Genome-Wide Identification and Expression Profiling of Cytokinin Oxidase/Dehydrogenase (CKX) Genes Reveal Likely Roles in Pod Development and Stress Responses in Oilseed Rape (Brassica napus L.)
Abstract
:1. Introduction
2. Materials and Methods
2.1 Identification of CKXs in B. napus, B. rapa, and B. oleracea
2.2. Multiple Sequence Alignment and Phylogenetic Analysis
2.3. Chromosomal Locations, Gene Structures and Protein Profiles of BnCKXs
2.4. Sequence Analysis of the BnCKXs Promoters
2.5. Plant Materials, Growth Conditions, and Treatments
2.6. RNA-seq Analysis
2.7. RNA Isolation and qRT-PCR Analysis
3. Results
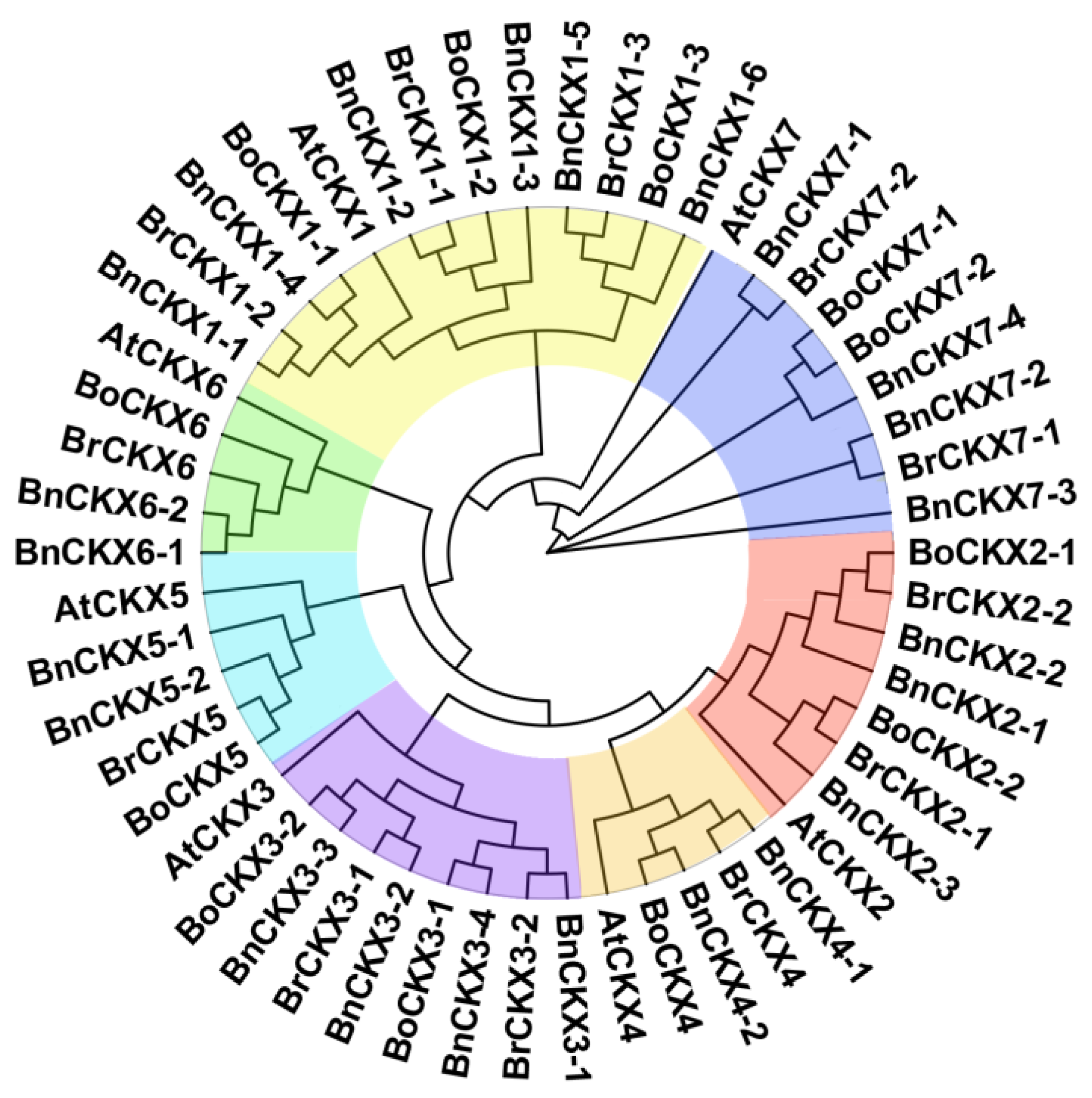
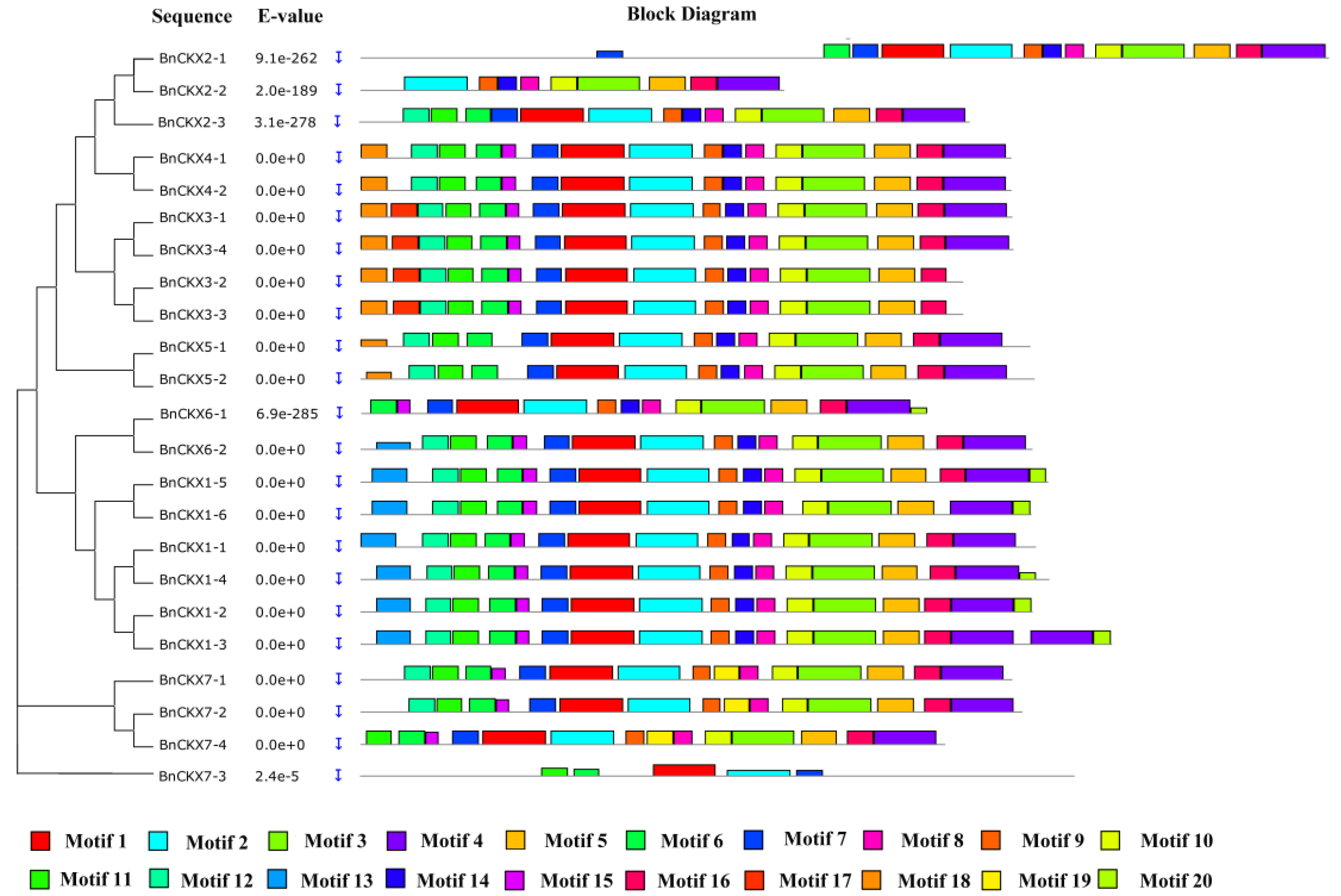
3.1. Identification and Phylogenetic Analysis of BnCKXs
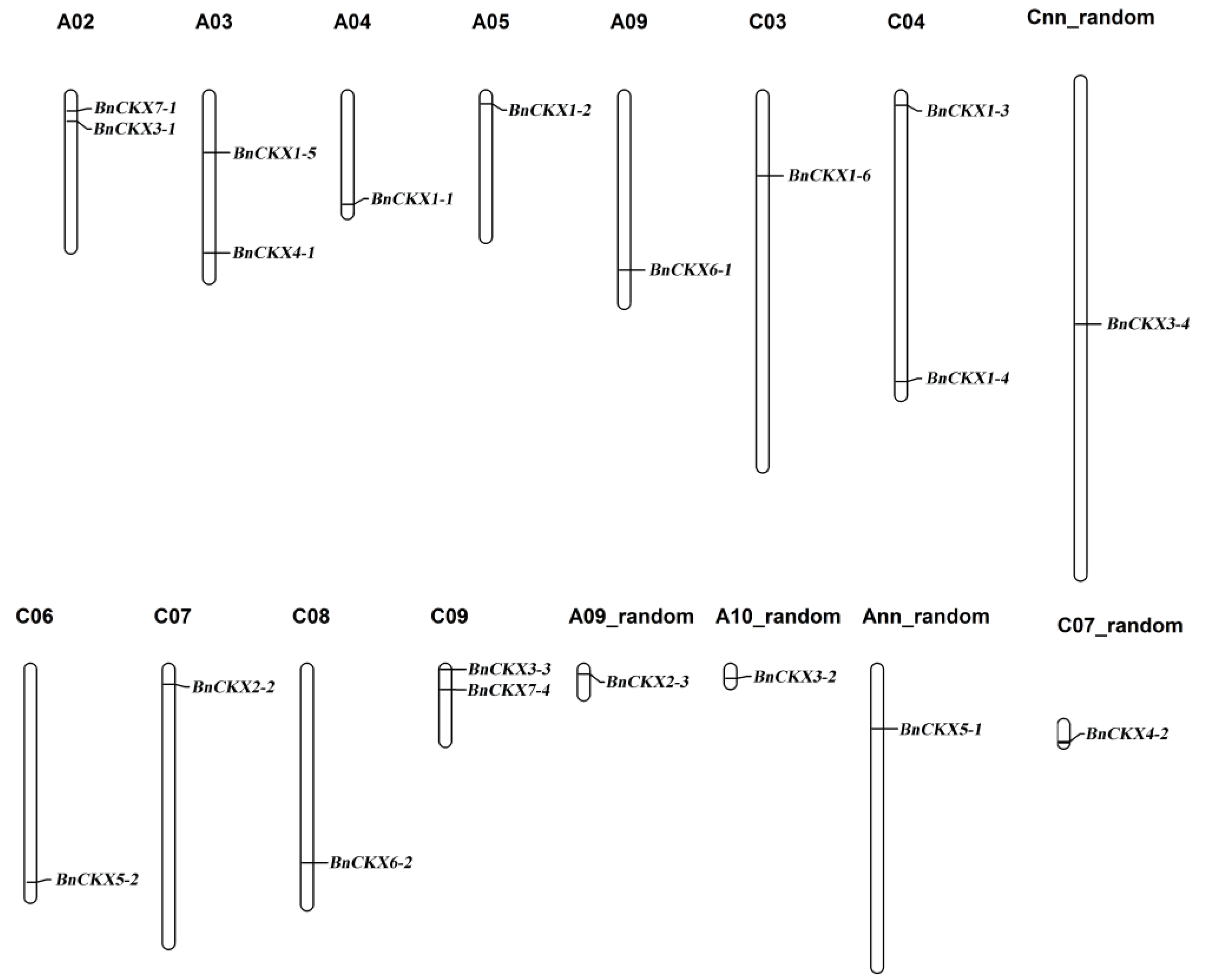
3.2. Chromosomal Localization
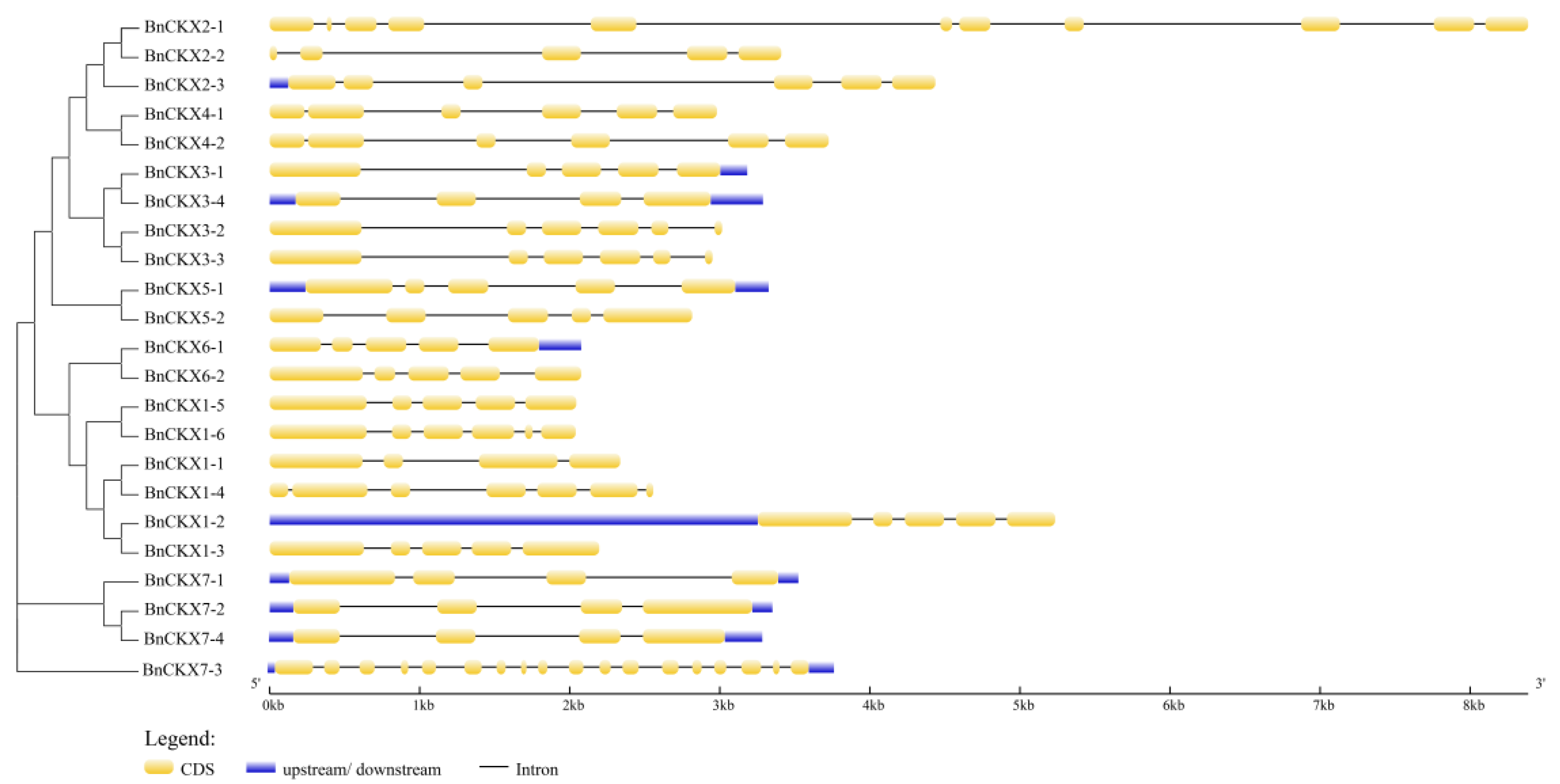
3.3. Gene Structures and Protein Profiles of BnCKXs
3.4. Cis-Acting Elements in the BnCKXs
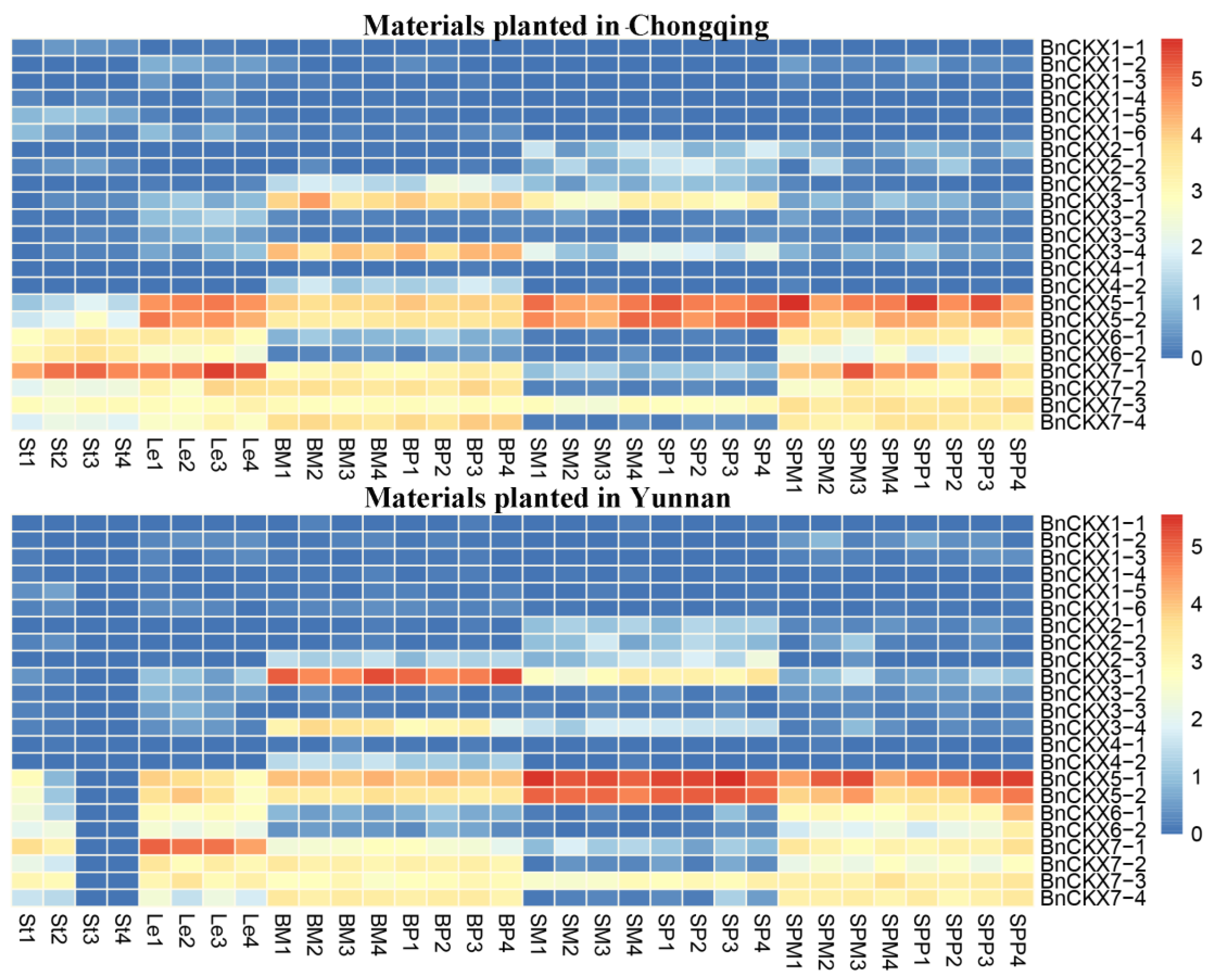
3.5. RNA-Seq Analysis
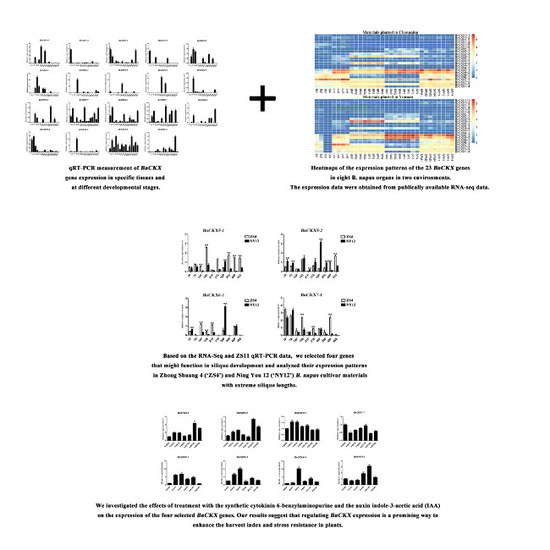
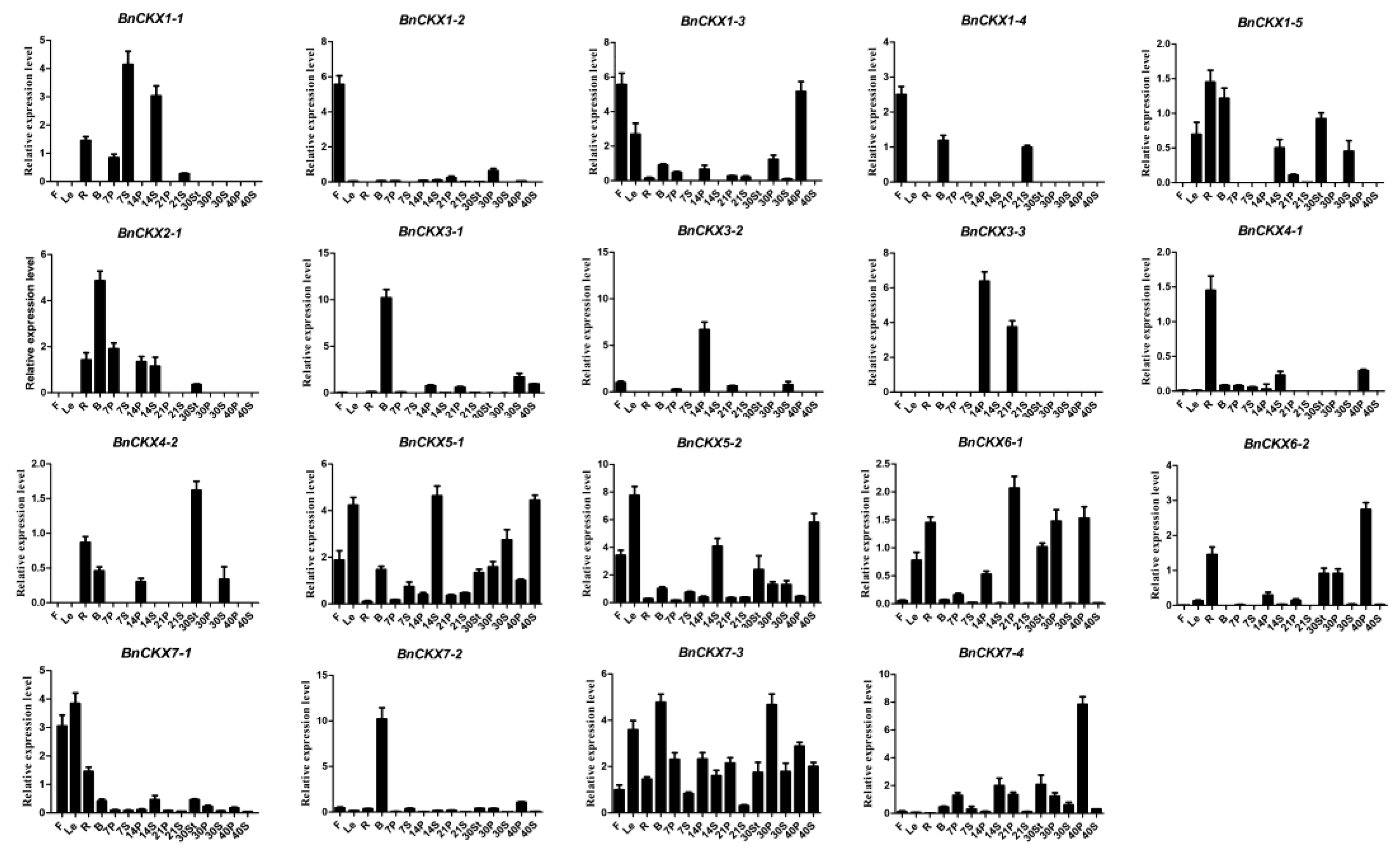
3.6. qRT-PCR Measurement of BnCKXs Gene Expression in Specific Tissues and at Different Developmental Stages
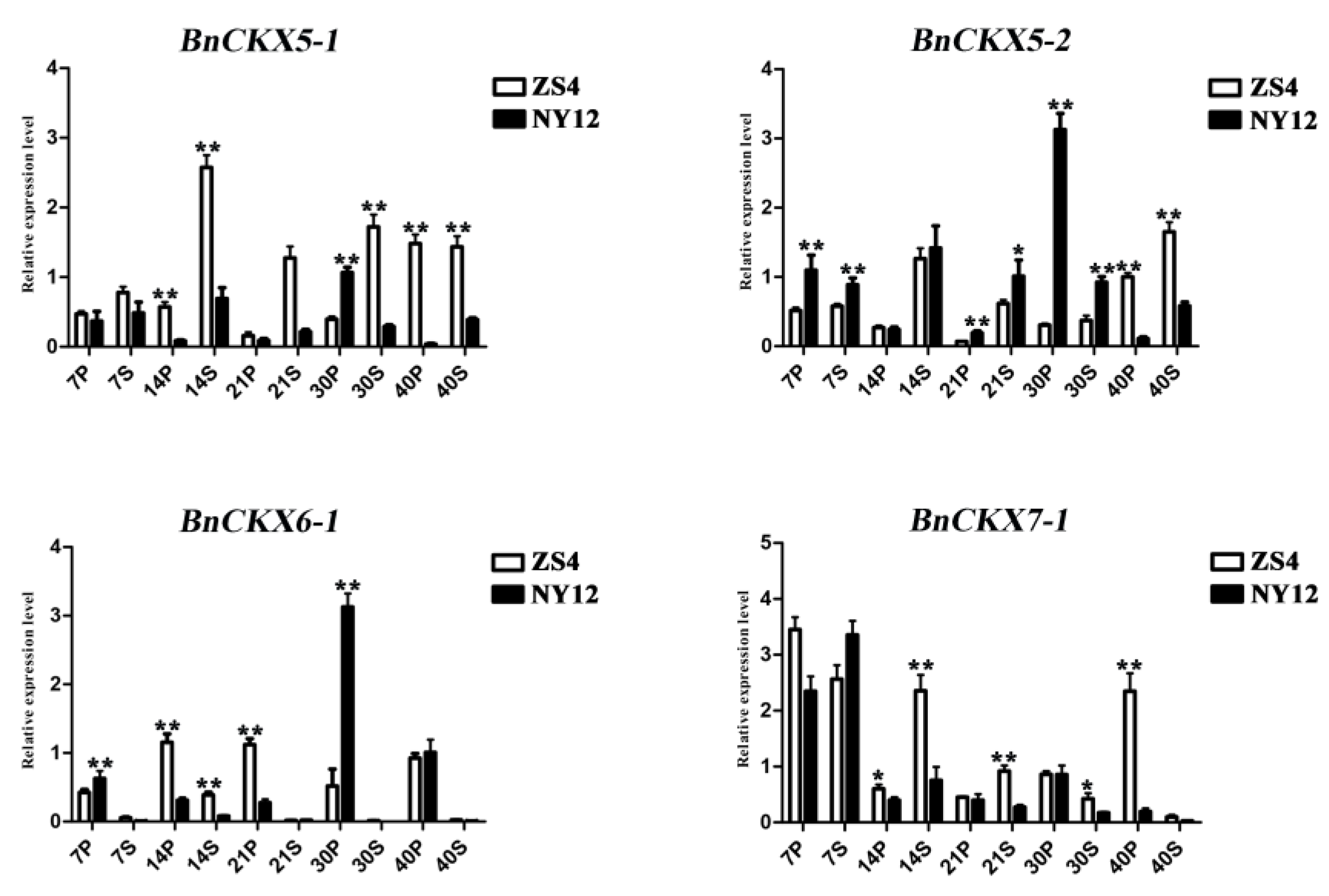
3.7. Expression of Selected BnCKX Genes in Different Plant Materials and in Response to IAA and 6-BA Treatment
4. Discussion
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Skoog, F.; Miller, C.O. Chemical regulation of growth and organ formation in plant tissues cultured in vitro. Cheminform 1996, 27, 118. [Google Scholar]
- Werner, T.; Motyka, V.; Strnad, M.; Schmülling, T. Regulation of plant growth by cytokinin. Proc. Natl. Acad. Sci. USA 2001, 98, 10487. [Google Scholar] [CrossRef] [PubMed]
- Eklöf, S.; Åstot, C.; Blackwell, J.; Moritz, T.; Olsson, O.; Sandberg, G. Auxin-cytokinin interactions in wild-type and transgenic tobacco. Plant Cell Physiol. 1997, 38, 225–235. [Google Scholar]
- Miller, C.O.; Skoog, F.; Saltza, M.H.V.; Strong, F.M. Kinetin, a cell division factor from deoxyribonucleic acid. J. Am. Chem. Soc. 1955, 77, 1392. [Google Scholar] [CrossRef]
- Werner, T.; Motyka, V.; Laucou, V.; Smets, R.; Onckelen, H.V.; Schmülling, T. Cytokinin-deficient transgenic Arabidopsis plants show multiple developmental alterations indicating opposite functions of cytokinins in the regulation of shoot and root meristem activity. Plant Cell 2003, 15, 2532–2550. [Google Scholar] [CrossRef] [PubMed]
- Mok, D.W.S.; Mok, M.C. Cytokinins: Chemistry, activity, and function; CRC Press: Boca Raton, FL, USA, 1994. [Google Scholar]
- Kim, H.J.; Ryu, H.; Hong, S.H.; Woo, H.R.; Lim, P.O.; Lee, I.C.; Sheen, J.; Nam, H.G.; Hwang, I. Cytokinin-mediated control of leaf longevity by AHK3 through phosphorylation of ARR2 in Arabidopsis. Proc. Natl. Acad. Sci. USA 2006, 103, 814–819. [Google Scholar] [CrossRef] [PubMed]
