Characterization of Microbials in the Lung Induced by Allergenic Platanus Pollen Protein (Pla a3) and Ambient Fine Particulate Matter
Abstract
1. Introduction
2. Materials and Methods
2.1. Preparation of Particulate Matter Samples
2.2. Preparation and Purification of Pla a3 Protein
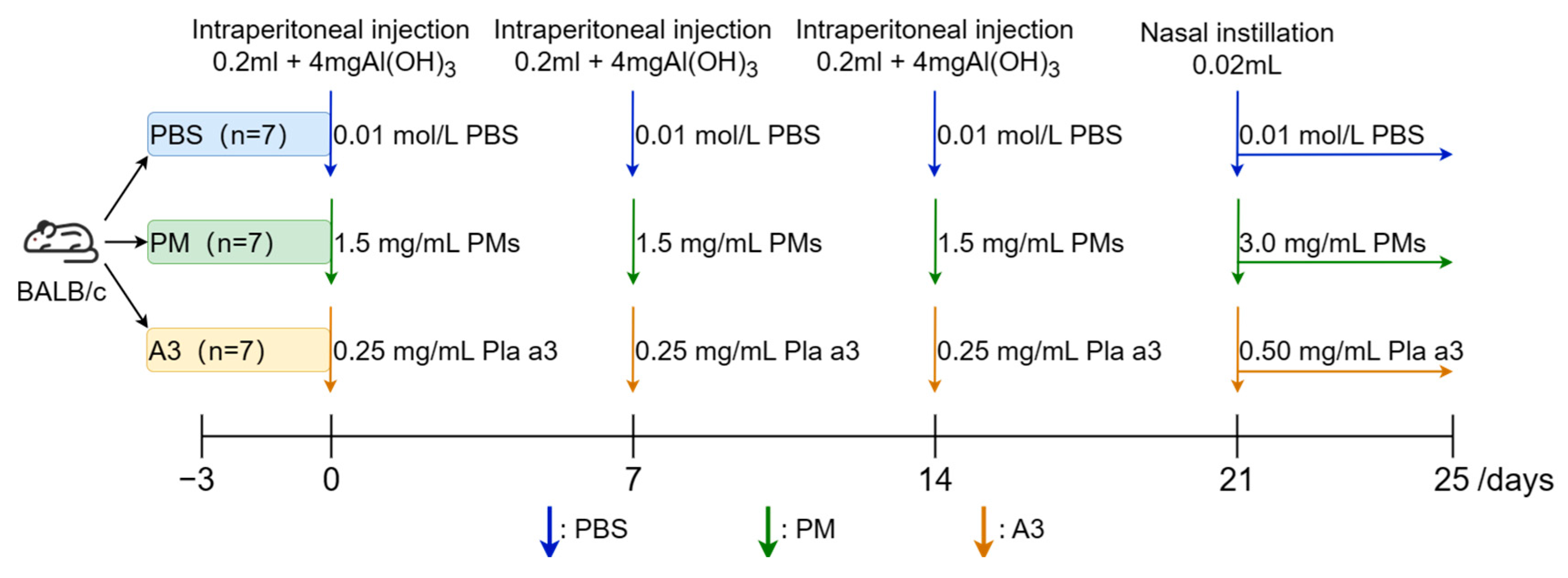
2.3. Establishment of Allergenic Animal Model
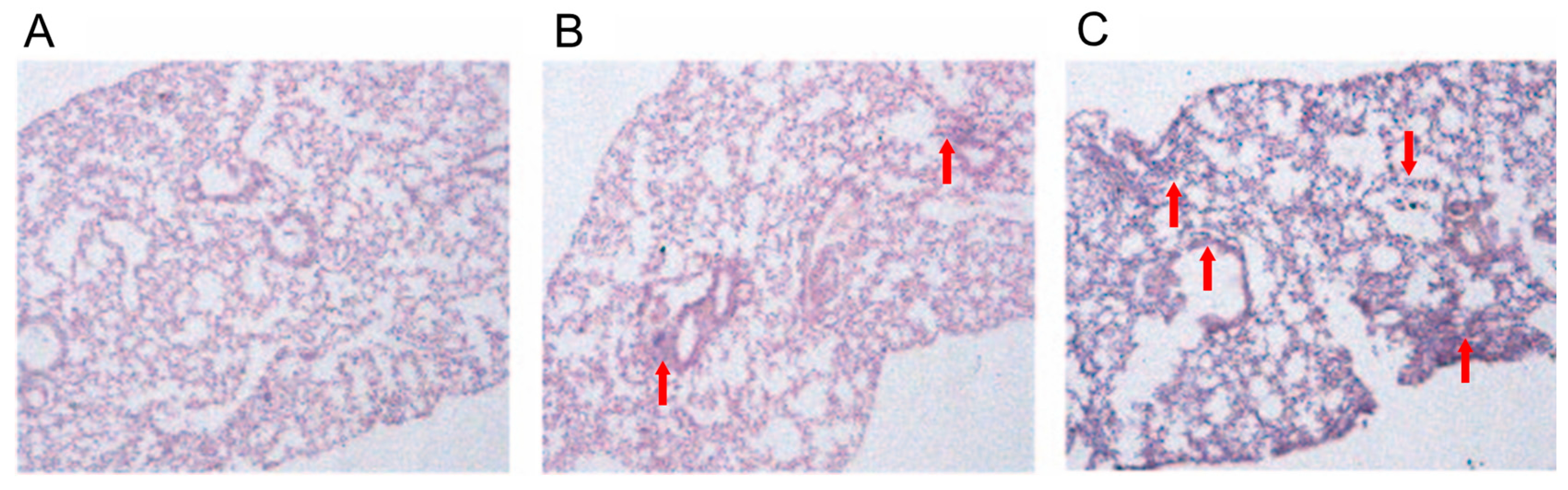
2.4. Histopathological Analysis
2.5. Immunoglobulin Assays
2.6. Detection of Inflammatory Factors from the Pulmonary Tissues
2.7. 16SrRNA Gene Sequence Analysis
2.8. Bioinformatic Analysis
2.9. Statistical Analysis
3. Results
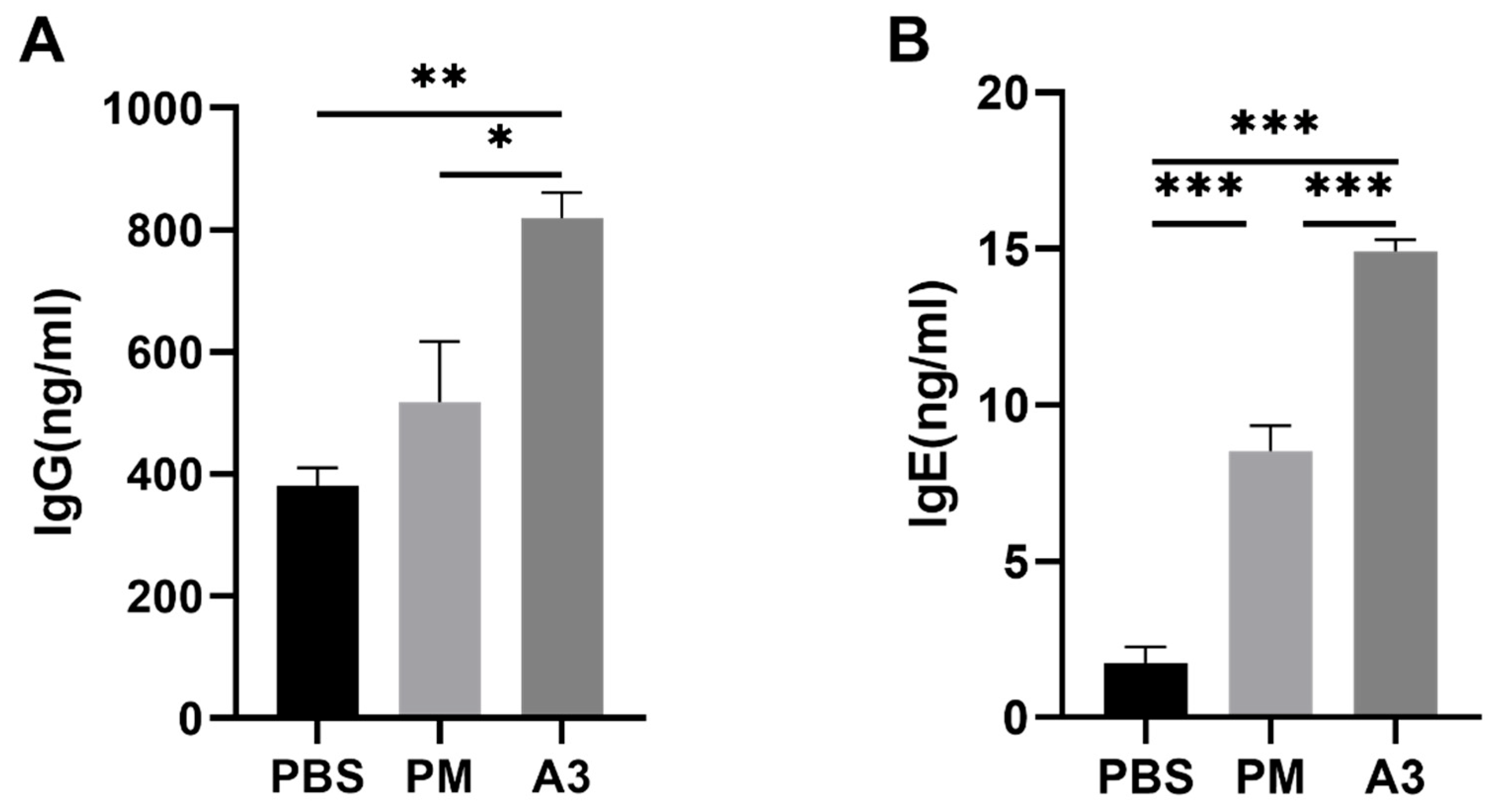
3.1. Expression Level of IgE and IgG Induced by Pla a3 Protein and PMs
3.2. Inflammatory Cytokines in the Lung
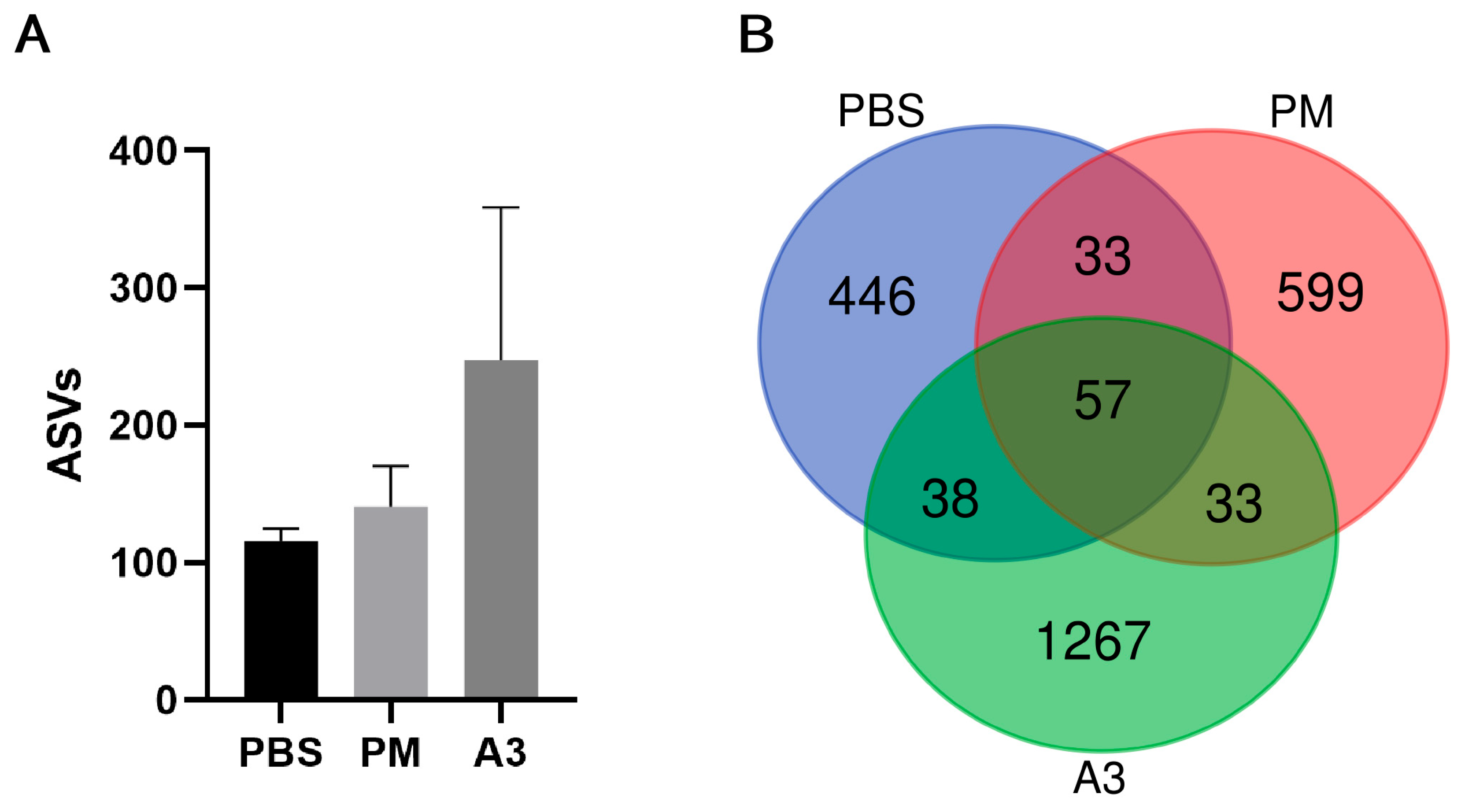
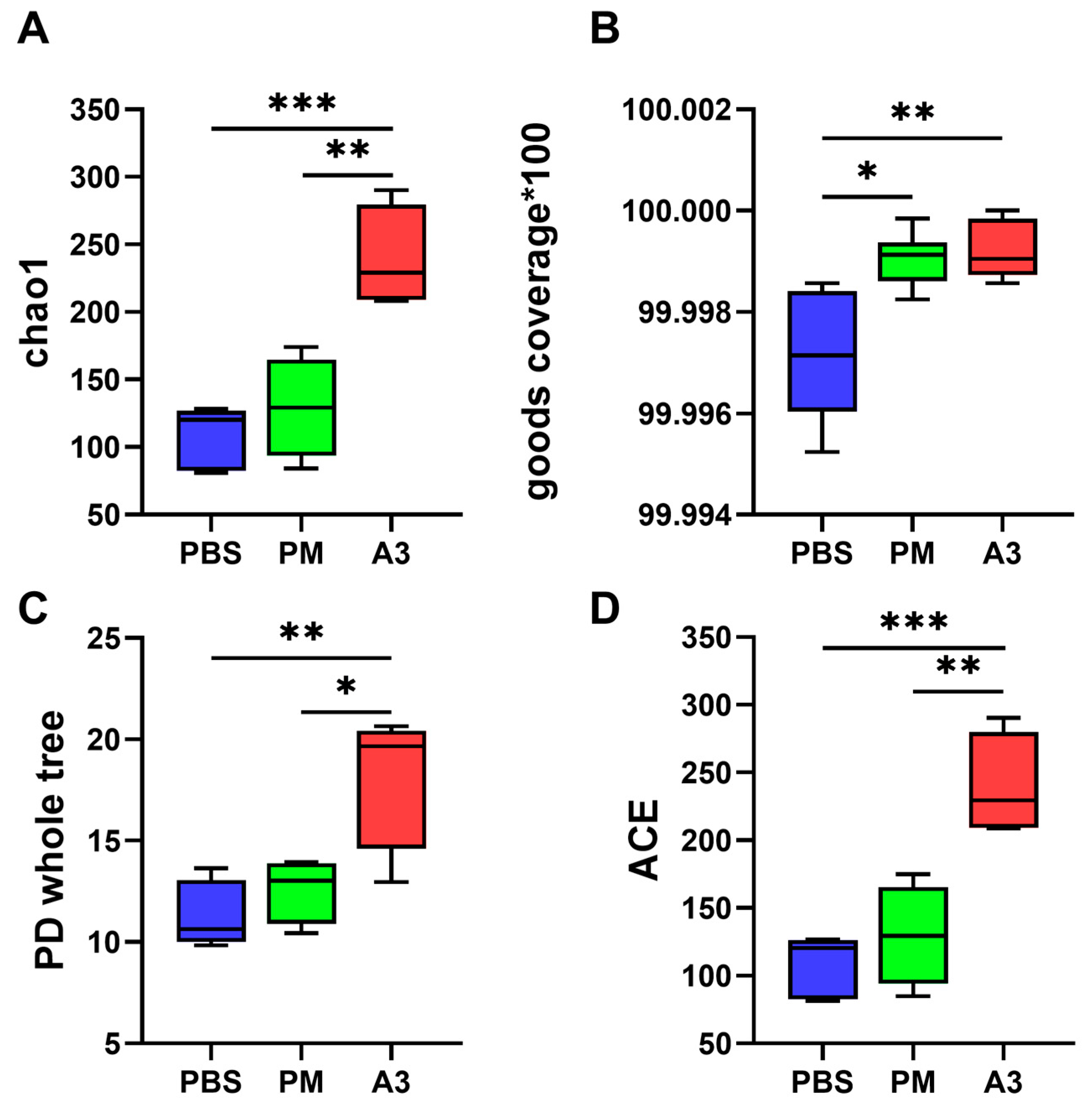
3.3. Microbial Diversity in Lung Tissue
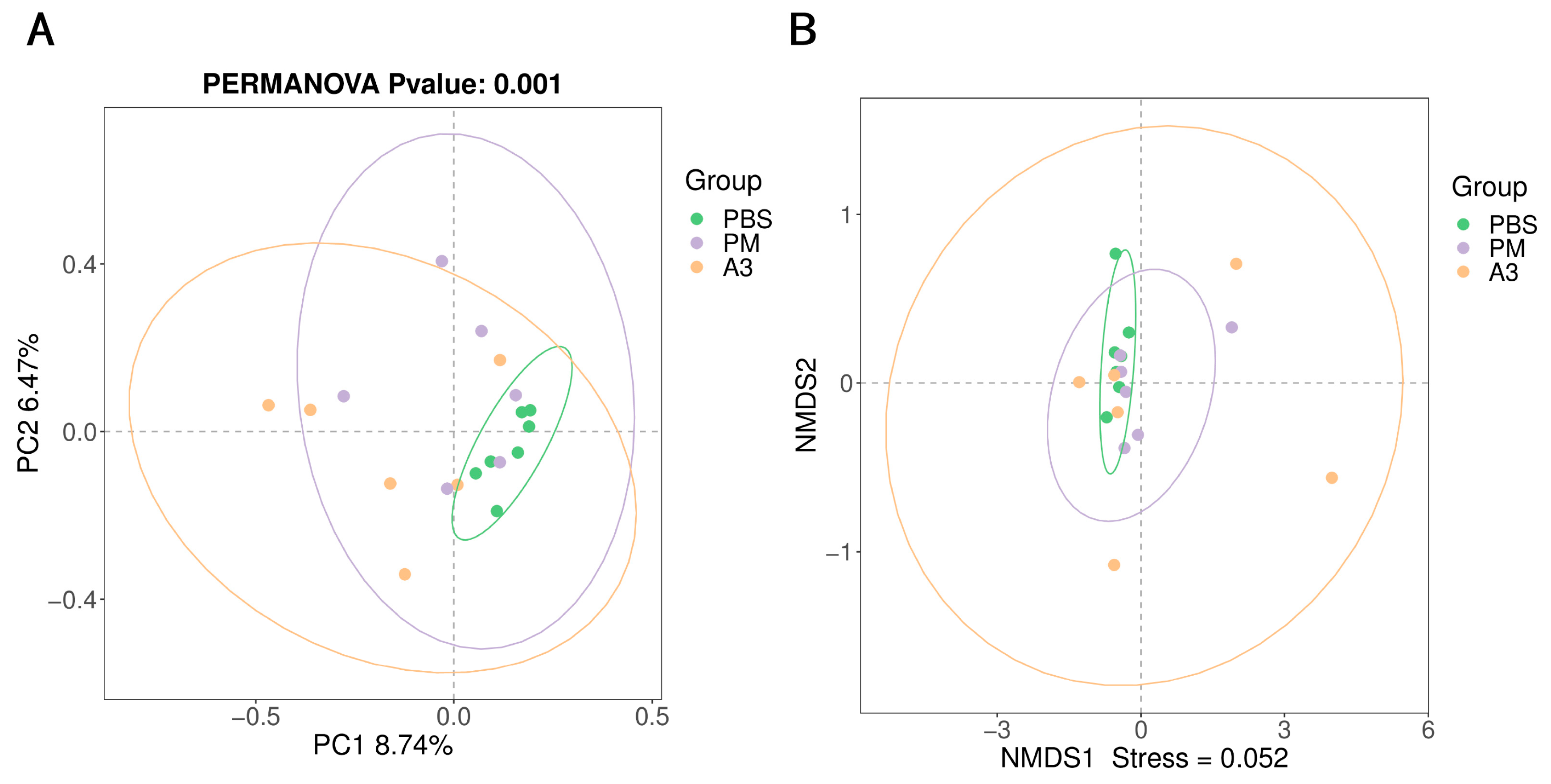
3.4. Microbial Community in the Lung Tissue
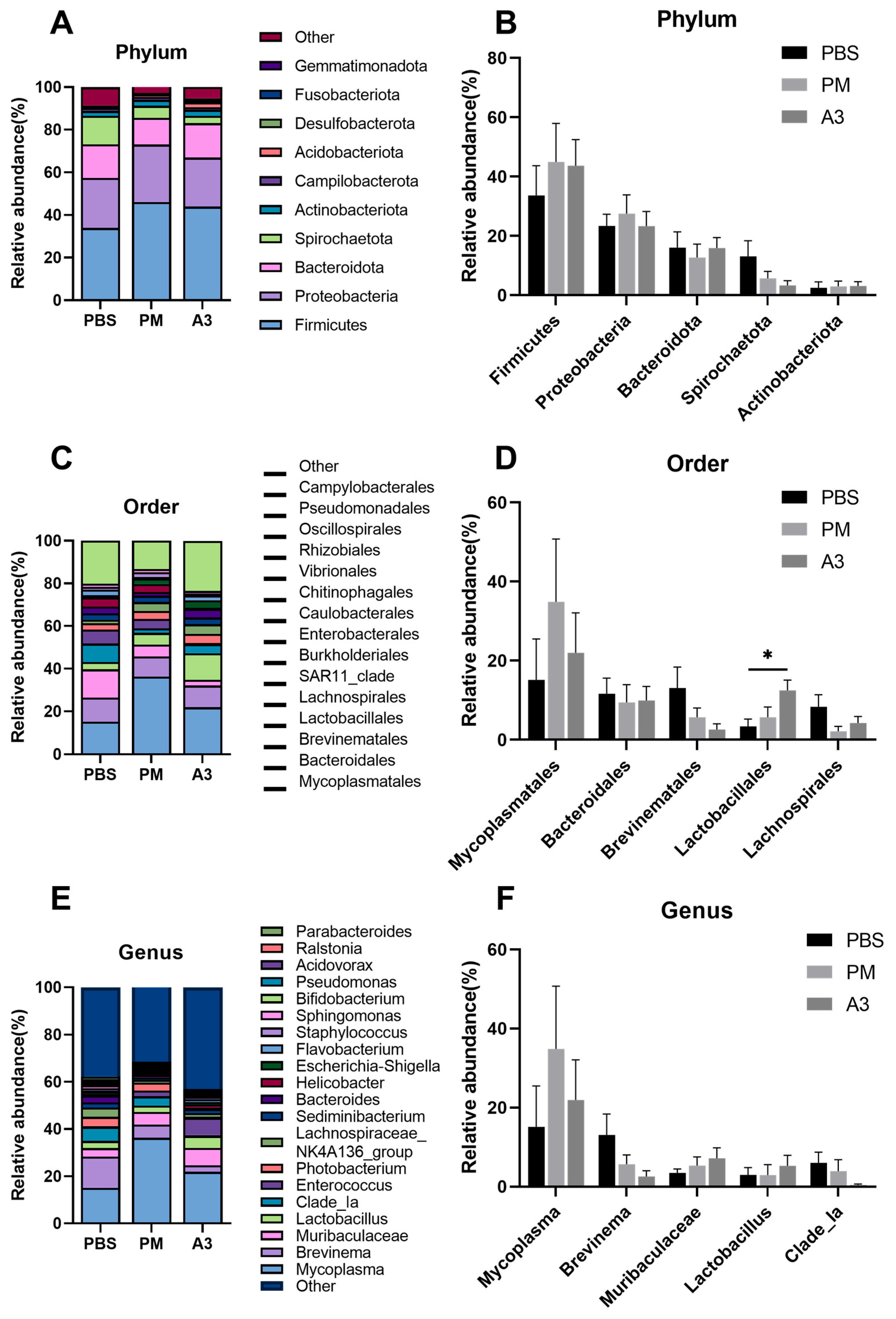
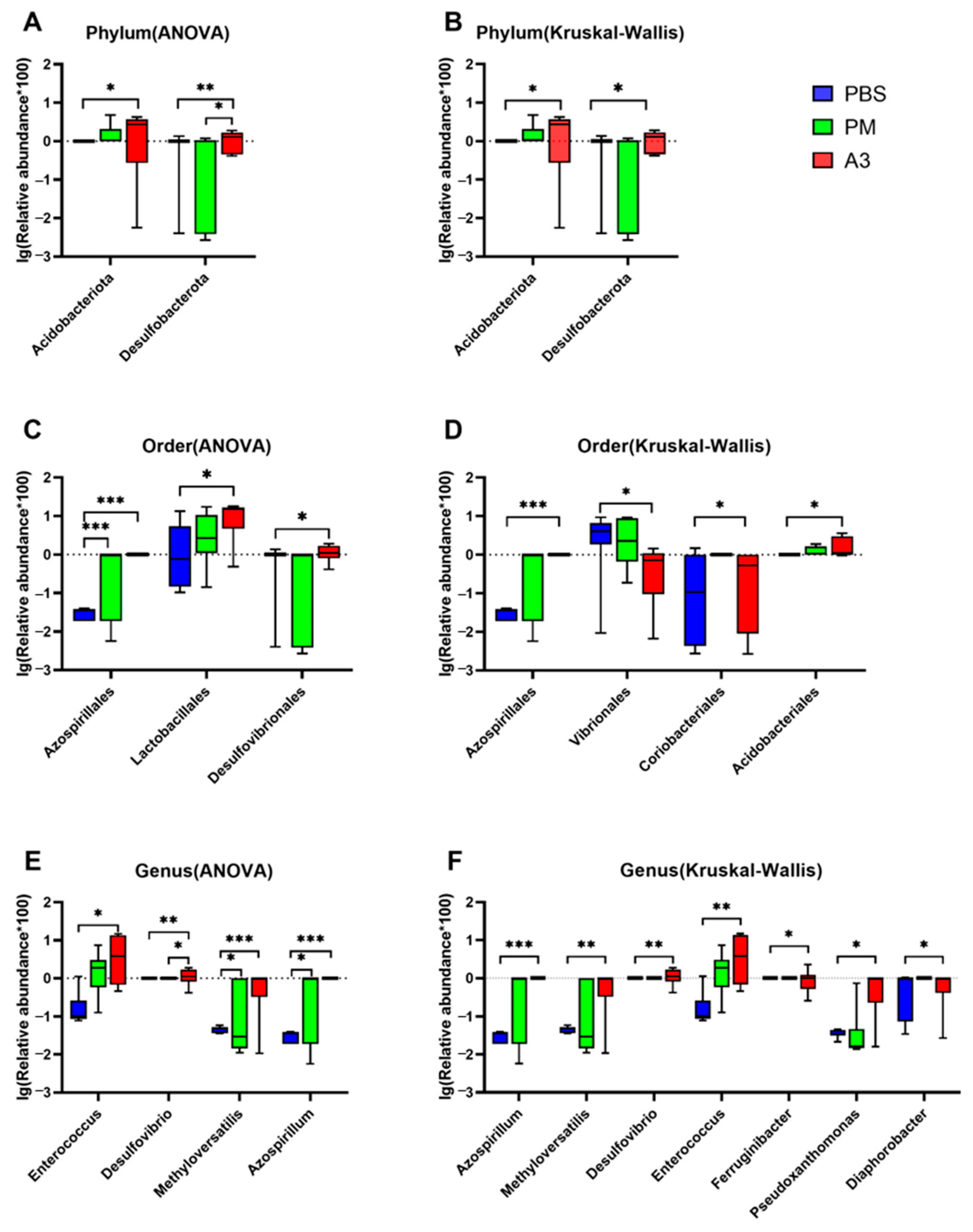
3.5. Comparison of Differences of the Microbiota at Phylum, Order, and Genus Levels in the Samples
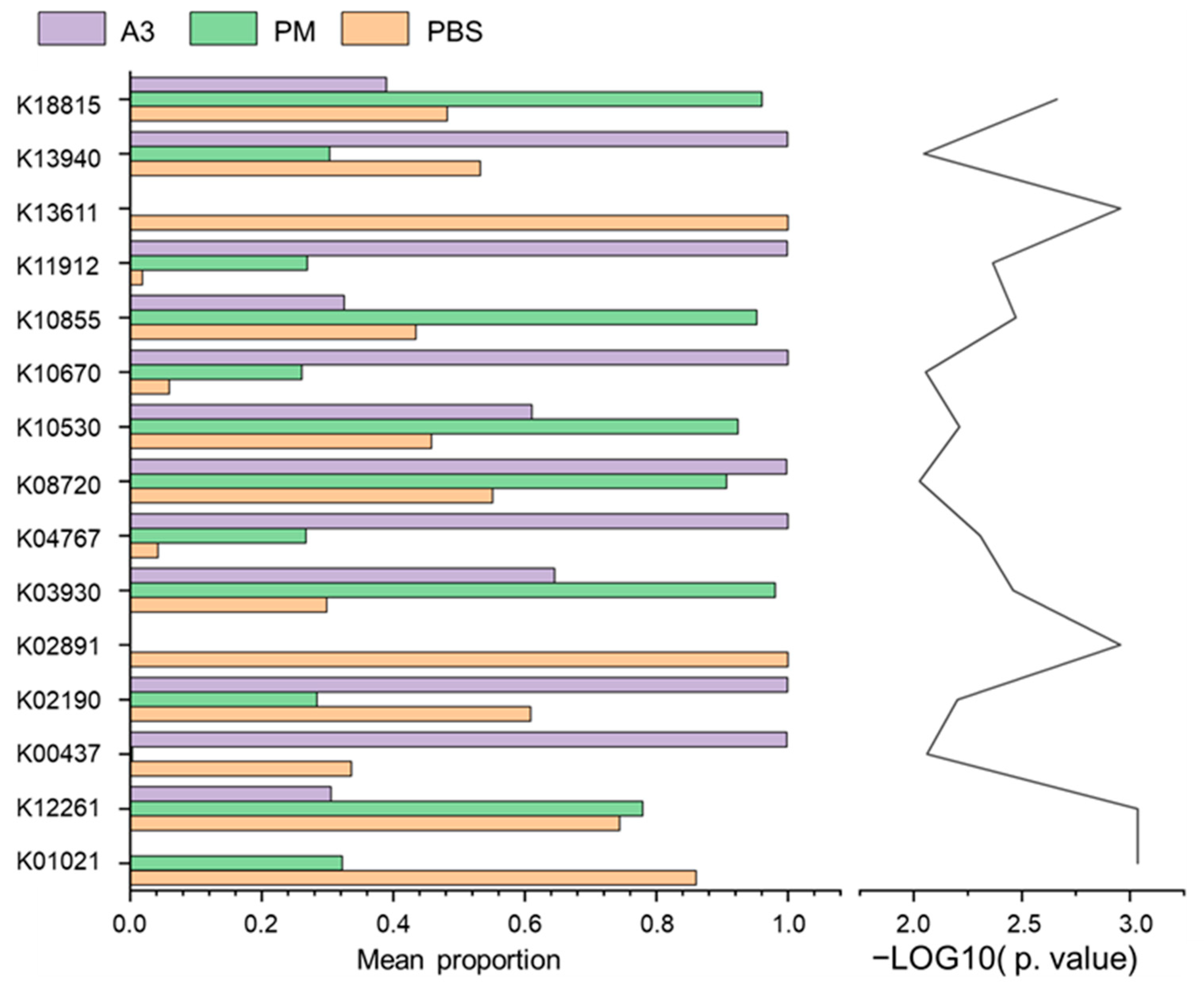
3.6. Functional Prediction of Lung Microbiota
3.7. The Relationship between Inflammatory Cytokines and Microbiota Alterations in Lung Tissue Samples
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Conflicts of Interest
References
- Wang, X.Y.; Ma, T.T.; Wang, X.Y.; Zhuang, Y.; Wang, X.D.; Ning, H.Y.; Shi, H.Y.; Yu, R.L.; Yan, D.; Huang, H.D.; et al. Prevalence of pollen-induced allergic rhinitis with high pollen exposure in grasslands of northern China. Allergy 2018, 73, 1232–1243. [Google Scholar] [CrossRef] [PubMed]
