Molecular Characterization of Near Full-Length Genomes of Hepatitis B Virus Isolated from Predominantly HIV Infected Individuals in Botswana
Abstract
:1. Introduction
2. Materials and Methods
2.1. Population
2.2. Ethical Considerations
2.3. Extraction of Plasma DNA
2.4. Amplification and Sequencing
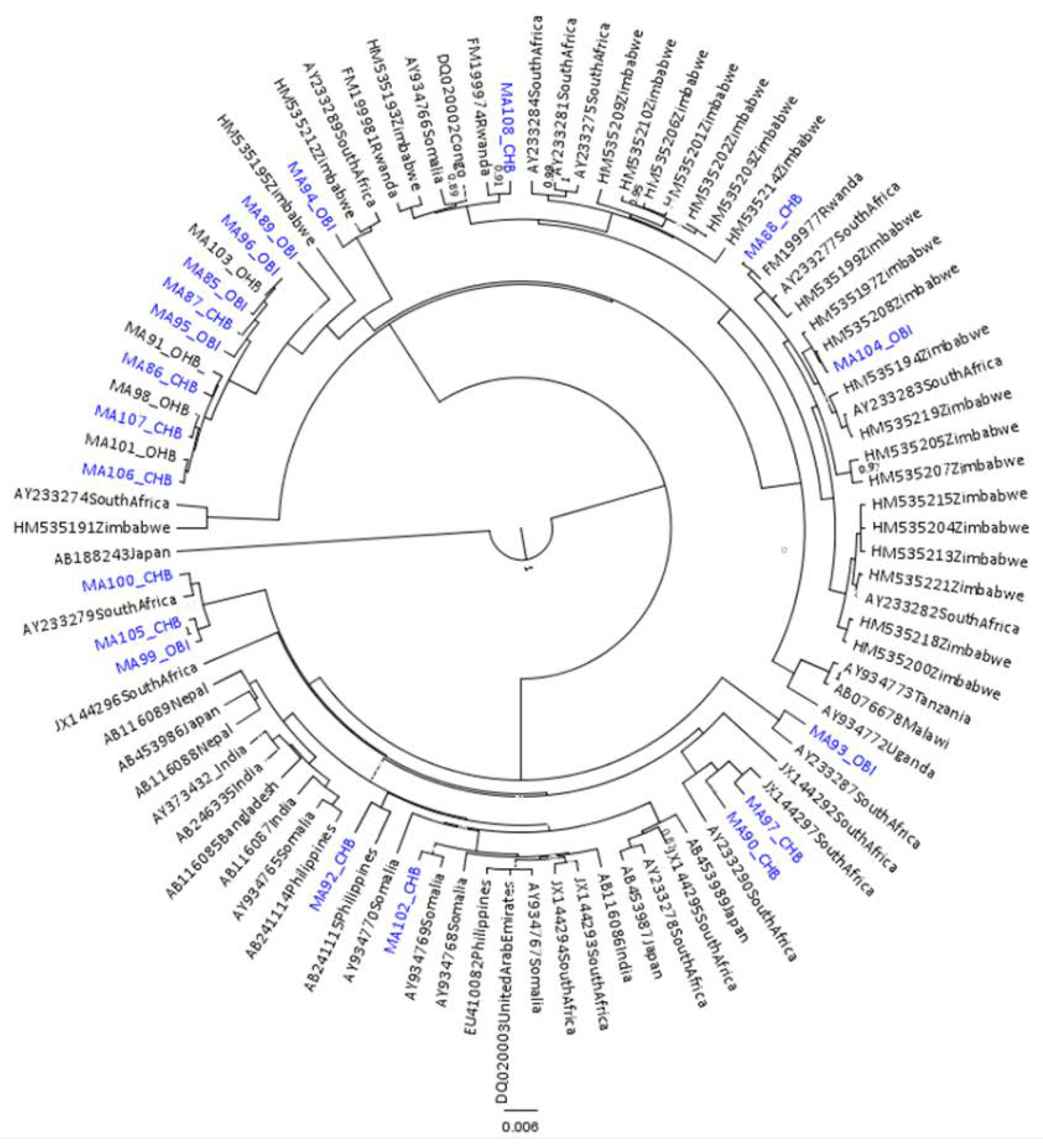
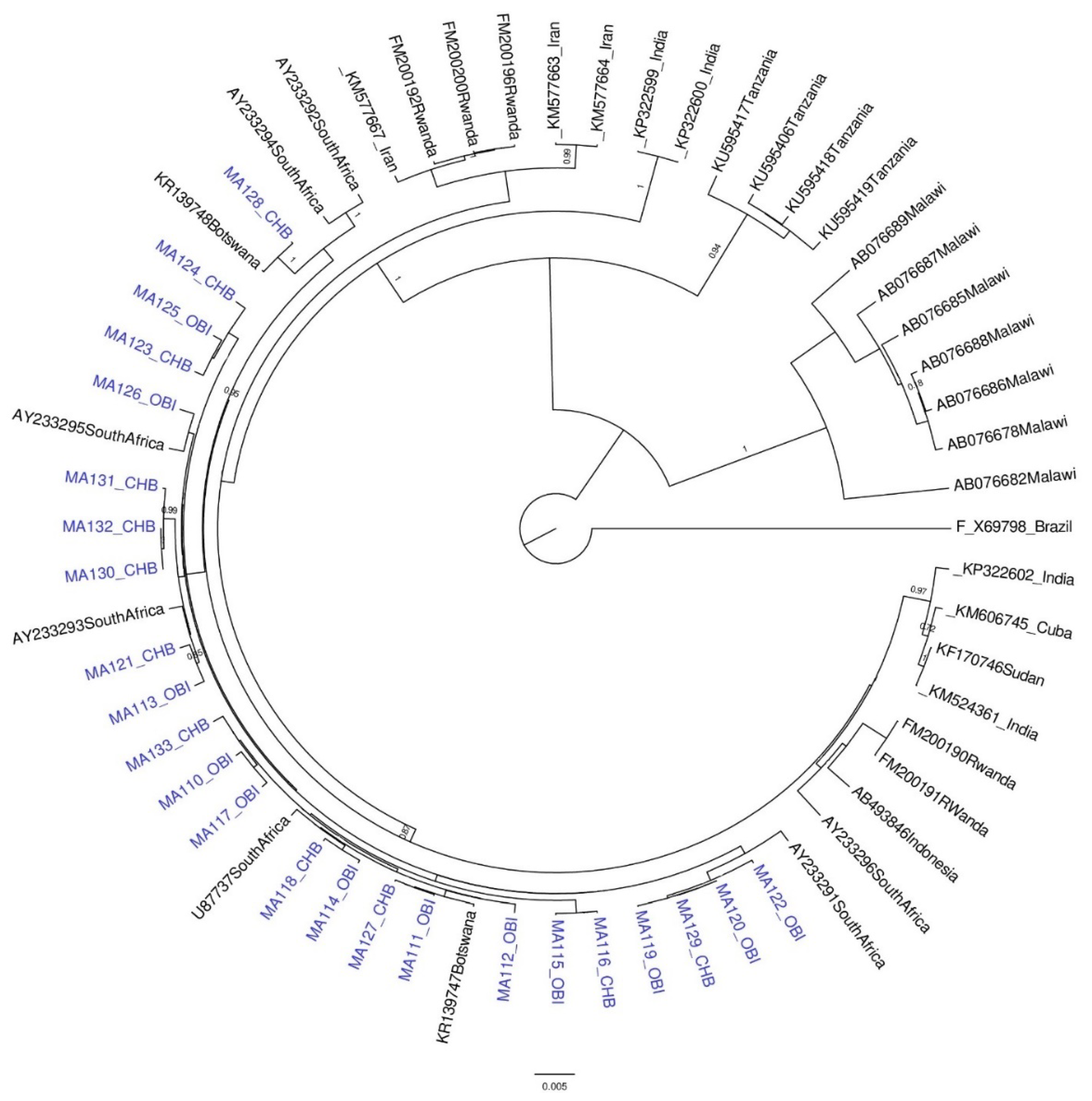
2.5. Phylogenetic Analysis
2.6. HBV Genotype Recombination Analysis
2.7. Analysis of Immune Selection Pressure, Signature Amino Acids, and Escape Mutations
2.8. Mutational Analysis
3. Results
3.1. Results for Subgenotypes A1 and D3
3.2. Nucleotide Divergence
3.3. HBV Escape Mutations in the S ORF of OBI Versus CHB Sequences
3.4. Occult-Unique Mutations
3.5. Signature Amino Acid Analysis
3.6. Immune Selection Pressure
4. Discussion
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- WHO. Hepatitis B. Available online: http://www.who.int/mediacentre/factsheets/fs204/en/ (accessed on 8 August 2017).