- Tanaka, M.; Takei, K.; Kojima, M.; Sakakibara, H.; Mori, H. Auxin controls local cytokinin biosynthesis in the nodal stem in apical dominance. Plant J. Cell Mol. Biol. 2006, 45, 1028. [Google Scholar] [CrossRef] [PubMed]
- Werner, T.; Schmülling, T. Cytokinin action in plant development. Curr. Opin. Plant Biol. 2009, 12, 527–538. [Google Scholar] [CrossRef] [PubMed]
- Takei, K.; Sakakibara, H.; Taniguchi, M.; Sugiyama, T. Nitrogen-dependent accumulation of cytokinins in root and thetranslocation to leaf: Implication of cytokinin species that induces geneexpression of maize responseregulator. Plant Cell Physiol. 2001, 42, 85. [Google Scholar] [CrossRef] [PubMed]
- Frébort, I.; Kowalska, M.; Hluska, T.; Frébortová, J.; Galuszka, P. Evolution of cytokinin biosynthesis and degradation. Journal of Experimental Botany 2011, 62, 2431. [Google Scholar] [CrossRef] [PubMed]
- Brownlee, B.G.; Hall, R.H.; Whitty, C.D. 3-Methyl-2-butenal: an enzymatic degradation product of the cytokinin, N-6-(delta-2 isopentenyl)adenine. Can. J. Biochem. 1975, 53, 37. [Google Scholar] [CrossRef] [PubMed]
- Armstrong, D.J. Cytokinin oxidase and the regulation of cytokinin degradation. 1994. [Google Scholar]
- Kakimoto, T. Identification of plant cytokinin biosynthetic enzymes as dimethylallyl diphosphate: ATP/ADP isopentenyltransferases. Plant Cell Physiol. 2001, 42, 677–685. [Google Scholar] [CrossRef] [PubMed]
- Motyka, V.; Vaňková, R.; Čapková, V.; Petrášek, J.; Kamínek, M.; Schmülling, T. Cytokinin-induced upregulation of cytokinin oxidase activity in tobacco includes changes in enzyme glycosylation and secretion. Physiol. Plant. 2003, 117, 11–21. [Google Scholar] [CrossRef]
- Galuszka, P.; Frébort, I.; Šebela, M.; Peč, P. Degradation of cytokinins by cytokinin oxidases in plants. Plant Growth Regul. 2000, 32, 315–327. [Google Scholar] [CrossRef]
- Frebortova, J.; Fraaije, M.P.; Sebela, M.; Pec, P.; Hrbac, J. Catalytic reaction of cytokinin dehydrogenase: preference for quinones as electron acceptors. Biochem. J. 2004, 380, 121. [Google Scholar] [CrossRef] [PubMed]
- Kopecný, D.; Pethe, C.; Sebela, M.; Houbahérin, N.; Madzak, C.; Majira, A.; Laloue, M. High-level expression and characterization of Zea mays cytokinin oxidase/dehydrogenase in Yarrowia lipolytica. Biochimie 2005, 87, 1011–1022. [Google Scholar] [CrossRef] [PubMed]
- Esparza, T.J.; Morris, R.O. Molecular and biochemical characterization of a cytokinin oxidase from maize. Plant Physiol. 2001, 125, 378–386. [Google Scholar]
- Werner, T.; Köllmer, I.; Bartrina, I.; Holst, K.; Schmülling, T. New insights into the biology of cytokinin degradation. Plant Biol. 2006, 8, 371. [Google Scholar] [CrossRef] [PubMed]