- Després, V.R.; Huffman, J.A.; Burrows, S.M.; Hoose, C.; Safatov, A.S.; Buryak, G.; Fröhlich-Nowoisky, J.; Elbert, W.; Andreae, M.O.; Pöschl, U.; et al. Primary biological aerosol particles in the atmosphere: A review. Tellus B Chem. Phys. Meteorol. 2012, 64, 15598. [Google Scholar] [CrossRef]
- Ravindra, K.; Goyal, A.; Mor, S. Does airborne pollen influence COVID-19 outbreak? Sustain. Cities Soc. 2021, 70, 102887. [Google Scholar] [CrossRef] [PubMed]
- Lou, H.; Ma, S.; Zhao, Y.; Cao, F.; He, F.; Liu, Z.; Bousquet, J.; Wang, C.; Zhang, L.; Bachert, C. Sensitization patterns and minimum screening panels for aeroallergens in self-reported allergic rhinitis in China. Sci. Rep. 2017, 7, 9286. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Sun, Y.; An, Y.; Wang, R.; Lin, H.; Liu, M.; Li, S.; Ma, M.; Xiao, C. Air pollution during the winter period and respiratory tract microbial imbalance in a healthy young population in Northeastern China. Environ. Pollut. 2019, 246, 972–979. [Google Scholar] [CrossRef] [PubMed]
- Hosgood, H.D., 3rd; Sapkota, A.R.; Rothman, N.; Rohan, T.; Hu, W.; Xu, J.; Vermeulen, R.; He, X.; White, J.R.; Wu, G.; et al. The potential role of lung microbiota in lung cancer attributed to household coal burning exposures. Environ. Mol. Mutagen. 2014, 55, 643–651. [Google Scholar] [CrossRef] [PubMed]
- Han, P.; Li, L.S.; Wang, Z.X.; Xi, L.; Yu, H.; Cong, L.; Zhang, Z.W.; Fu, J.; Peng, R.; Pan, L.B.; et al. Multi-Omics Analysis Provides Insight into the Possible Molecular Mechanism of Hay Fever Based on Gut Microbiota. Engineering 2022, 15, 115–125. [Google Scholar] [CrossRef]
- Thibeault, C.; Suttorp, N.; Opitz, B. The microbiota in pneumonia: From protection to predisposition. Sci. Transl. Med. 2021, 13, eaba0501. [Google Scholar] [CrossRef]
- Zhou, S.M.; Zhao, H.; Peng, J.X.; Hong, Q.; Xiao, K.; Shang, Y.; Lu, S.L.; Zhang, W.; Wu, M.H.; Li, S.J.; et al. Size distribution of allergen 3 (Pla a3) in Shanghai ambient size-resolved particles and its allergenic effects. Atmos. Environ. 2019, 198, 324–334. [Google Scholar] [CrossRef]