- Stanaway, J.D.; Flaxman, A.D.; Naghavi, M.; Fitzmaurice, C.; Vos, T.; Abubakar, I.; Abu-Raddad, L.J.; Assadi, R.; Bhala, N.; Cowie, B.; et al. The global burden of viral hepatitis from 1990 to 2013: Findings from the global burden of disease Study 2013. Lancet 2016, 388, 1081–1088. [Google Scholar] [CrossRef]
- WHO. Global Hepatitis Report. 2017. Available online: https://www.afro.who.int/sites/default/files/2017-06/9789241565455-eng.pdf (accessed on 12 February 2018).
- Matthews, P.C.; Beloukas, A.; Malik, A.; Carlson, J.M.; Jooste, P.; Ogwu, A.; Shapiro, R.; Riddel, L.; Chen, F.; Luzzi, G.; et al. Prevalence and characteristics of hepatitis B virus (HBV) coinfection among HIV-positive women in South Africa and Botswana. PLoS ONE 2015, 10. [Google Scholar] [CrossRef] [PubMed]
- Patel, P.; Davis, S.; Tolle, M.; Mabikwa, V.; Anabwani, G. Prevalence of hepatitis B and hepatitis C coinfections in an adult HIV centre population in Gaborone, Botswana. Am. J. Trop. Med. Hyg. 2011, 85, 390–394. [Google Scholar] [CrossRef] [PubMed]
- Anderson, M.; Gaseitsiwe, S.; Moyo, S.; Thami, K.P.; Mohammed, T.; Setlhare, D.; Sebunya, T.K.; Powell, E.A.; Makhema, J.; Blackard, J.T.; et al. Slow CD4+ T-Cell recovery in human immunodeficiency virus/hepatitis B virus-coinfected patients initiating truvada-based combination antiretroviral therapy in Botswana. Open Forum Infect. Dis. 2016, 3, ofw140. [Google Scholar] [CrossRef] [PubMed]
- Wester, C.W.; Bussmann, H.; Moyo, S.; Avalos, A.; Gaolathe, T.; Ndwapi, N.; Essex, M.; MacGregor, R.R.; Marlink, R. Serological evidence of HIV-associated infection among HIV-1-infected adults in Botswana. Clin. Infect. Dis. 2006, 43, 1612–1615. [Google Scholar] [CrossRef] [PubMed]
- Seeger, C.; Mason, W.S. Hepatitis B virus biology. Microbiol. Mol. Biol. Rev. 2000, 64, 51–68. [Google Scholar] [CrossRef] [PubMed]
- Chen, M.; Sälberg, M.; Hughes, J.; Jones, J.; Guidotti, L.G.; Chisari, F.V.; Billaud, J.N.; Milich, D.R. Immune tolerance split between hepatitis B virus precore and core proteins. J. Virol. 2005, 79, 3016–3027. [Google Scholar] [CrossRef] [PubMed]
- Glebe, D.; Urban, S.; Knoop, E.V.; Cag, N.; Krass, P.; Grün, S.; Bulavaite, A.; Sasnauskas, K.; Gerlich, W.H. Mapping of the hepatitis B virus attachment site by use of infection-inhibiting preS1 lipopeptides and tupaia hepatocytes. Gastroenterology 2005, 129, 234–245. [Google Scholar] [CrossRef] [PubMed]
- Pang, R.; Tse, E.; Poon, R.T. Molecular pathways in hepatocellular carcinoma. Cancer Lett. 2006, 240, 157–169. [Google Scholar] [CrossRef] [PubMed]
- Beck, J.; Nassal, M. Hepatitis B virus replication. World J. Gastroenterol. 2007, 13, 48–64. [Google Scholar] [CrossRef] [PubMed]
- Buti, M.; Rodriguez-Frias, F.; Jardi, R.; Esteban, R. Hepatitis B virus genome variability and disease progression: The impact of pre-core mutants and HBV genotypes. J. Clin. Virol. 2005, 34, S79–S82. [Google Scholar] [CrossRef]
- Orito, E.; Mizokami, M.; Ina, Y.; Moriyama, E.N.; Kameshima, N.; Yamamoto, M.; Gojobori, T. Host-independent evolution and a genetic classification of the hepadnavirus family based on nucleotide sequences. Proc. Natl. Acad. Sci. USA 1989, 86, 7059–7062. [Google Scholar] [CrossRef] [PubMed]
- Kramvis, A.; Kew, M.; François, G. Hepatitis B virus genotypes. Vaccine 2005, 23, 2409–2423. [Google Scholar] [CrossRef] [PubMed]
- Norder, H.; Courouce, A.M.; Coursaget, P.; Echevarria, J.M.; Lee, S.D.; Mushahwar, I.K.; Robertson, B.H.; Locarnini, S.; Magnius, L.O. Genetic diversity of hepatitis B virus strains derived worldwide: Genotypes, subgenotypes, and HBsAg subtypes. Intervirology 2004, 47, 289–309. [Google Scholar] [CrossRef] [PubMed]
- Tatematsu, K.; Tanaka, Y.; Kurbanov, F.; Sugauchi, F.; Mano, S.; Maeshiro, T.; Nakayoshi, T.; Miyakawa, Y.; Mizokami, M. A genetic variant of hepatitis B virus divergent from known human and ape genotypes isolated from a Japanese patient and provisionally assigned to new genotype J. J. Virol. 2009, 83, 10538–10547. [Google Scholar] [CrossRef] [PubMed]
- Kramvis, A. Genotypes and genetic variability of hepatitis B virus. Intervirology 2014, 57, 141–150. [Google Scholar] [CrossRef] [PubMed]
- Raimondo, G.; Allain, J.P.; Brunetto, M.R.; Buendia, M.A.; Chen, D.S.; Colombo, M.; Craxì, A.; Donato, F.; Ferrari, C.; Gaeta, G.B.; et al. Statements from the Taormina expert meeting on occult hepatitis B virus infection. J. Hepatol. 2008, 49, 652–657. [Google Scholar] [CrossRef] [PubMed]
- Liu, C.J.; Lo, S.C.; Kao, J.H.; Tseng, P.T.; Lai, M.Y.; Ni, Y.H.; Yeh, S.H.; Chen, P.J.; Chen, D.S. Transmission of occult hepatitis B virus by transfusion to adult and pediatric recipients in Taiwan. J. Hepatol. 2006, 44, 39–46. [Google Scholar] [CrossRef] [PubMed]
- Hoofnagle, J.H.; Seeff, L.B.; Bales, Z.B.; Zimmerman, H.J. Type B hepatitis after transfusion with blood containing antibody to hepatitis B core antigen. N. Engl. J. Med. 1978, 298, 1379–1383. [Google Scholar] [CrossRef] [PubMed]
- Shahmoradi, S.; Yahyapour, Y.; Mahmoodi, M.; Alavian, S.M.; Fazeli, Z.; Jazayeri, S.M. High prevalence of occult hepatitis B virus infection in children born to HBsAg-positive mothers despite prophylaxis with hepatitis B vaccination and HBIG. J. Hepatol. 2012, 57, 515–521. [Google Scholar] [CrossRef] [PubMed]
- Hu, L.P.; Liu, D.P.; Chen, Q.Y.; Harrison, T.J.; He, X.; Wang, X.Y.; Li, H.; Tan, C.; Yang, Q.L.; Li, K.W.; et al. Occult HBV Infection may be transmitted through close contact and manifest as an overt infection. PLoS ONE 2015, 10. [Google Scholar] [CrossRef] [PubMed]