- Ashikari, M.; Sakakibara, H.; Lin, S.; Yamamoto, T.; Takashi, T.; Nishimura, A.; Angeles, E.R.; Kitano, H.; Matsuoka, M. Cytokinin oxidase regulates rice grain production. Science 2005, 309, 741. [Google Scholar] [CrossRef] [PubMed]
- Motyka, V.; Faiss, M.; Strand, M.; Kaminek, M.; Schmulling, T. Changes in cytokinin content and cytokinin oxidase activity in response to derepression of ipt gene transcription in transgenic tobacco calli and plants. Plant Physiol. 1996, 112, 1035. [Google Scholar] [CrossRef] [PubMed]
- Houbahérin, N.; Pethe, C.; D'Alayer, J.; Laloue, M. Cytokinin oxidase from Zea mays: purification, cDNA cloning and expression in moss protoplasts. Plant J. Cell Mol. Biol. 1999, 17, 615–626. [Google Scholar] [CrossRef]
- Morris, R.O.; Bilyeu, K.D.; Laskey, J.G.; Cheikh, N.N. Isolation of a gene encoding a glycosylated cytokinin oxidase from maize. Biochem. Biophys. Res. Commun. 1999, 255, 328–333. [Google Scholar] [CrossRef] [PubMed]
- Galuszka, P.; Frébortová, J.; Werner, T.; Yamada, M.; Strnad, M.; Schmülling, T.; Frébort, I. Cytokinin oxidase/dehydrogenase genes in barley and wheat: cloning and heterologous expression. Eur. J. Biochem. 2004, 271, 3990. [Google Scholar] [CrossRef] [PubMed]
- Mameaux, S.; Cockram, J.; Thiel, T.; Steuernagel, B.; Stein, N.; Taudien, S.; Jack, P.; Werner, P.; Gray, J.C.; Greenland, A.J. Molecular, phylogenetic and comparative genomic analysis of the cytokinin oxidase/dehydrogenase gene family in the Poaceae. Plant Biotechnol. J. 2012, 10, 67–82. [Google Scholar] [CrossRef] [PubMed]
- Liu, Z.; Lv, Y.; Zhang, M.; Liu, Y.; Kong, L.; Zou, M.; Lu, G.; Cao, J.; Yu, X. Identification, expression, and comparative genomic analysis of the ipt and ckx gene families in chinese cabbage (Brassica rapa ssp. Pekinensis). Bmc Genom. 2013, 14, 594. [Google Scholar] [CrossRef] [PubMed]
- Zeng, Q.W.; Qin, S.; Song, S.Q.; Zhang, M.; Xiao, Y.H.; Luo, M.; Hou, L.; Pei, Y. Molecular cloning and characterization of a cytokinin dehydrogenase gene from upland cotton (Gossypium hirsutum L.). Plant Mol. Biol. Report. 2012, 30, 1–9. [Google Scholar] [CrossRef]
- Wang, Y.; Liu, H.; Xin, Q. Genome-wide analysis and identification of cytokinin oxidase/dehydrogenase (CKX) gene family in foxtail millet (Setaria italica). Crop J. 2014, 2, 244–254. [Google Scholar] [CrossRef]
- Jiang, Y.; Mi, X.; Lin, Y.; Wu, H.; Gu, T.; Ding, J.; Li, Y. Evolution and expression patterns of cytokinin oxidase genes in Fragaria vesca. Sci. Hortic. 2016, 212, 115–125. [Google Scholar] [CrossRef]
- Yang, S.H.; Yu, H.; Chong, J.G. Functional characterisation of a cytokinin oxidase gene dsckx1 in Dendrobium orchid. Plant Mol. Biol. 2003, 51, 237–248. [Google Scholar] [CrossRef] [PubMed]