- Zhou, S.; Wang, X.; Lu, S.; Yao, C.; Zhang, L.; Rao, L.; Liu, X.; Zhang, W.; Li, S.; Wang, W.; et al. Characterization of allergenicity of Platanus pollen allergen a 3 (Pla a 3) after exposure to NO2 and O3. Environ. Pollut. 2021, 278, 116913. [Google Scholar] [CrossRef]
- Zhou, S.; Hong, Q.; Li, Y.; Li, Q.; Wang, M. Autophagy contributes to regulate the ROS levels and PCD progress in TMV-infected tomatoes. Plant Sci. 2018, 269, 12–19. [Google Scholar] [CrossRef] [PubMed]
- Hong, Q.; Zhou, S.; Zhao, H.; Peng, J.; Li, Y.; Shang, Y.; Wu, M.; Zhang, W.; Lu, S.; Li, S.; et al. Allergenicity of recombinant Humulus japonicus pollen allergen 1 after combined exposure to ozone and nitrogen dioxide. Environ. Pollut. 2018, 234, 707–715. [Google Scholar] [CrossRef] [PubMed]
- Chao, A. Nonparametric Estimation of the Number of Classes in a Population. Scand. J. Stat. 1984, 11, 265–270. [Google Scholar]
- Esty, W.W. The Efficiency of Good’s Nonparametric Coverage Estimator. Ann. Stat. 1986, 14, 1257–1260. [Google Scholar] [CrossRef]
- Douglas, G.M.; Maffei, V.J.; Zaneveld, J.; Yurgel, S.N.; Brown, J.R.; Taylor, C.M.; Huttenhower, C.; Langille, M.G.I. PICRUSt2: An improved and customizable approach for metagenome inference. BioRxiv 2019, 672295. [Google Scholar] [CrossRef]
- Saravia, J.; You, D.; Thevenot, P.; Lee, G.I.; Shrestha, B.; Lomnicki, S.; Cormier, S.A. Early-life exposure to combustion-derived particulate matter causes pulmonary immunosuppression. Mucosal Immunol. 2014, 7, 694–704. [Google Scholar] [CrossRef] [PubMed]
- Moelling, K.; Broecker, F. Air Microbiome and Pollution: Composition and Potential Effects on Human Health, Including SARS Coronavirus Infection. J. Environ. Public. Health 2020, 2020, 1646943. [Google Scholar] [CrossRef] [PubMed]