- Rai, R.R.; Mathur, A.; Udawat, H.P.; Nepalia, S.; NijHawan, S.; Mathud, A. Prevalence of occult hepatitis B & C in HIV patients infected through sexual transmission. Trop. Gastroenterol. 2007, 28, 19–23. [Google Scholar] [PubMed]
- Singh, S.P.; Singh, S.K.; Misra, B.; Panigrahi, M.K.; Misra, D. A prospective study of prevalence of occult HBV infection and assessment of risk factors for HBV transmission in persons with occult HBV infection. J. Clin. Exp. Hepatol. 2013, 3, S69. [Google Scholar] [CrossRef]
- Blaich, A.; Manz, M.; Dumoulin, A.; Schüttler, C.G.; Hirsch, H.H.; Gerlich, W.H.; Frei, R. Reactivation of hepatitis B virus with mutated hepatitis B surface antigen in a liver transplant recipient receiving a graft from an antibody to hepatitis B surface antigen- and antibody to hepatitis B core antigen-positive donor. Transfusion 2012, 52, 1999–2006. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pollicino, T.; Squadrito, G.; Cerenzia, G.; Cacciola, I.; Raffa, G.; Craxi, A.; Farinati, F.; Missale, G.; Smedile, A.; Tiribelli, C.; et al. Hepatitis B virus maintains its pro-oncogenic properties in the case of occult HBV infection. Gastroenterology 2004, 126, 102–110. [Google Scholar] [CrossRef] [PubMed]
- Raimondo, G.; Caccamo, G.; Filomia, R.; Pollicino, T. Occult HBV infection. Semin. Immunopathol. 2013, 35, 39–52. [Google Scholar] [CrossRef] [PubMed]
- Samal, J.; Kandpal, M.; Vivekanandan, P. Molecular mechanisms underlying occult hepatitis B virus infection. Clin. Microbiol. Rev. 2012, 25, 142–163. [Google Scholar] [CrossRef] [PubMed]
- Mak, D.; de Villiers, C.B.; Chasela, C.; Urban, M.I.; Kramvis, A. Viral and non-viral risk factors associated with the development of hepatocellular carcinoma in black South Africans: 2000-2012. PLoS ONE 2018. [Google Scholar] [CrossRef] [PubMed]
- Blackberg, J.; Kidd-Ljunggren, K. Occult hepatitis B virus after acute self-limited infection persisting for 30 years without sequence variation. J. Hepatol. 2000, 33, 992–997. [Google Scholar] [CrossRef]
- Askari, A.; Hassanshahi, G.H.; Ghalebi, S.R.; Jafarzadeh, A.; Mohit, M.; Hajghani, M.; Arababadi, M.K. Intensity of HLA-A2 Expression Significantly Decreased in Occult Hepatitis B Infection. Jundishapur J. Microbiol. 2014, 7. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sagnelli, E.; Coppola, N.; Scolastico, C.; Filippini, P.; Santantonio, T.; Stroffolini, T.; Piccinino, F. Virologic and clinical expressions of reciprocal inhibitory effect of hepatitis B, C, and delta viruses in patients with chronic hepatitis. Hepatology 2000, 32, 1106–1110. [Google Scholar] [CrossRef] [PubMed]
- Mphahlele, M.J.; Lukhwareni, A.; Burnett, R.J.; Moropeng, L.M.; Ngobeni, J.M. High risk of occult hepatitis B virus infection in HIV-positive patients from South Africa. J. Clin. Virol. 2006, 35, 14–20. [Google Scholar] [CrossRef] [PubMed]
- Huang, F.Y.; Wong, D.K.H.; Seto, W.K.; Zhang, A.Y.; Lee, C.K.; Lin, C.K.; Fung, J.; Lai, C.L.; Yuen, M.F. Sequence variations of full-length hepatitis B virus genomes in Chinese patients with HBsAg-negative hepatitis B infection. PLoS ONE 2014, 9. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kim, H.; Gong, J.R.; Lee, S.A.; Kim, B.J. Discovery of a novel mutation (X8Del) resulting in an 8-bp deletion in the hepatitis B virus X gene associated with occult infection in Korean vaccinated individuals. PLoS ONE 2015, 10. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; Zhang, L.; Dai, Y.; Zhang, Y.; Li, J.; Li, X. Occult hepatitis B virus infection: Influence of S protein variants. Virol. J. 2016, 13, 10. [Google Scholar] [CrossRef] [PubMed]
- Kim, H.; Kim, B.J. Association of preS/S mutations with occult hepatitis B virus (HBV) infection in South Korea: Transmission potential of distinct occult HBV variants. Int. J. Mol. Sci. 2015, 16, 13595–13609. [Google Scholar] [CrossRef] [PubMed]
- Kim, H.; Lee, S.A.; Wom, Y.S.; Lee, H.J.; Kim, B.J. Occult infection related hepatitis B surface antigen variants showing lowered secretion capacity. World J. Gastroenterol. 2015, 21, 1794–1803. [Google Scholar] [CrossRef] [PubMed]
- Powell, E.A.; Boyce, C.L.; Gededzha, M.P.; Selabe, S.G.; Mphahlele, M.J.; Blackard, J.T. Functional analysis of “a” determinant mutations associated with occult HBV in HIV-positive South Africans. J. Gen. Virol. 2016, 1615–1624. [Google Scholar] [CrossRef] [PubMed]
- Powell, E.A.; Gededzha, M.P.; Rentz, M.; Rakgole, N.J.; Selabe, S.G.; Seleise, T.A.; Mphahlele, M.J.; Blackard, J.T. Mutations associated with occult hepatitis B in HIV-positive South Africans. J. Med. Virol. 2015, 87, 388–400. [Google Scholar] [CrossRef] [PubMed]
- Martin, C.M.; Welge, J.A.; Rouster, S.D.; Shata, M.T.; Sherman, K.E.; Blackard, J.T. Mutations associated with occult hepatitis B virus infection result in decreased surface antigen expression in vitro. J. Viral Hepat. 2012, 19, 716–723. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zoulim, F.; Locarnini, S. Hepatitis B virus resistance to nucleos(t)ide analogues. Gastroenterology 2009, 137, 1593–1608. [Google Scholar] [CrossRef] [PubMed]
- Fang, Y.; Teng, X.; Xu, W.Z.; Li, D.; Zhao, H.W.; Fu, L.J.; Zhang, F.M.; Gu, H.X. Molecular characterization and functional analysis of occult hepatitis B virus infection in Chinese patients infected with genotype C. J. Med. Virol. 2009, 81, 826–835. [Google Scholar] [CrossRef] [PubMed]
- WHO. Draft Global Health Sector Strategies. Available online: http://apps.who.int/gb/ebwha/pdf_files/WHA69/A69_32-en.pdf?ua=1 (accessed on 20 March 2018).