- Gaudinová, A.; Dobrev, P.I.; Šolcová, B.; Novák, O.; Strnad, M.; Friedecký, D.; Motyka, V. The involvement of cytokinin oxidase/dehydrogenase and zeatin reductase in regulation of cytokinin levels in pea (Pisum sativum L.) leaves. J. Plant Growth Regul. 2005, 24, 188–200. [Google Scholar] [CrossRef]
- Le, D.T.; Nishiyama, R.; Watanabe, Y.; Vankova, R.; Tanaka, M.; Seki, M.; Le, H.H.; Yamaguchishinozaki, K.; Shinozaki, K.; Tran, L.S.P. Identification and expression analysis of cytokinin metabolic genes in soybean under normal and drought conditions in relation to cytokinin levels. PloS ONE 2012, 7, e42411. [Google Scholar] [CrossRef] [PubMed]
- Galuszka, P.; Popelková, H.; Werner, T.; Frébortová, J.; Pospíšilová, H. Biochemical characterization and histochemical localization of cytokinin oxidases/dehydrogenases from Arabidopsis thaliana expressed in Nicotiana tabaccum L. J. Plant Growth Regul. 2007. [Google Scholar] [CrossRef]
- Trifunović, M.; Cingel, A.; Simonović, A.; Jevremović, S.; Petrić, M.; Dragićević, I.Č.; Motyka, V.; Dobrev, P.I.; Zahajská, L.; Subotić, A. Overexpression of Arabidopsis cytokinin oxidase/dehydrogenase genes Atckx1 and Atckx2 in transgenic Centaurium erythraea Rafn. Plant Cell Tissue Organ Cult. 2013, 115, 139–150. [Google Scholar] [CrossRef]
- Zalewski, W.; Galuszka, P.; Gasparis, S.; Orczyk, W.; Nadolskaorczyk, A. Silencing of the HvCKX1 gene decreases the cytokinin oxidase/dehydrogenase level in barley and leads to higher plant productivity. J. Exp. Bot. 2010, 61, 1839. [Google Scholar] [CrossRef] [PubMed]
- Chen, W.; Zhang, Y.; Liu, X.; Chen, B.; Tu, J.; Tingdong, F. Detection of QTL for six yield-related traits in oilseed rape (Brassica napus) using DH and immortalized F(2) populations. TAG. Theor. Appl. Genet. 2007, 115, 849–858. [Google Scholar] [CrossRef] [PubMed]
- Diepenbrock, W. Yield analysis of winter oilseed rape (Brassica napus L.): A review. Field Crops Res. 2000, 67, 35–49. [Google Scholar] [CrossRef]
- Liu, J.; Hua, W.; Hu, Z.; Yang, H.; Zhang, L.; Li, R.; Deng, L.; Sun, X.; Wang, X.; Wang, H. Natural variation in ARF18 gene simultaneously affects seed weight and silique length in polyploid rapeseed. Proc. Natl. Acad. Sci. USA 2015, 112, 5123–5132. [Google Scholar] [CrossRef] [PubMed]
- Tran, L.S.; Urao, T.; Qin, F.; Maruyama, K.; Kakimoto, T.; Shinozaki, K.; Yamaguchishinozaki, K. Functional analysis of AHK1/ATHK1 and cytokinin receptor histidine kinases in response to abscisic acid, drought, and salt stress in Arabidopsis. Proc. Natl. Acad. Sci. USA 2007, 104, 20623–20628. [Google Scholar] [CrossRef] [PubMed]