- Rylance, J.; Kankwatira, A.; Nelson, D.E.; Toh, E.; Day, R.B.; Lin, H.; Gao, X.; Dong, Q.; Sodergren, E.; Weinstock, G.M.; et al. Household air pollution and the lung microbiome of healthy adults in Malawi: A cross-sectional study. BMC Microbiol. 2016, 16, 182. [Google Scholar] [CrossRef] [PubMed]
- Chen, D.; Xiao, C.; Jin, H.; Yang, B.; Niu, J.; Yan, S.; Sun, Y.; Zhou, Y.; Wang, X. Exposure to atmospheric pollutants is associated with alterations of gut microbiota in spontaneously hypertensive rats. Exp. Ther. Med. 2019, 18, 3484–3492. [Google Scholar] [CrossRef]
- Li, N.; He, F.; Liao, B.; Zhou, Y.; Li, B.; Ran, P. Exposure to ambient particulate matter alters the microbial composition and induces immune changes in rat lung. Respir. Res. 2017, 18, 143. [Google Scholar] [CrossRef]
- Beerens, H.; Romond, C. Sulfate-reducing anaerobic bacteria in human feces. Am. J. Clin. Nutr. 1977, 30, 1770–1776. [Google Scholar] [CrossRef] [PubMed]
- Goldstein, E.J.; Citron, D.M.; Peraino, V.A.; Cross, S.A. Desulfovibrio desulfuricans bacteremia and review of human Desulfovibrio infections. J. Clin. Microbiol. 2003, 41, 2752–2754. [Google Scholar] [CrossRef]
- Lu, H.Z.; Weng, X.H.; Li, H.; Yin, Y.K.; Pang, M.Y.; Tang, Y.W. Enterococcus faecium-related outbreak with molecular evidence of transmission from pigs to humans. J. Clin. Microbiol. 2002, 40, 913–917. [Google Scholar] [CrossRef] [PubMed]
- Sava, I.G.; Heikens, E.; Huebner, J. Pathogenesis and immunity in enterococcal infections. Clin. Microbiol. Infect. 2010, 16, 533–540. [Google Scholar] [CrossRef]
- Doronina, N.V.; Kaparullina, E.N.; Trotsenko, Y.A. Methyloversatilis thermotolerans sp. nov., a novel thermotolerant facultative methylotroph isolated from a hot spring. Int. J. Syst. Evol. Microbiol. 2014, 64, 158–164. [Google Scholar] [CrossRef]