- Yuan, Q.; Ou, S.H.; Chen, C.R.; Ge, S.X.; Pei, B.; Chen, Q.R.; Yan, Q.; Lin, Y.C.; Ni, H.Y.; Huang, C.H.; et al. Molecular characteristics of occult hepatitis B virus from blood donors in Southeast China. J. Clin. Microbiol. 2010, 48, 357–362. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chaudhuri, V.; Tayal, R.; Nayak, B.; Acharya, S.K.; Panda, S.K. Occult hepatitis B virus infection in chronic liver disease: Full-length genome and analysis of mutant surface promoter. Gastroenterology 2004, 127, 1356–1371. [Google Scholar] [CrossRef] [PubMed]
- Ryan, K.; Anderson, M.; Gyurova, I.; Ambroggio, L.; Moyo, S.; Sebunya, T.; Makhema, J.; Marlink, R.; Essex, M.; Musonda, R.; et al. High rates of occult hepatitis B virus infection in HIV-positive individuals initiating antiretroviral therapy in Botswana. Open Forum Infect. Dis. 2017, 4. [Google Scholar] [CrossRef] [PubMed]
- Chaudhury, S.; Williams, P.L.; Mayondi, G.K.; Leidner, J.; Holding, P.; Tepper, V.; Nichols, S.; Magetse, J.; Sakoi, M.; Moabi, K.; et al. Neurodevelopment of HIV-exposed and HIV-unexposed uninfected children at 24 months. Pediatrics 2017, 140. [Google Scholar] [CrossRef] [PubMed]
- Mbangiwa, T.; Kasvosve, I.; Anderson, M.; Thami, P.K.; Choga, W.T.; Needleman, A.; Phinius, B.B.; Moyo, S.; Leteane, M.; Leidner, J.; et al. Chronic and occult hepatitis B virus infection in pregnant women in Botswana. Genes 2018, 9, 259. [Google Scholar] [CrossRef] [PubMed]
- Gunther, S.; Li, B.C.; Miska, S.; Krüger, D.H.; Meisel, H.; Will, H. A novel method for efficient amplification of whole hepatitis B virus genomes permits rapid functional analysis and reveals deletion mutants in immunosuppressed patients. J. Virol. 1995, 69, 5437–5444. [Google Scholar] [PubMed]
- Zahn, A.; Li, C.; Danso, K.; Candotti, D.; Owusu-Ofori, S.; Temple, J.; Allain, J.P. Molecular characterization of occult hepatitis B virus in genotype E-infected subjects. J. Gen. Virol. 2008, 89, 409–418. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Candotti, D.; Opare-Sem, O.; Rezvan, H.; Sarkodie, F.; Allain, J.P. Molecular and serological characterization of hepatitis B virus in deferred Ghanaian blood donors with and without elevated alanine aminotransferase. J. Viral. Hepat. 2006, 13, 715–724. [Google Scholar] [CrossRef] [PubMed]
- Kuwahara, R.; Kumashiro, R.; Murashima, S.; Ogata, K.; Tanaka, K.; Hisamochi, A.; Hino, T.; Ide, T.; Tanaka, E.; Koga, Y.; et al. Genetic heterogeneity of the precore and the core promoter region of genotype C hepatitis B virus during lamivudine therapy. J. Med. Virol. 2004, 72, 26–34. [Google Scholar] [CrossRef] [PubMed]
- Maponga, T.G. An investigation of hepatitis B virus in antenatal women tested for human immunodeficiency virus, in the Western Cape Province of South Africa. Master’s Thesis, University of Stellenbosch, Stellenbosch, South Africa, 2012. [Google Scholar]
- Werle, B.; Cinquin, K.; Marcellin, P.; Pol, S.; Maynard, M.; Trépo, C.; Zoulim, F. Evolution of hepatitis B viral load and viral genome sequence during adefovir dipivoxil therapy. J. Viral. Hepat. 2004, 11, 74–83. [Google Scholar] [CrossRef] [PubMed]
- Drummond, A.J.; Rambaut, A. BEAST: Bayesian evolutionary analysis by sampling trees. BMC Evol. Biol. 2007, 7. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lole, K.S.; Bollinger, R.C.; Paranjape, R.S.; Gadkari, D.; Kulkarni, S.S.; Novak, N.G.; Ingersoll, R.; Sheppard, H.W.; Ray, S.C. Full-length human immunodeficiency virus type 1 genomes from subtype C-infected seroconverters in India, with evidence of intersubtype recombination. J. Virol. 1999, 73, 152–160. [Google Scholar] [PubMed]
- Poon, A.F.; Frost, S.D.; Pond, S.L. Detecting signatures of selection from DNA sequences using Datamonkey. Methods Mol. Biol. 2009, 537, 163–183. [Google Scholar] [PubMed]
- Los Alamos National Laboratoy, VESPA. Available online: https://www.hiv.lanl.gov/content/sequence/VESPA/vespa.html (accessed on 30 May 2017).