- Altschul, S.F.; Madden, T.L.; Schäffer, A.A.; Zhang, J.; Zhang, Z.; Miller, W.; Lipman, D.J. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucl. Acid. Res. 1997; 25, 3389–3402. [Google Scholar]
- Cheng, F.; Liu, S.; Wu, J.; Fang, L.; Sun, S.; Liu, B.; Li, P.; Hua, W.; Wang, X. BRAD, the genetics and genomics database for Brassica plants. Bmc Plant Biology 2011, 11, 1–6. [Google Scholar] [CrossRef] [PubMed]
- Quevillon, E.; Silventoinen, V.; Pillai, S.; Harte, N.; Mulder, N.; Apweiler, R.; Lopez, R. Interproscan: Protein domains identifier. Nucleic Acids Res. 2005, 33, W116. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Stecher, G.; Tamura, K. Mega7: Molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Bio.Evol. 2016, 33, 1870. [Google Scholar] [CrossRef] [PubMed]
- Capellagutiérrez, S.; Sillamartínez, J.M.; Gabaldón, T. Trimal: A tool for automated alignment trimming in large-scale phylogenetic analyses. Bioinformatics 2009, 25, 1972. [Google Scholar] [CrossRef] [PubMed]
- Guindon, S.; Dufayard, J.F.; Lefort, V.; Anisimova, M.; Hordijk, W.; Gascuel, O. New algorithms and methods to estimate maximum-likelihood phylogenies: Assessing the performance of PhyML 3.0. Syst. Biol. 2010, 59, 307–321. [Google Scholar] [CrossRef] [PubMed]
- Huelsenbeck, J.P.; Ronquist, F. MRBAYES: Bayesian inference of phylogenetic trees. Bioinformatics 2001, 17, 754–755. [Google Scholar] [CrossRef] [PubMed]
- Schmidt, H.A.; Strimmer, K.; Vingron, M.; Von, H.A. Tree-puzzle: Maximum likelihood phylogenetic analysis using quartets and parallel computing. Bioinformatics 2002, 18, 502. [Google Scholar] [CrossRef] [PubMed]
- Voorrips, R.E. MapChart: Software for the graphical presentation of linkage maps and QTLs. J. Hered. 2002, 93, 77–78. [Google Scholar] [CrossRef] [PubMed]
- Guo, A.Y.; Zhu, Q.H.; Chen, X.; Luo, J.C. GSDS: A gene structure display server. Hereditas 2007, 29, 1023. [Google Scholar] [CrossRef] [PubMed]
- Bailey, T.L.; Williams, N.; Misleh, C.; Li, W.W. Meme: Discovering and analyzing DNA and protein sequence motifs. Nucleic Acids Res. 2006, 34, W369. [Google Scholar] [CrossRef] [PubMed]
- Gasteiger, E.; Gattiker, A.; Hoogland, C.; Ivanyi, I.; Appel, R.D.; Bairoch, A. Expasy: The proteomics server for in-depth protein knowledge and analysis. Nucleic Acids Res. 2003, 31, 3784–3788. [Google Scholar] [CrossRef] [PubMed]
- Paul, H.; Keun-Joon, P.; Takeshi, O.; Naoya, F.; Hajime, H.; Adams-Collier, C.J.; Kenta, N. WoLF PSORT: Protein localization predictor. Nucleic Acids Res. 2007, 35, 585–587. [Google Scholar]
- Chalhoub, B.; Denoeud, F.; Liu, S.; Parkin, I.A.P.; Tang, H.; Wang, X.; Chiquet, J.; Belcram, H.; Tong, C.; Samans, B. Early allopolyploid evolution in the post-neolithic Brassica napus oilseed genome. Science 2014, 345, 950. [Google Scholar] [CrossRef] [PubMed]