- Foulon, V.; Antonenkov, V.D.; Croes, K.; Waelkens, E.; Mannaerts, G.P.; Van Veldhoven, P.P.; Casteels, M. Purification, molecular cloning, and expression of 2-hydroxyphytanoyl-CoA lyase, a peroxisomal thiamine pyrophosphate-dependent enzyme that catalyzes the carbon-carbon bond cleavage during alpha-oxidation of 3-methyl-branched fatty acids. Proc. Natl. Acad. Sci. USA 1999, 96, 10039–10044. [Google Scholar] [CrossRef] [PubMed]
- Casteels, M.; Sniekers, M.; Fraccascia, P.; Mannaerts, G.P.; Van Veldhoven, P.P. The role of 2-hydroxyacyl-CoA lyase, a thiamin pyrophosphate-dependent enzyme, in the peroxisomal metabolism of 3-methyl-branched fatty acids and 2-hydroxy straight-chain fatty acids. Biochem. Soc. Trans. 2007, 35, 876–880. [Google Scholar] [CrossRef]
- Straight, P.D.; Fischbach, M.A.; Walsh, C.T.; Rudner, D.Z.; Kolter, R. A singular enzymatic megacomplex from Bacillus subtilis. Proc. Natl. Acad. Sci. USA 2007, 104, 305–310. [Google Scholar] [CrossRef]
- Filloux, A.; Hachani, A.; Bleves, S. The bacterial type VI secretion machine: Yet another player for protein transport across membranes. Microbiology 2008, 154, 1570–1583. [Google Scholar] [CrossRef]
- Coulthurst, S.J. The Type VI secretion system—A widespread and versatile cell targeting system. Res. Microbiol. 2013, 164, 640–654. [Google Scholar] [CrossRef]
- Deckers, H.M.; Wilson, F.R.; Voordouw, G. Cloning and sequencing of a [NiFe] hydrogenase operon from Desulfovibrio vulgaris Miyazaki F. J. Gen. Microbiol. 1990, 136, 2021–2028. [Google Scholar] [CrossRef] [PubMed]
- Maeda-Yorita, K.; Aki, K.; Sagai, H.; Misaki, H.; Massey, V. L-lactate oxidase and L-lactate monooxygenase: Mechanistic variations on a common structural theme. Biochimie 1995, 77, 631–642. [Google Scholar] [CrossRef] [PubMed]
- Wagner, M.; Sonntag, D.; Grimm, R.; Pich, A.; Eckerskorn, C.; Sohling, B.; Andreesen, J.R. Substrate-specific selenoprotein B of glycine reductase from Eubacterium acidaminophilum. Biochemical and molecular analysis. Eur. J. Biochem. 1999, 260, 38–49. [Google Scholar] [CrossRef] [PubMed]