- Bell, T.G.; Yousif, M.; Kramvis, A. Bioinformatic curation and alignment of genotyped hepatitis B virus (HBV) sequence data from the GenBank public database. Springerplus 2016, 5. [Google Scholar] [CrossRef] [PubMed]
- Bell, T.G.; Kramvis, A. Bioinformatics tools for small genomes, such as hepatitis B virus. Viruses 2015, 7, 781–797. [Google Scholar] [CrossRef] [PubMed]
- Yousif, M.; Kramvis, A. Genotype D of hepatitis B virus and its subgenotypes: An update. Hepatol. Res. 2013, 43, 355–364. [Google Scholar] [CrossRef] [PubMed]
- Zhu, H.L.; Li, X.; Zhang, Z.H. Genetic variation of occult hepatitis B virus infection. World J. Gastroenterol. 2016, 22, 3531–3546. [Google Scholar] [CrossRef] [PubMed]
- Martin, C.M.; Welge, J.A.; Shire, N.J.; Rouster, S.D.; Shata, M.T.; Sherman, K.E.; Blackard, J.T. Genomic variability associated with the presence of occult hepatitis B virus in HIV co-infected individuals. J. Viral Hepat. 2010, 17, 588–597. [Google Scholar] [CrossRef] [PubMed]
- Panigrahi, R.; Biswas, A.; Datta, S.; Banerjee, A.; Chandra, P.K.; Mahapatra, P.K.; Patnaik, B.; Chakrabarti, S.; Chakravarty, R. Anti-hepatitis B core antigen testing with detection and characterization of occult hepatitis B virus by an in-house nucleic acid testing among blood donors in Behrampur, Ganjam, Orissa in southeastern India: Implications for transfusion. Virol. J. 2010, 7. [Google Scholar] [CrossRef] [PubMed]
- Chen, S.J.; Zhao, Y.X.; Fang, Y.; Xu, W.Z.; Ma, Y.X.; Song, Z.W.; Teng, X.; Gu, H.X. Viral deletions among healthy young Chinese adults with occult hepatitis B virus infection. Virus Res. 2012, 163, 197–201. [Google Scholar] [CrossRef] [PubMed]
- Hass, M.; Hannoun, C.; Kalinina, T.; Sommer, G.; Manegold, C.; Günther, S. Functional analysis of hepatitis B virus reactivating in hepatitis B surface antigen-negative individuals. Hepatology 2005, 42, 93–103. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Garcia-Montalvo, B.M.; Ventura-Zapata, L.P. Molecular and serological characterization of occult hepatitis B infection in blood donors from Mexico. Ann. Hepatol. 2011, 10, 133–141. [Google Scholar] [PubMed]
- Kimbi, G.C.; Kramvis, A.; Kew, M.C. Distinctive sequence characteristics of subgenotype A1 isolates of hepatitis B virus from South Africa. J. Gen. Virol. 2004, 85, 1211–1220. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bowyer, S.M.; van Staden, L.; Kew, M.C.; Sim, J.G. A unique segment of the hepatitis B virus group A genotype identified in isolates from South Africa. J. Gen. Virol. 1997, 78, 1719–1729. [Google Scholar] [CrossRef] [PubMed]
- Tamura, K.; Kumar, S. Evolutionary distance estimation under heterogeneous substitution pattern among lineages. Mol. Biol. Evol. 2002, 19, 1727–1736. [Google Scholar] [CrossRef] [PubMed]
- Echevarria, J.M.; Avellón, A. Hepatitis B virus genetic diversity. J. Med. Virol. 2006, 78, S36–S42. [Google Scholar] [CrossRef] [PubMed]
- Yong-Lin, Y.; Qiang, F.; Ming-Shun, Z.; Jie, C.; Gui-Ming, M.; Zu-Hu, H.; Xu-Bing, C. Hepatitis B surface antigen variants in voluntary blood donors in Nanjing, China. Virol. J. 2012, 9. [Google Scholar] [CrossRef] [PubMed]
- Huang, C.H.; Yuan, Q.; Chen, P.J.; Zhang, Y.L.; Chen, C.R.; Zheng, Q.B.; Yeh, S.H.; Yu, H.; Xue, Y.; Chen, Y.X.; et al. Influence of mutations in hepatitis B virus surface protein on viral antigenicity and phenotype in occult HBV strains from blood donors. J. Hepatol. 2012, 57, 720–729. [Google Scholar] [CrossRef] [PubMed]
- Kwei, K.; Tang, X.; Lok, A.S.; Sureau, C.; Tamako, G.; Li, J.; Wands, J.; Tong, S. Impaired virion secretion by hepatitis B virus immune escape mutants and its rescue by wild-type envelope proteins or a second-site mutation. J. Virol. 2013, 87, 2352–2357. [Google Scholar] [CrossRef] [PubMed]
- Wu, C.; Zhang, X.; Tian, Y.; Song, J.; Yang, D.; Roggendorf, M.; Lu, M.; Chen, X. Biological significance of amino acid substitutions in hepatitis B surface antigen (HBsAg) for glycosylation, secretion, antigenicity and immunogenicity of HBsAg and hepatitis B virus replication. J. Gen. Virol. 2010, 91, 483–492. [Google Scholar] [CrossRef] [PubMed]
- Ma, Q.; Wang, Y. Comprehensive analysis of the prevalence of hepatitis B virus escape mutations in the major hydrophilic region of surface antigen. J. Med. Virol. 2012, 84, 198–206. [Google Scholar] [CrossRef] [PubMed]
- Avellón, A.; Echevarria, J.M. Frequency of hepatitis B virus ’a’ determinant variants in unselected Spanish chronic carriers. J. Med. Virol. 2006, 78, 24–36. [Google Scholar] [CrossRef] [PubMed]
- Anderson, M.; Gaseitsiwe, S.; Moyo, S.; Wessels, M.J.C.; Mohammed, T.; Sebunya, T.K.; Powell, E.A.; Makhema, J.; Blackard, J.T.; Marlink, R.; et al. Molecular characterisation of hepatitis B virus in HIV-1 subtype C infected patients in Botswana. BMC Infect. Dis. 2015, 15. [Google Scholar] [CrossRef] [PubMed]
- Makondo, E.; Bell, T.G.; Kramvis, A. Genotyping and molecular characterization of hepatitis B virus from human immunodeficiency virus-infected individuals in southern Africa. PLoS ONE 2012, 7. [Google Scholar] [CrossRef] [PubMed]
- Taffon, S.; Genovese, D.; Blasi, M.; Pierotti, P.; Degli Esposti, A.; Catone, S.; Chionne, P.; Pulimanti, B.; Candido, A.; Dettori, S. HBV whole-genome mutation profile in HIV-1/HBV coinfected patients in a long-term follow-up study. Infection 2014, 42, 675–687. [Google Scholar] [CrossRef] [PubMed]
- Allain, J.P.; Belkhiri, D.; Vermeulen, M.; Crookes, R.; Cable, R.; Amiri, A.; Reddy, R.; Bird, A.; Candotti, D. Characterization of occult hepatitis B virus strains in South African blood donors. Hepatology 2009, 49, 1868–1876. [Google Scholar] [CrossRef] [PubMed]
- Mondal, R.K.; Khatun, M.; Banerjee, P.; Ghosh, A.; Sarkar, S.; Santra, A.; Das, K.; Chowdhury, A.; Banerjee, S.; Datta, S. Synergistic impact of mutations in Hepatitis B Virus genome contribute to its occult phenotype in chronic hepatitis C virus carriers. Sci. Rep. 2017, 7. [Google Scholar] [CrossRef] [PubMed]