- Postel, D.; Vanlemmens, P.; Gode, P.; Ronco, G.; Villa, P. Plantcare, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Res. 2002, 30, 325. [Google Scholar]
- Lu, K.; Guo, W.; Lu, J.; Yu, H.; Qu, C.; Tang, Z.; Li, J.; Chai, Y.; Liang, Y. Genome-wide survey and expression profile analysis of the mitogen-activated protein kinase (MAPK) gene family in Brassica rapa. Plos ONE 2015, 10, e0132051. [Google Scholar] [CrossRef] [PubMed]
- Lu, K.; Xiao, Z.; Jian, H.; Peng, L.; Qu, C.; Fu, M.; He, B.; Tie, L.; Liang, Y.; Xu, X. A combination of genome-wide association and transcriptome analysis reveals candidate genes controlling harvest index-related traits in Brassica napus. Sci. rep. 2016, 6, 36452. [Google Scholar] [CrossRef] [PubMed]
- Ma, J.Q.; Jian, H.J.; Yang, B.; Lu, K.; Zhang, A.X.; Liu, P.; Li, J.N. Genome-wide analysis and expression profiling of the grf gene family in oilseed rape (Brassica napus L.). Gene 2017, 620, 36–45. [Google Scholar] [CrossRef] [PubMed]
- Lalitha, S. Primer premier 5. Biotech Softw. Internet Rep. 2000, 1, 270–272. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]
- Lysak, M.A.; Koch, M.A.; Pecinka, A.; Schubert, I. Chromosome triplication found across the tribe Brassiceae. Genome Res. 2005, 15, 516. [Google Scholar] [CrossRef] [PubMed]
- Mun, J.H.; Kwon, S.J.; Yang, T.J.; Seol, Y.J.; Jin, M.; Kim, J.A.; Lim, M.H.; Kim, J.S.; Baek, S.; Choi, B.S. Genome-wide comparative analysis of the Brassica rapa gene space reveals genome shrinkage and differential loss of duplicated genes after whole genome triplication. Genome Biol. 2009, 10, R111. [Google Scholar] [CrossRef] [PubMed]
- Brugière, N.; Jiao, S.; Hantke, S.; Zinselmeier, C.; Roessler, J.A.; Niu, X.; Jones, R.J.; Habben, J.E. Cytokinin oxidase gene expression in maize is localized to the vasculature, and is induced by cytokinins, abscisic acid, and abiotic stress. Plant physiol. 2003, 132, 1228. [Google Scholar] [CrossRef] [PubMed]
- Todorova, D.; Vaseva, I.; Malbeck, J.; Trávníčková, A.; Macháčková, I.; Karanov, E. Cytokinin oxidase/dehydrogenase activity as a tool in gibberellic acid/cytokinin cross talk. Biol. Plant. 2007, 51, 579–583. [Google Scholar] [CrossRef]
- Gao, S.; Chu, C. Cytokinin oxidase/dehydrogenase4 integrates cytokinin and auxin signaling to control rice crown root formation. Plant Physiol. 2014, 165, 1035–1046. [Google Scholar] [CrossRef] [PubMed]
- Werner, T.; Nehnevajova, E.; Köllmer, I.; Novák, O.; Strnad, M.; Krämer, U.; Schmülling, T. Root-specific reduction of cytokinin causes enhanced root growth, drought tolerance, and leaf mineral enrichment in Arabidopsis and tobacco. Plant Cell 2010, 22, 3905–3920. [Google Scholar] [CrossRef] [PubMed]