- Skorupski, K.; Taylor, R.K. Control of the ToxR virulence regulon in Vibrio cholerae by environmental stimuli. Mol. Microbiol. 1997, 25, 1003–1009. [Google Scholar] [CrossRef] [PubMed]
- Sperandio, V.; Bailey, C.; Giron, J.A.; DiRita, V.J.; Silveira, W.D.; Vettore, A.L.; Kaper, J.B. Cloning and characterization of the gene encoding the OmpU outer membrane protein of Vibrio cholerae. Infect. Immun. 1996, 64, 5406–5409. [Google Scholar] [CrossRef] [PubMed]
- Mishra, V.; Banga, J.; Silveyra, P. Oxidative stress and cellular pathways of asthma and inflammation: Therapeutic strategies and pharmacological targets. Pharmacol. Ther. 2018, 181, 169–182. [Google Scholar] [CrossRef] [PubMed]
- Rizzatti, G.; Lopetuso, L.R.; Gibiino, G.; Binda, C.; Gasbarrini, A. Proteobacteria: A Common Factor in Human Diseases. Biomed. Res. Int. 2017, 2017, 9351507. [Google Scholar] [CrossRef] [PubMed]
- Saint-Georges-Chaumet, Y.; Edeas, M. Microbiota-mitochondria inter-talk: Consequence for microbiota-host interaction. Pathog. Dis. 2016, 74, ftv096. [Google Scholar] [CrossRef]
- Marsland, B.J.; Gollwitzer, E.S. Host-microorganism interactions in lung diseases. Nat. Rev. Immunol. 2014, 14, 827–835. [Google Scholar] [CrossRef]
- Duncan, S.H.; Louis, P.; Thomson, J.M.; Flint, H.J. The role of pH in determining the species composition of the human colonic microbiota. Environ. Microbiol. 2009, 11, 2112–2122. [Google Scholar] [CrossRef]
- Lodovici, M.; Bigagli, E. Oxidative stress and air pollution exposure. J. Toxicol. 2011, 2011, 487074. [Google Scholar] [CrossRef] [PubMed]
- Zhao, H.; Chen, S.; Yang, F.; Wu, H.; Ba, Y.; Cui, L.; Chen, R.; Zhu, J. Alternation of nasopharyngeal microbiota in healthy youth is associated with environmental factors: Implication for respiratory diseases. Int. J. Environ. Health Res. 2022, 32, 952–962. [Google Scholar] [CrossRef] [PubMed]