- Qiu, J.; Quin, B.; Rayner, S.; Wu, C.C.; Pei, R.J.; Xu, S.; Wang, Y.; Chen, X.W. Novel evidence suggests hepatitis B virus surface proteins participate in regulation of HBV genome replication. Virol. Sin. 2011, 26, 131–138. [Google Scholar] [CrossRef] [PubMed]
- Wu, C.; Deng, W.; Deng, L.; Cao, L.; Qin, B.; Li, S.; Wang, Y.; Pei, R.; Yang, D.; Lu, M.; et al. Amino acid substitutions at positions 122 and 145 of hepatitis B virus surface antigen (HBsAg) determine the antigenicity and immunogenicity of HBsAg and influence in vivo HBsAg clearance. J. Virol. 2012, 86, 4658–4669. [Google Scholar] [CrossRef] [PubMed]
- Tian, Y.; Xu, Y.; Zhang, Z.; Meng, Z.; Qin, L.; Lu, M.; Yang, D. The amino Acid residues at positions 120 to 123 are crucial for the antigenicity of hepatitis B surface antigen. J. Clin. Microbiol. 2007, 45, 2971–2978. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.H.; Jung, Y.K.; Joo, M.K.; Kim, J.H.; Yim, H.J.; Park, J.J.; Kim, J.S.; Bak, Y.T.; Yeon, J.E.; Byun, K.S. Hepatitis B viral surface mutations in patients with adefovir resistant chronic hepatitis B with A181T/V polymerase mutations. J. Korean Med. Sci. 2010, 25, 257–264. [Google Scholar] [CrossRef] [PubMed]
- Reuter, S.; Oette, M.; Wilhelm, F.C.; Beggel, B.; Kaiser, R.; Balduin, M.; Schweitzer, F.; Verheyen, J.; Adams, O.; Lengauer, T.; et al. Prevalence and characteristics of hepatitis B and C virus infections in treatment-naive HIV-infected patients. Med. Microbiol. Immunol. 2011, 200, 39–49. [Google Scholar] [CrossRef] [PubMed]
- Coppola, N.; Onorato, L.; Iodice, V.; Starace, M.; Minichini, C.; Farella, N.; Liorre, G.; Filippini, P.; Sagnelli, E.; de Stefano, G. Occult HBV infection in HCC and cirrhotic tissue of HBsAg-negative patients: A virological and clinical study. Oncotarget 2016, 7, 62706–62714. [Google Scholar] [CrossRef] [PubMed]
- Jeantet, D.; Chemin, I.; Mandrand, B.; Zoulin, F.; Trepo, C.; Kay, A. Characterization of two hepatitis B virus populations isolated from a hepatitis B surface antigen-negative patient. Hepatology 2002, 35, 1215–1224. [Google Scholar] [CrossRef] [PubMed]
- Amponsah-Dacosta, E.; Lebelo, R.L.; Rakgole, J.N.; Selabe, S.G.; Gededzha, M.P.; Mayaphi, S.H.; Powell, E.A.; Blackard, J.T.; Mphahlele, M.J. Hepatitis B virus infection in post-vaccination South Africa: Occult HBV infection and circulating surface gene variants. J. Clin. Virol. 2015, 63, 12–17. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lazarevic, I. Clinical implications of hepatitis B virus mutations: Recent advances. World J. Gastroenterol. 2014, 20, 7653–7664. [Google Scholar] [CrossRef] [PubMed]
- Gerlich, W.H.; Glebe, D.; Schüttler, C.G. Deficiencies in the standardization and sensitivity of diagnostic tests for hepatitis B virus. J. Viral. Hepat. 2007, 14, 16–21. [Google Scholar] [CrossRef] [PubMed]
- Ireland, J.H.; O’Donnell, B.; Basuni, A.A.; Kean, J.D.; Wallace, L.A.; Lau, G.K.; Carman, W.F. Reactivity of 13 in vitro expressed hepatitis B surface antigen variants in 7 commercial diagnostic assays. Hepatology 2000, 31, 1176–1182. [Google Scholar] [CrossRef] [PubMed]
- Pinarbasi, B.; Onel, D.; Cosan, F.; Akyuz, F.; Dirlik, N.; Cakaloglu, Y.; Badur, S.; Besisik, F.; Demir, K.; Okten, A.; et al. Prevalence and virological features of occult hepatitis B virus infection in female sex workers who work uncontrolled in Turkey. Liver Int. 2009, 29, 227–230. [Google Scholar] [CrossRef] [PubMed]
- Sengupta, S.; Rehman, S.; Durgapal, H.; Acharya, S.K.; Panda, S.K. Role of surface promoter mutations in hepatitis B surface antigen production and secretion in occult hepatitis B virus infection. J. Med. Virol. 2007, 79, 220–228. [Google Scholar] [CrossRef] [PubMed]
- Kim, H.; Lee, S.A.; Kim, D.W.; Lee, S.H.; Kim, B.J. Naturally occurring mutations in large surface genes related to occult infection of hepatitis B virus genotype C. PLoS ONE 2013, 8. [Google Scholar] [CrossRef] [PubMed]
- Pollicino, T.; Raffa, G.; Costantino, L.; Lisa, A.; Campello, C.; Squadrito, G.; Levrero, M.; Raimondo, G. Molecular and functional analysis of occult hepatitis B virus isolates from patients with hepatocellular carcinoma. Hepatology 2007, 45, 277–285. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gopalakrishnan, D.; Keyter, M.; Shenoy, K.T.; Leena, K.B.; Thayumanavan, T.; Thomas, V.; Vinayakumar, K.R.; Panackel, C.; Korah, A.T.; Nair, R.; et al. Hepatitis B virus subgenotype A1 predominates in liver disease patients from Kerala, India. World J. Gastroenterol. 2013, 19, 9294–9306. [Google Scholar] [CrossRef] [PubMed]
- Hao, Z.; Zheng, L.; Kluwe, L.; Huang, W. Ferritin light chain and squamous cell carcinoma antigen 1 are coreceptors for cellular attachment and entry of hepatitis B virus. Int. J. Nanomed. 2012, 7, 827–834. [Google Scholar] [CrossRef] [PubMed]
- Pollicino, T.; Cacciola, I.; Saffoti, F.; Raimondo, G. Hepatitis B virus PreS/S gene variants: Pathobiology and clinical implications. J. Hepatol. 2014, 61, 408–417. [Google Scholar] [CrossRef] [PubMed]
- Burley, S.K.; Petsko, G.A. Aromatic-aromatic interaction: A mechanism of protein structure stabilization. Science 1985, 229, 23–28. [Google Scholar] [CrossRef] [PubMed]
- Datta, S.; Ghosh, A.; Dasgupta, D.; Gosh, A.; Roychoudhury, S.; Roy, G.; Das, S.; Gupta, S.; Basu, K.; Basu, A.; et al. Novel point and combo-mutations in the genome of hepatitis B virus-genotype D: Characterization and impact on liver disease progression to hepatocellular carcinoma. PLoS ONE 2014, 9. [Google Scholar] [CrossRef] [PubMed]
- Zhand, S.; Tabarraei, A.; Nazari, A.; Moradi, A. Cytotoxic T lymphocytes and CD4 epitope mutations in the pre-core/core region of hepatitis B virus in chronic hepatitis B carriers in Northeast Iran. Indian J. Gastroenterol. 2017, 36, 253–257. [Google Scholar] [CrossRef] [PubMed]