- Macková, H.; Vanková, R. Enhanced drought and heat stress tolerance of tobacco plants with ectopically enhanced cytokinin oxidase/dehydrogenase gene expression. J. Exp. Bot. 2013, 64, 2805. [Google Scholar] [CrossRef] [PubMed]
- Bartrina, I.; Otto, E.; Strnad, M.; Werner, T.; Schmülling, T. Cytokinin regulates the activity of reproductive meristems, flower organ size, ovule formation, and thus seed yield in Arabidopsis thaliana. Plant Cell 2011, 23, 69–80. [Google Scholar] [CrossRef] [PubMed]
- Köllmer, I.; Novák, O.; Strnad, M.; Schmülling, T.; Werner, T. Overexpression of the cytosolic cytokinin oxidase/dehydrogenase (CKX7) from Arabidopsis causes specific changes in root growth and xylem differentiation. Plant J. Cell Mol. Biol. 2014, 78, 359–371. [Google Scholar] [CrossRef] [PubMed]

| Gene Name | Locus Name | Chr. | Location | ORF Length (bp) | Protein | Subcellular Location | ||
|---|---|---|---|---|---|---|---|---|
| Length (aa) | Mw (Da) | pI | ||||||
| BnCKX1-1 | BnaA04g23980D | A04 | 17680020…17682356 | 1611 | 536 | 60,681.24 | 8.58 | chlo: 6, extr: 3, vacu: 2, nucl: 1, plas: 1 |
| BnCKX1-2 | BnaA05g02240D | A05 | 1254144...1259378 | 1602 | 533 | 60,411.94 | 8.72 | mito: 7, chlo_mito: 6.33333, chlo: 4.5, cyto_mito: 4.33333 |
| BnCKX1-3 | BnaC04g01930D | C04 | 1500877…1503073 | 1791 | 596 | 68,227.7 | 8.53 | nucl: 4, vacu: 4, chlo: 3, plas: 1, extr: 1 |
| BnCKX1-4 | BnaC04g47740D | C04 | 46681907…46684463 | 1641 | 546 | 61,665.39 | 9.2 | mito: 7.5, cyto_mito: 4.5, chlo: 3, nucl: 2 |
| BnCKX1-5 | BnaA03g19500D | A03 | 9235344...9237387 | 1638 | 545 | 61,439.11 | 8.72 | mito:4,chlo: 4, cyto: 3, nucl: 2 |
| BnCKX1-6 | BnaC03g23380D | C03 | 13022610...13024649 | 1599 | 532 | 60,320.83 | 8.8 | mito: 5, chlo: 4, nucl: 2, cyto: 2 |
| BnCKX2-1 | BnaA07g35940D | A07_random | 5099…13485 | 2307 | 768 | 87,023.55 | 5.86 | nucl: 7, cyto: 4, chlo: 2 |
| BnCKX2-2 | BnaC07g01710D | C07 | 2422452...2425861 | 1011 | 336 | 38,171.83 | 5.83 | chlo: 4, cyto: 2, extr: 2, vacu: 2, mito: 1.5, mito_plas: 1.5, nucl: 1 |
| BnCKX2-3 | BnaA09g52930D | A09_random | 753807...758243 | 1455 | 484 | 53,519.55 | 5.96 | vacu: 4, chlo: 3, extr: 3, plas: 2, cyto: 1 |
| BnCKX3-1 | BnaA02g08420D | A02 | 4,079,790…4,082,972 | 1554 | 517 | 58,722.84 | 5.7 | vacu: 6, mito: 2, extr: 2, golg: 2, plas: 1 |
| BnCKX3-2 | BnaA10g28940D | A10_random | 1464876…1467891 | 1437 | 478 | 54,173.56 | 5.93 | vacu: 5, extr: 3, mito: 2, plas: 2, chlo: 1 |
| BnCKX3-3 | BnaC09g33450D | C09 | 36760027…36762978 | 1437 | 478 | 54,183.47 | 5.78 | vacu: 7, extr: 4, cyto: 1, mito: 1 |
| BnCKX3-4 | BnaCnng41060D | Cnn_random | 39653407…39656550 | 1557 | 518 | 58,855.91 | 5.83 | vacu: 6, extr: 3, golg: 2, mito: 1, plas: 1 |
| BnCKX4-1 | BnaA03g49660D | A03 | 25641300…25644280 | 1551 | 516 | 57,072.39 | 5.51 | chlo: 5, cyto: 2, mito: 2, vacu: 2, E.R.: 2 |