- Yu, C.X.; Zhang, P.; Liu, S.J.; Niu, Y.M.; Fu, L. SESN2 ablation weakens exercise benefits on resilience of gut microbiota following high-fat diet consumption in mice. Food Sci. Human. Wellness 2023, 12, 1961–1968. [Google Scholar] [CrossRef]
- Dong, P.; Yang, Y.; Wang, W.P. The role of intestinal bifidobacteria on immune system development in young rats. Early Hum. Dev. 2010, 86, 51–58. [Google Scholar] [CrossRef] [PubMed]
- Lopez, P.; Gueimonde, M.; Margolles, A.; Suarez, A. Distinct Bifidobacterium strains drive different immune responses in vitro. Int. J. Food Microbiol. 2010, 138, 157–165. [Google Scholar] [CrossRef] [PubMed]
- Menard, O.; Butel, M.J.; Gaboriau-Routhiau, V.; Waligora-Dupriet, A.J. Gnotobiotic mouse immune response induced by Bifidobacterium sp. strains isolated from infants. Appl. Environ. Microbiol. 2008, 74, 660–666. [Google Scholar] [CrossRef] [PubMed]
- Degois, J.; Simon, X.; Clerc, F.; Bontemps, C.; Leblond, P.; Duquenne, P. One-year follow-up of microbial diversity in bioaerosols emitted in a waste sorting plant in France. Waste Manag. 2021, 120, 257–268. [Google Scholar] [CrossRef] [PubMed]
- Ley, R.E. Gut microbiota in 2015: Prevotella in the gut: Choose carefully. Nat. Rev. Gastroenterol. Hepatol. 2016, 13, 69–70. [Google Scholar] [CrossRef] [PubMed]
- Hu, J.; Bao, Y.; Zhu, Y.; Osman, R.; Shen, M.; Zhang, Z.; Wang, L.; Cao, S.; Li, L.; Wu, Q. The Preliminary Study on the Association between PAHs and Air Pollutants and Microbiota Diversity. Arch. Environ. Contam. Toxicol. 2020, 79, 321–332. [Google Scholar] [CrossRef]
- Larsen, J.M. The immune response to Prevotella bacteria in chronic inflammatory disease. Immunology 2017, 151, 363–374. [Google Scholar] [CrossRef]