- Berke, J.M.; Tan, Y.; Verbinnen, T.; Dehertogh, P.; Vergauwen, K.; Vos, A.; Lenz, O.; Pauwels, F. Antiviral profiling of the capsid assembly modulator BAY41-4109 on full-length HBV genotype A-H clinical isolates and core site-directed mutants in vitro. Antivir. Res. 2017, 144, 205–215. [Google Scholar] [CrossRef] [PubMed]
- Banerjee, A.; Banarjee, S.; Chowdhury, A.; Santra, A.; Chowdhury, S.; Roychowdhury, S.; Panda, C.K.; Bhattacharya, S.K.; Chakravarty, R. Nucleic acid sequence analysis of basal core promoter/precore/core region of hepatitis B virus isolated from chronic carriers of the virus from Kolkata, eastern India: Low frequency of mutation in the precore region. Intervirology 2005, 48, 389–399. [Google Scholar] [CrossRef] [PubMed]
- Ponsel, D.; Bruss, V. Mapping of amino acid side chains on the surface of hepatitis B virus capsids required for envelopment and virion formation. J. Virol. 2003, 77, 416–422. [Google Scholar] [CrossRef] [PubMed]
- Wu, S.; Zhao, Q.; Zhang, P.; Kulp, J.; Hu, L.; Hwang, N.; Zhang, J.; Block, T.M.; Xu, X.; Du, Y.; et al. Discovery and Mechanistic Study of Benzamide Derivatives That Modulate Hepatitis B Virus Capsid Assembly. J. Virol. 2017, 91. [Google Scholar] [CrossRef] [PubMed]
- Utama, A.; Purwantomo, S.; Siburian, M.D.; Dhenni, R.; Gani, R.A.; Hasan, I.; Sanityoso, A.; Miskad, U.A.; Akil, F.; Yusuf, I.; et al. Hepatitis B virus subgenotypes and basal core promoter mutations in Indonesia. World J. Gastroenterol. 2009, 15, 4028–4036. [Google Scholar] [CrossRef] [PubMed]
- Khattar, E.; Mukherji, A.; Kumar, V. Akt augments the oncogenic potential of the HBx protein of hepatitis B virus by phosphorylation. FEBS J. 2012, 279, 1220–1230. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kim, H.; Jee, Y.M.; Song, B.C.; Hyun, J.W.; Mun, H.S.; Kim, H.J.; Oh, E.J.; Yoon, J.H.; Kim, Y.J.; Lee, H.S.; et al. Analysis of hepatitis B virus quasispecies distribution in a Korean chronic patient based on the full genome sequences. J. Med. Virol. 2007, 79, 212–219. [Google Scholar] [CrossRef] [PubMed]
- Saha, D.; Pal, A.; Sarkar, N.; Das, D.; Blackard, J.T.; Guha, S.K.; Saha, B.; Chakravarty, R. Occult hepatitis B virus infection in HIV positive patients at a tertiary healthcare unit in eastern India. PLoS ONE 2017, 12. [Google Scholar] [CrossRef] [PubMed]
- Sheldon, J.; Soriano, V. Hepatitis B virus escape mutants induced by antiviral therapy. J. Antimicrob. Chemother. 2008, 61, 766–768. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Selabe, S.G.; Lukhwareni, A.; Song, E.; Leeuw, Y.G.; Burnett, R.J.; Mphahlele, M.J. Mutations associated with lamivudine-resistance in therapy-naive hepatitis B virus (HBV) infected patients with and without HIV co-infection: Implications for antiretroviral therapy in HBV and HIV co-infected South African patients. J. Med. Virol. 2007, 79, 1650–1654. [Google Scholar] [CrossRef] [PubMed]
- Mahabadi, M.; Norouzi, M.; Alavian, S.M.; Samimirad, K.; Azad, T.M.; Saberfar, E.; Mahmoodi, M.; Ramezani, F.; Karimzadeh, H.; Malekzadeh, R.; et al. Drug-related mutational patterns in hepatitis B virus (HBV) reverse transcriptase proteins from Iranian treatment-naive chronic HBV patients. Hepat. Mon. 2013, 13. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vincenti, D.; Solmone, M.; Garbuglia, A.R.; Iacomi, F.; Capobianchi, M.R. A sensitive direct sequencing assay based on nested PCR for the detection of HBV polymerase and surface glycoprotein mutations. J. Virol. Methods. 2009, 159, 53–57. [Google Scholar] [CrossRef] [PubMed]
- Panigrahi, R.; Biswas, A.; De, B.K.; Chakrabarti, S.; Chakravarty, R. Characterization of antiviral resistance mutations among the Eastern Indian Hepatitis B virus infected population. Virol. J. 2013, 10. [Google Scholar] [CrossRef] [PubMed]
- Santantonio, T.; Fasano, M.; Durantel, S.; Barraud, L.; Heichen, M.; Guastadisegni, A.; Pastore, G.; Zoulim, F. Adefovir dipivoxil resistance patterns in patients with lamivudine-resistant chronic hepatitis B. Antivir. Ther. 2009, 14, 557–565. [Google Scholar] [PubMed]
- Ciftci, S.; Keskin, F.; Cakiris, A.; Akyuz, F.; Pinarbasi, B.; Abaci, N.; Dincer, E.; Badur, S.; Kaymakogly, S.; Ustek, D. Analysis of potential antiviral resistance mutation profiles within the HBV reverse transcriptase in untreated chronic hepatitis B patients using an ultra-deep pyrosequencing method. Diagn. Microbiol. Infect. Dis. 2014, 79, 25–30. [Google Scholar] [CrossRef] [PubMed]
- Villet, S.; Pichoud, C.; Villeneuve, J.P.; Trépo, C.; Zoulim, F. Selection of a multiple drug-resistant hepatitis B virus strain in a liver-transplanted patient. Gastroenterology 2006, 131, 1253–1261. [Google Scholar] [CrossRef] [PubMed]
- Sunbul, M. Hepatitis B virus genotypes: Global distribution and clinical importance. World J. Gastroenterol. 2014, 20, 5427–5434. [Google Scholar] [CrossRef] [PubMed]
- Stemler, M.; Weimer, T.; Tu, Z.X.; Wan, D.F.; Levrero, M.; Jung, C.; Pape, G.R.; Will, H. Mapping of B-cell epitopes of the human hepatitis B virus X protein. J. Virol. 1990, 64, 2802–2809. [Google Scholar] [PubMed]
- Jung, M.C.; Diepolder, H.M.; Pape, G.R. T cell recognition of hepatitis B and C viral antigens. Eur. J. Clin. Investig. 1994, 24, 641–650. [Google Scholar] [CrossRef] [Green Version]
- Mizukoshi, E.; Sidney, J.; Livingston, B.; Ghany, M.; Hoofnagle, J.H.; Sette, A.; Rehermann, B. Cellular immune responses to the hepatitis B virus polymerase. J. Immunol. 2004, 173, 5863–5871. [Google Scholar] [CrossRef] [PubMed]
- Zu Putlitz, J.; Landford, R.E.; Carlson, R.I.; Notvall, L.; de la Monte, S.M.; Wands, J.R. Properties of monoclonal antibodies directed against hepatitis B virus polymerase protein. J. Virol. 1999, 73, 4188–4196. [Google Scholar] [PubMed]
- Lin, Y.M.; Jow, G.M.; Mu, S.C.; Chen, B.F. Naturally occurring hepatitis B virus B-cell and T-cell epitope mutants in hepatitis B vaccinated children. Sci. World J. 2013, 2013. [Google Scholar] [CrossRef] [PubMed]
| Primer Name | Primer Sequence | Coordinates from EcoRI Site | Reference |