| BnCKX4-2 | BnaC07g50850D | C07_random | 2732541…2736266 | 1551 | 516 | 57,301.75 | 5.62 | vacu: 4, chlo: 2, mito: 2, extr: 2, golg: 2, cyto: 1 |
| BnCKX5-1 | BnaAnng09190D | Ann_random | 9695112...9698437 | 1596 | 531 | 59,613.56 | 5.62 | extr: 5, chlo: 3, vacu: 3, cyto: 1, mito: 1 |
| BnCKX5-2 | BnaC06g36220D | C06 | 34801906…34804721 | 1608 | 535 | 60,144.10 | 5.53 | vacu: 8, extr: 3, cyto: 1, mito: 1 |
| BnCKX6-1 | BnaA09g40400D | A09 | 28427957...28430033 | 1350 | 449 | 50,468.31 | 7.3 | mito: 7, nucl: 4, chlo: 2 |
| BnCKX6-2 | BnaC08g32840D | C08 | 31616324...31618400 | 1602 | 533 | 60,109.91 | 8.55 | mito: 7, chlo: 1, nucl: 1, cyto: 1, plas: 1, extr: 1, E.R.: 1 |
| BnCKX7-1 | BnaA02g05340D | A02 | 2433263…2436786 | 1554 | 517 | 57,559.29 | 5.2 | cyto: 6, nucl: 3, cysk: 2, chlo: 1, plas: 1 |
| BnCKX7-2 | BnaA10g14370D | A10 | 11435086…11438436 | 1578 | 525 | 57,864.41 | 4.92 | cyto: 11, chlo: 1, plas: 1 |
| BnCKX7-3 | BnaA10g24380D | A10 | 15919508...15923282 | 1704 | 567 | 61,909.23 | 7.93 | chlo: 7.5, chlo_mito: 7, mito: 5.5 |
| BnCKX7-4 | BnaC09g36710D | C09 | 40060399…40063686 | 1395 | 464 | 51,901.91 | 5.45 | chlo: 8, cyto: 4, nucl: 1 |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liu, P.; Zhang, C.; Ma, J.-Q.; Zhang, L.-Y.; Yang, B.; Tang, X.-Y.; Huang, L.; Zhou, X.-T.; Lu, K.; Li, J.-N. Genome-Wide Identification and Expression Profiling of Cytokinin Oxidase/Dehydrogenase (CKX) Genes Reveal Likely Roles in Pod Development and Stress Responses in Oilseed Rape (Brassica napus L.). Genes 2018, 9, 168. https://doi.org/10.3390/genes9030168
Liu P, Zhang C, Ma J-Q, Zhang L-Y, Yang B, Tang X-Y, Huang L, Zhou X-T, Lu K, Li J-N. Genome-Wide Identification and Expression Profiling of Cytokinin Oxidase/Dehydrogenase (CKX) Genes Reveal Likely Roles in Pod Development and Stress Responses in Oilseed Rape (Brassica napus L.). Genes. 2018; 9(3):168. https://doi.org/10.3390/genes9030168
Chicago/Turabian StyleLiu, Pu, Chao Zhang, Jin-Qi Ma, Li-Yuan Zhang, Bo Yang, Xin-Yu Tang, Ling Huang, Xin-Tong Zhou, Kun Lu, and Jia-Na Li. 2018. "Genome-Wide Identification and Expression Profiling of Cytokinin Oxidase/Dehydrogenase (CKX) Genes Reveal Likely Roles in Pod Development and Stress Responses in Oilseed Rape (Brassica napus L.)" Genes 9, no. 3: 168. https://doi.org/10.3390/genes9030168
APA StyleLiu, P., Zhang, C., Ma, J.-Q., Zhang, L.-Y., Yang, B., Tang, X.-Y., Huang, L., Zhou, X.-T., Lu, K., & Li, J.-N. (2018). Genome-Wide Identification and Expression Profiling of Cytokinin Oxidase/Dehydrogenase (CKX) Genes Reveal Likely Roles in Pod Development and Stress Responses in Oilseed Rape (Brassica napus L.). Genes, 9(3), 168. https://doi.org/10.3390/genes9030168