference, parameter learning, and structure learning that combines the data and prior knowledge [
41] that are necessary for Bayesian network modeling. It is capable of handling discrete Bayesian networks, Gaussian Bayesian networks, and conditional linear Gaussian Bayesian networks using real data [
41].
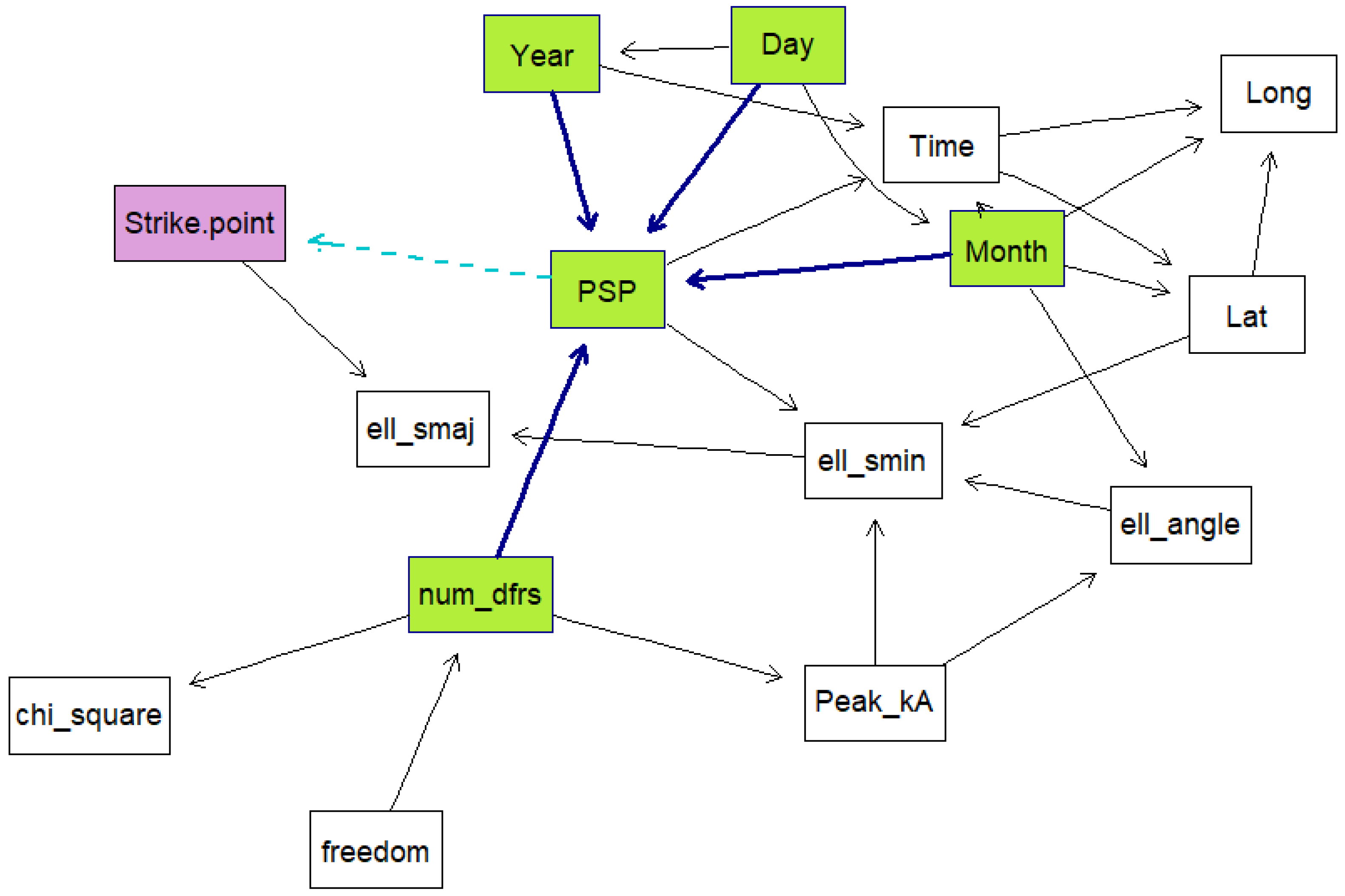
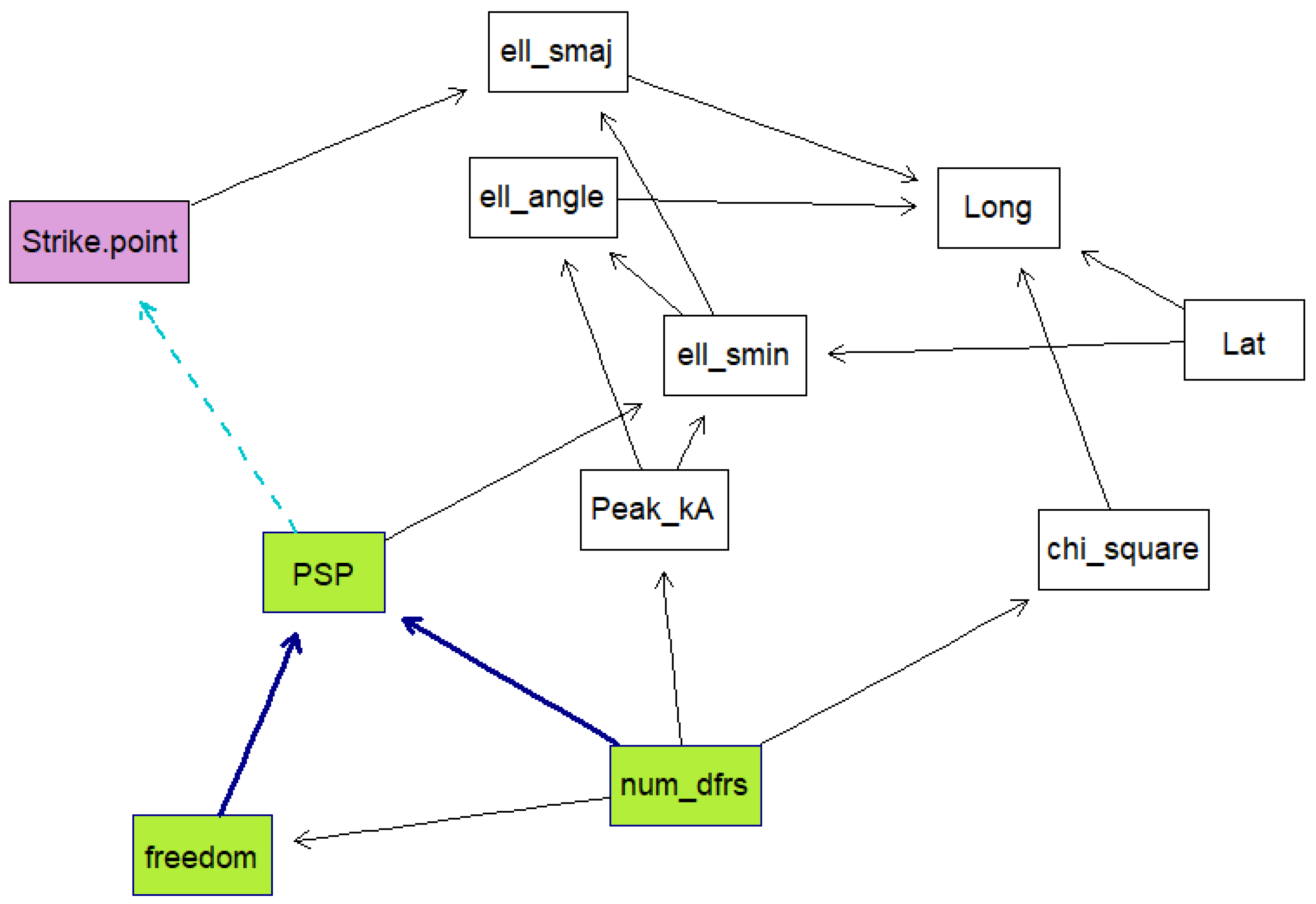
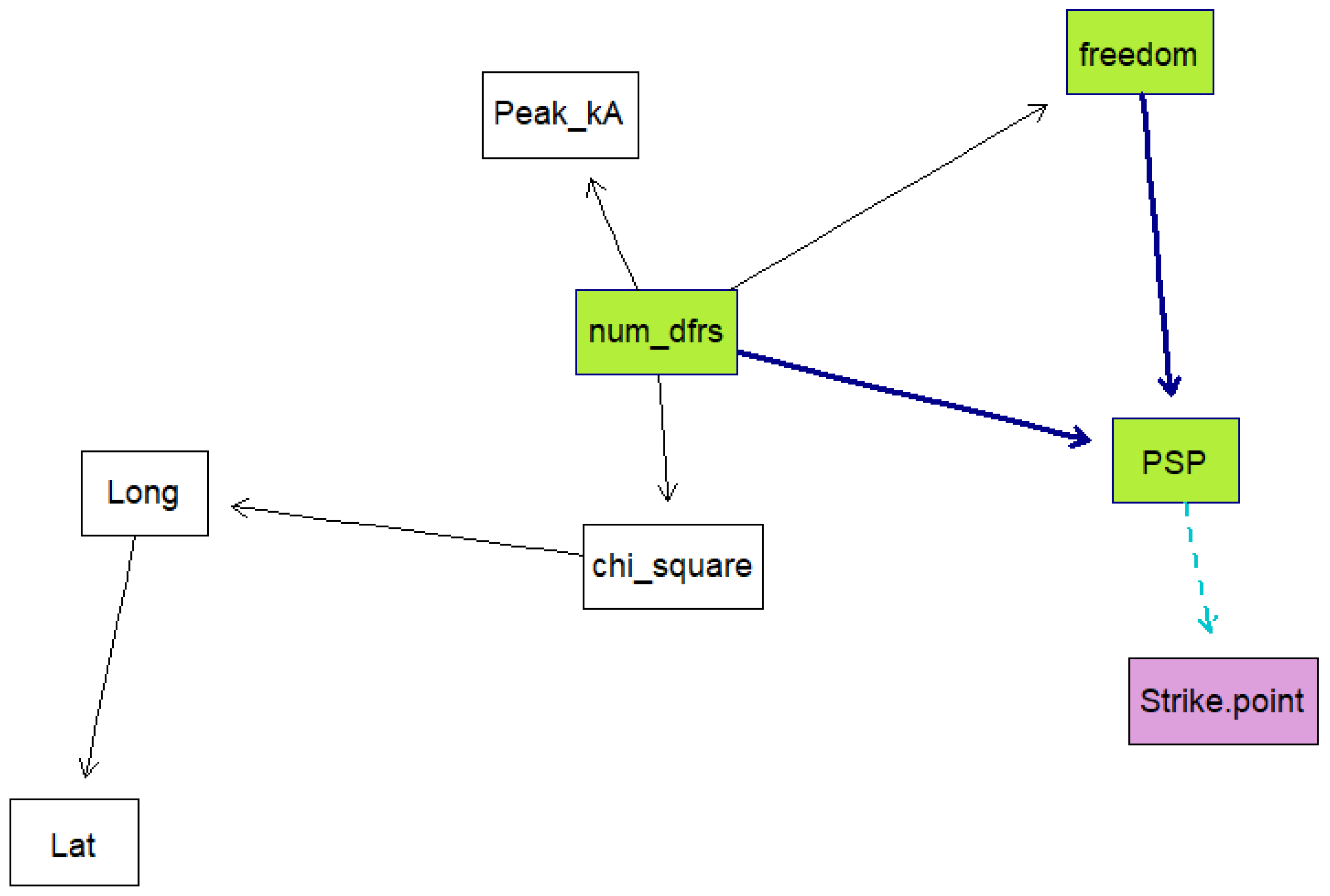
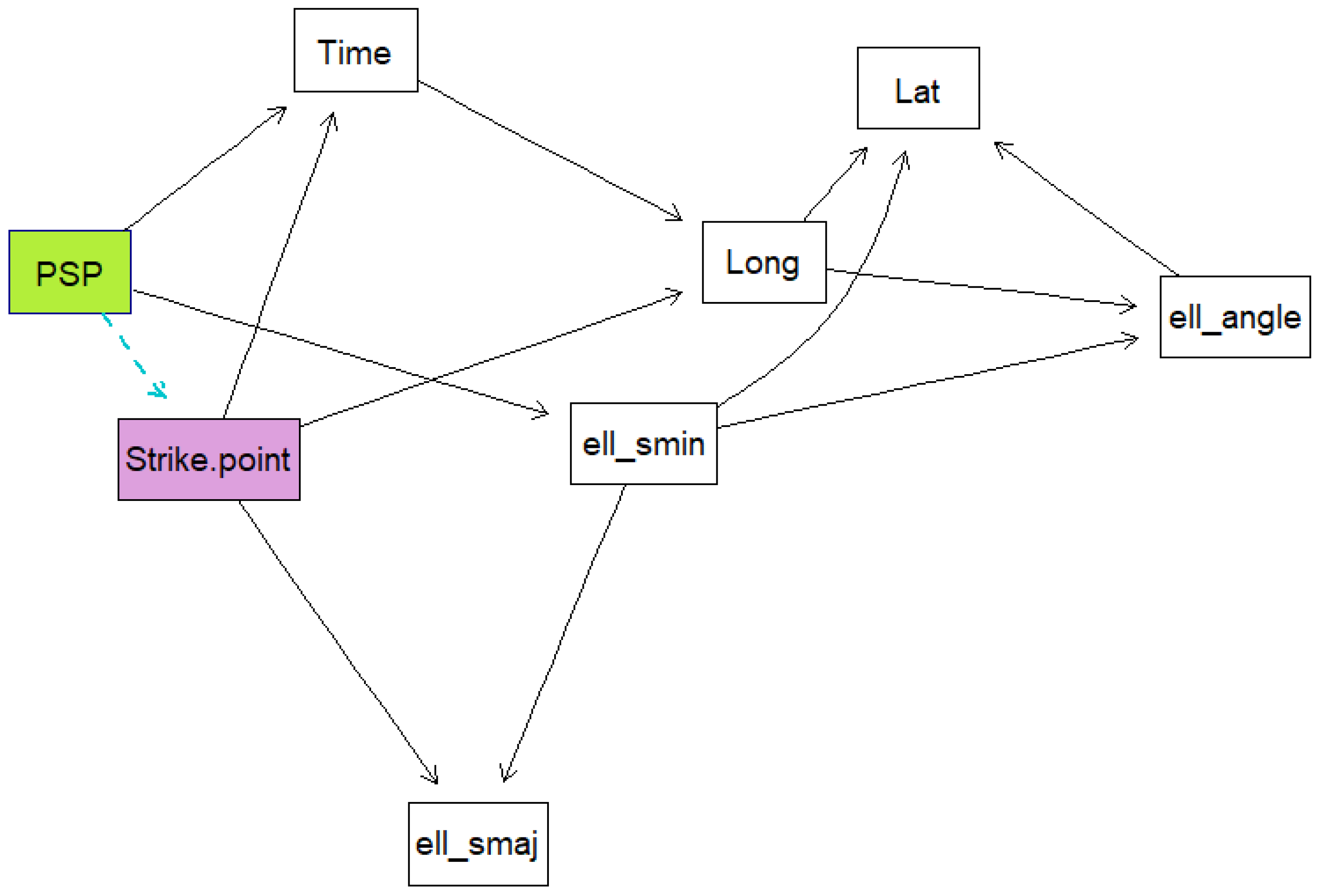
Five Bayesian network models were created using the data. For each model, a k-fold cross validation was run 