|---|---|---|---|
| P1 | 5′CCGGAAAGCTTGAGCTCTTCTTTTTCACCTCTGCCTAATCA 3′ | 1821–1841 | [51] |
| P2 | 5′CCGGAAAGCTTGAGCTCTTCAAAAAGTTGCATGGTGCTGG 3′ | 1823–1806 | [51] |
| P3WRS | 5′CTACTGTTCAAGCCTCCAAGC 3′ | [52] | |
| P4WRS | 5′CGCAGACCAATTTATGCCTAC 3′ | [52] | |
| KU1 | 5′CATAAGAGGACTCTTGGACT 3′ | 1653–1672 | [54] |
| KU2 | 5′AATGTCAACGACCGACCTTG 3′ | 1679–1698 | [54] |
| MA3 | 5′GAAAGAAGTCAGAAGGCAAA 3′ | 1973–1954 | [53] |
| HBV Z | 5′AGCCCTCAGGCTCAGGGCATA 3′ | 1179–1199 | [55] |
| HBV 3 | 5′CGTTGCCKDGCAACSGGGTAAAGG 3′ | 2478–2455 | [55] |
| HBV M | 5′GACACACTTTCCAATCAATNGG 3′ | 2306–2287 | [55] |
| HBV P | 5′TCATCCTCAGGCCATGCAGT 3′ | 1292–1311 | [55] |
| HBV H | 5′TATCAAGGAATTCTGCCCGTTTGTCCT 3′ | 1767–1793 | [55] |
| HBV N | 5′ACTGAGCCAGGAGAAACGGACTGAGGC 3′ | 1991–1965 | [55] |
| Werle AS | 5′CGTCAGCAAACACTTGGC 3′ | 1175–1192 | [56] |
| CoreF | 5′GTGTGGATTCGCACTCCT 3′ | 2269–2287 | [41] |
| P6 | 5′GGCAGGTCCCCTAGAAGAAGAACT 3′ | 2363–2386 | [51] |
| ORF | CHB n = 12 | OBI n = 12 | p-Value | CHB n = 12 | OBI n = 12 | p-Value |
|---|---|---|---|---|---|---|
| Subgenotype A1 (n = 24) Median (Q1, Q3) | Subgenotype D3 (n = 24) Median (Q1, Q3) | |||||
| Pol | 2.8 (1.8, 3.4) | 0.5 (0.0, 1.4) | <0.001 | 0.7 (0.4, 0.9) | 0.4 (0.3, 0.5) | <0.001 |
| S | 1.2 (0.6, 2.1) | 0.2 (0.1, 0.8) | <0.001 | 0.3 (0.1, 0.9) | 0.1 (0.0, 0.3) | <0.001 |
| PreS1 | 3.2 (2.0, 5.5) | 0.9 (0.0, 2.7) | <0.001 | 1.1 (0.6, 1.1) | 0.3 (0.0, 1.7) | 0.001 |
| PreS2 | 0.6 (0.0, 0.6) | 0.0 (0.0, 1.2) | 0.387 | 0.6 (0.0, 0.6) | 0.0 (0.0, 1.2) | 0.387 |
| X | 1.5 (1.1, 1.8) | 11.8 (8.9, 23.3) | <0.001 | 11.5 (1.9, 13.4) | 8.6 (2.0,11.5) | 0.103 |
| Core | 4.0 (2.3, 5.8) | 6.7 (2.8, 15.5) | <0.001 | 0.9 (0.5–2.7) | 1.1 (0.7, 1.7) | <0.001 |
| Mutation | HBV Type | Reported Impact | References |
|---|---|---|---|
| SP120S | OBI | Vaccine, detection escape | [73] |
| SK122R | OBI | Decreased HBsAg expression, diagnostic escape | [42,74] |
| SC124Y | OBI | Immunoglobulin therapy escape, decreases antigenicity and viral secretion | [73,75] |
| SQ129H | OBI | Vaccine and immunoglobulin therapy escape, decreases viral secretion | [73,76] |
| SK160N | OBI/CHB | Decreases antigenicity and viral secretion | [77] |
| SM103I | CHB | Decreases antigenicity | [42] |
| SQ129R | CHB | Vaccine and detection escape and decrease viral secretion | [73,75] |
| SG130N | CHB | Diagnostic and vaccine escape | [78,79] |
| ST140I | CHB | Decreases viral secretion | [75] |
| SG145R | CHB | Vaccine, detection and immunoglobulin therapy escape, decreased antigenicity and viral secretion | [75] |
| ORF/Region | Variant | Subgenotype |
|---|---|---|
| PreS1 | preS1S78N | D3 |
| PreS2 | preS2F22P, preS2F22H | D3 |
| S | sL97P, sT114I, sC124Y, sN131K, sP217L | A1 |
| S | sQ129H | D3 |
| Pol | tpN120Y, tpK155R, spS91T, spS133G, rtL140I, rtT225A, rtA329T, rhI81M | A1 |
| Pol | spW64R, spP103S, rtT128I, rtY257F | D3 |
| X | xS11A, xV15I | A1 |
| X | xP11S, xS31A, xQ87L, xS101P, xL116V | D3 |
| Core | cS26P, cD32G, cP45S, cE46D, cI59L, cE117Stop, cR127H | A1 |
| Core | cD2A, cI3R, cD4Y, cE64K, cV74N, cS87N, cF97I, cW102V, cF103V | D3 |
| PID | Subgenotype | Variant |
|---|---|---|
| MA121 | D3 | preS2F22H |
| MA122 | D3 | preS1S78N, preS2F22P, spW64R, spP103S |
| MA123 | D3 | cD2A, cE64K |
| MA127 | D3 | rtT128I, cW102V, cF103V |
| MA128 | D3 | xP11S, xS31A, xS101P, xL116V, cV74N, cS87N, cF97I |
| MA125 | D3 | cI13R, cD4Y |
| MA107 | A1 | sN131K, rtL140I |
| MA98 | A1 | sL97P, sC124Y, tpN120Y, tpK155R, xV15I |
| MA103 | A1 | sP217L, spS91T, spS133G, cS26P, cD32G, cP45S, cE46D, cI59L |
| MA102 | A1 | rtT225A |
| MA99 | A1 | cE117Stop, cR127H |
| MA126 | D3 | sQ129H, rtY257F, xQ87L |
| MA100 | A1 | rhI81M, xS11A |
| MA101 | A1 | sT114I, rtA329T |
| ORF | Sub Genotype | Signature Amino Acid | Number in OBI Participants * | Number in CHB Participants * |
|---|---|---|---|---|
| PreS1 | D3 | preS1P65L | 11 | 5 |
| PreS1 | A1 | PreS1P94T | 9 | 6 |
| Surface | A1 | SN131T | 7 | 4 |
| Surface | A1 | SA194V | 10 | 5 |
| Pol | D3 | rhI54V | 6 | 4 |
| Pol | A1 | tpH87Q | 9 | 5 |
| Pol | A1 | spT11A | 9 | 5 |
| Pol | A1 | spA68P | 10 | 5 |
| Pol | A1 | spS97H | 9 | 5 |
| Pol | A1 | rtI103V | 8 | 4 |
| Pol | A1 | rtQ139H | 7 | 4 |
| Pol | A1 | rtS332N | 10 | 5 |
| Pol | A1 | rtK333Q | 10 | 5 |
| Pol | A1 | rhP2S | 9 | 4 |
| X | D3 | xG27R | 6 | 3 |
| X | D3 | xF30L | 7 | 3 |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Anderson, M.; Choga, W.T.; Moyo, S.; Bell, T.G.; Mbangiwa, T.; Phinius, B.B.; Bhebhe, L.; Sebunya, T.K.; Lockman, S.; Marlink, R.; et al. Molecular Characterization of Near Full-Length Genomes of Hepatitis B Virus Isolated from Predominantly HIV Infected Individuals in Botswana. Genes 2018, 9, 453. https://doi.org/10.3390/genes9090453
Anderson M, Choga WT, Moyo S, Bell TG, Mbangiwa T, Phinius BB, Bhebhe L, Sebunya TK, Lockman S, Marlink R, et al. Molecular Characterization of Near Full-Length Genomes of Hepatitis B Virus Isolated from Predominantly HIV Infected Individuals in Botswana. Genes. 2018; 9(9):453. https://doi.org/10.3390/genes9090453
Chicago/Turabian StyleAnderson, Motswedi, Wonderful Tatenda Choga, Sikhulile Moyo, Trevor Graham Bell, Tshepiso Mbangiwa, Bonolo Bonita Phinius, Lynnette Bhebhe, Theresa Kibirige Sebunya, Shahin Lockman, Richard Marlink, and et al. 2018. "Molecular Characterization of Near Full-Length Genomes of Hepatitis B Virus Isolated from Predominantly HIV Infected Individuals in Botswana" Genes 9, no. 9: 453. https://doi.org/10.3390/genes9090453
APA StyleAnderson, M., Choga, W. T., Moyo, S., Bell, T. G., Mbangiwa, T., Phinius, B. B., Bhebhe, L., Sebunya, T. K., Lockman, S., Marlink, R., Kramvis, A., Essex, M., Musonda, R. M., Blackard, J. T., & Gaseitsiwe, S. (2018). Molecular Characterization of Near Full-Length Genomes of Hepatitis B Virus Isolated from Predominantly HIV Infected Individuals in Botswana. Genes, 9(9), 453. https://doi.org/10.3390/genes9090453