- Signat, B.; Roques, C.; Poulet, P.; Duffaut, D. Fusobacterium nucleatum in periodontal health and disease. Curr. Issues Mol. Biol. 2011, 13, 25–36. [Google Scholar] [PubMed]
- Castellarin, M.; Warren, R.L.; Freeman, J.D.; Dreolini, L.; Krzywinski, M.; Strauss, J.; Barnes, R.; Watson, P.; Allen-Vercoe, E.; Moore, R.A.; et al. Fusobacterium nucleatum infection is prevalent in human colorectal carcinoma. Genome Res. 2012, 22, 299–306. [Google Scholar] [CrossRef] [PubMed]
- Brazier, J.S. Human infections with Fusobacterium necrophorum. Anaerobe 2006, 12, 165–172. [Google Scholar] [CrossRef] [PubMed]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liu, J.; Lu, S.; Hou, G.; Hu, W.; Zhao, J.; Zhang, W.; Liu, X.; Ebere, E.C.; Wang, W.; Wang, Q. Characterization of Microbials in the Lung Induced by Allergenic Platanus Pollen Protein (Pla a3) and Ambient Fine Particulate Matter. Atmosphere 2024, 15, 503. https://doi.org/10.3390/atmos15040503
Liu J, Lu S, Hou G, Hu W, Zhao J, Zhang W, Liu X, Ebere EC, Wang W, Wang Q. Characterization of Microbials in the Lung Induced by Allergenic Platanus Pollen Protein (Pla a3) and Ambient Fine Particulate Matter. Atmosphere. 2024; 15(4):503. https://doi.org/10.3390/atmos15040503
Chicago/Turabian StyleLiu, Jin, Senlin Lu, Guoqing Hou, Wenwen Hu, Jiumei Zhao, Wei Zhang, Xinchun Liu, Enyoh Christian Ebere, Weiqian Wang, and Qingyue Wang. 2024. "Characterization of Microbials in the Lung Induced by Allergenic Platanus Pollen Protein (Pla a3) and Ambient Fine Particulate Matter" Atmosphere 15, no. 4: 503. https://doi.org/10.3390/atmos15040503
APA StyleLiu, J., Lu, S., Hou, G., Hu, W., Zhao, J., Zhang, W., Liu, X., Ebere, E. C., Wang, W., & Wang, Q. (2024). Characterization of Microbials in the Lung Induced by Allergenic Platanus Pollen Protein (Pla a3) and Ambient Fine Particulate Matter. Atmosphere, 15(4), 503. https://doi.org/10.3390/atmos15040503