50 times on the data to obtain the mean classification errors and their standard deviations. This is done to evaluate how well the models are expected to perform. K-fold cross validation is a training method whereby the training data are divided into samples, then samples are trained while the other sample is used as the testing sample. The algorithm is repeated until all the samples have been used as the testing sample, then the average loss is calculated as the performance measure. The models were trained using a score-based structure-learning procedure with tabu search. The first model included all the variables and was trained from the raw data. The second model was trained with all the variables and with the date information set as the conditional dependencies of PSP. The third model excluded dates and times, while the fourth model excluded dates, times, and ellipse information. Both these models had the number of sensors and the degrees of freedom set as parent nodes of PSP. The last model was created using the time, strike point, PSP, latitude and longitude and ellipse information.
The algorithm used for creating the models involved a structure-learning procedure with tabu search and the Bayesian information criterion (BIC). The full code is available on GitHub:
https://github.com/Lwano31/BN_models (accessed on 11 May 2024) [
40,
42]. The steps of the algorithm are summarized as follows:
Set up conditional dependencies from the data.
Learn the best-fit directed acyclic graph (DAG) from data with dependencies.
Perform cross validation on the DAG (data = DAG, runs = 50).
Calculate the loss with target = PSP.
Predict the output from the fitted DAG.
Plot the DAG.
Repeat the algorithm for the other models.
This is captured in the following code sample:
net = tabu(flash_data, score = ‘bic-g’)
graphviz.plot(net, shape = ‘rectangle’,
layout = ‘fdp’, highlight =
list(nodes = c (“PSP”, parents(net, “PSP”)),
arcs = incoming.arcs(net, ”PSP”),
col = “darkblue”, fill = "tomato", lwd = 3))
bn.cv(data=flash_data, net, runs=50
loss.args = list (target = ‘PSP’))
predicted = predict(bn.fit(net, flash_data), “PSP”, flash_data)
3.4. Analysis
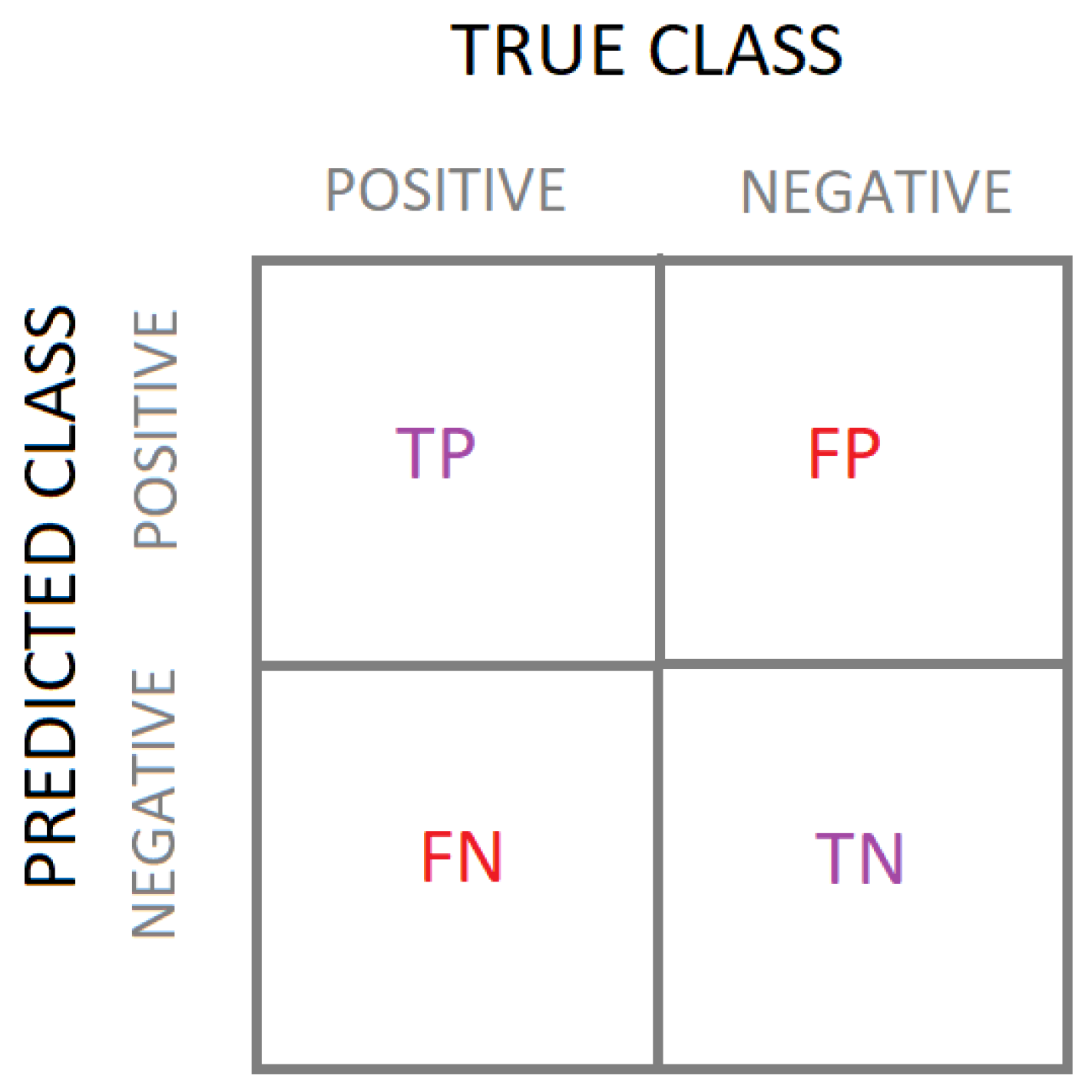
The performance of the models was evaluated using a confusion matrix, which is suitable for binary classification models [
43].
Figure 5 shows a schematic of a confusion matrix with the true and predicted class values. It computes the ratio of true positives (TPs), false positives (FPs), true negatives (TNs), and false negatives (FNs), allowing the calculation of performance measures such as accuracy, precision, recall, and F1-score.
Balanced accuracy and kappa statistics were also used to evaluate model performance while taking into account the class imbalance [
43,
45].
Baselines
The following baseline algorithms were used for comparison:
Naive Bayes: constructs a Bayesian probabilistic model assuming all variables are independent [
46];
Multilayer perceptron (MLP): a neural network that assigns weights to multiple variables to produce a binary output [
47];
Logistic regression: similar to MLP but uses an activation function like sigmoid to obtain an output [
47];
Random tree: a decision tree generated from random datasets [
48];
Random forest: a combination of tree classifiers with random sampling [
49];
Sequential minimal optimization (SMO): optimizes a support vector machine (SVM) using Lagrangian multipliers [
50].
The methods have various limitations due to the tools used and the type of data. The data are a hybrid of discrete and continuous data for which BN-learn has only one loss function that is suitable. BN-learn only has two algorithms for maxima and minima searches. Another limitation of BN-learn is that discrete nodes can only be parent nodes of continuous nodes: even with prior knowledge of a continuous node being a parent of a discrete node, the library does not allow that.