Integration of lncRNA and mRNA Transcriptome Analyses Reveals Genes and Pathways Potentially Involved in Calf Intestinal Growth and Development during the Early Weeks of Life
Abstract
1. Introduction
2. Materials and Methods
2.1. Animals and Management
2.2. RNA Isolation
2.3. Library Preparation and RNA-Sequencing
2.4. Sequence Data Processing, Alignment and Identification of Genes
2.5. Differential mRNA and lncRNA Expression Analyses
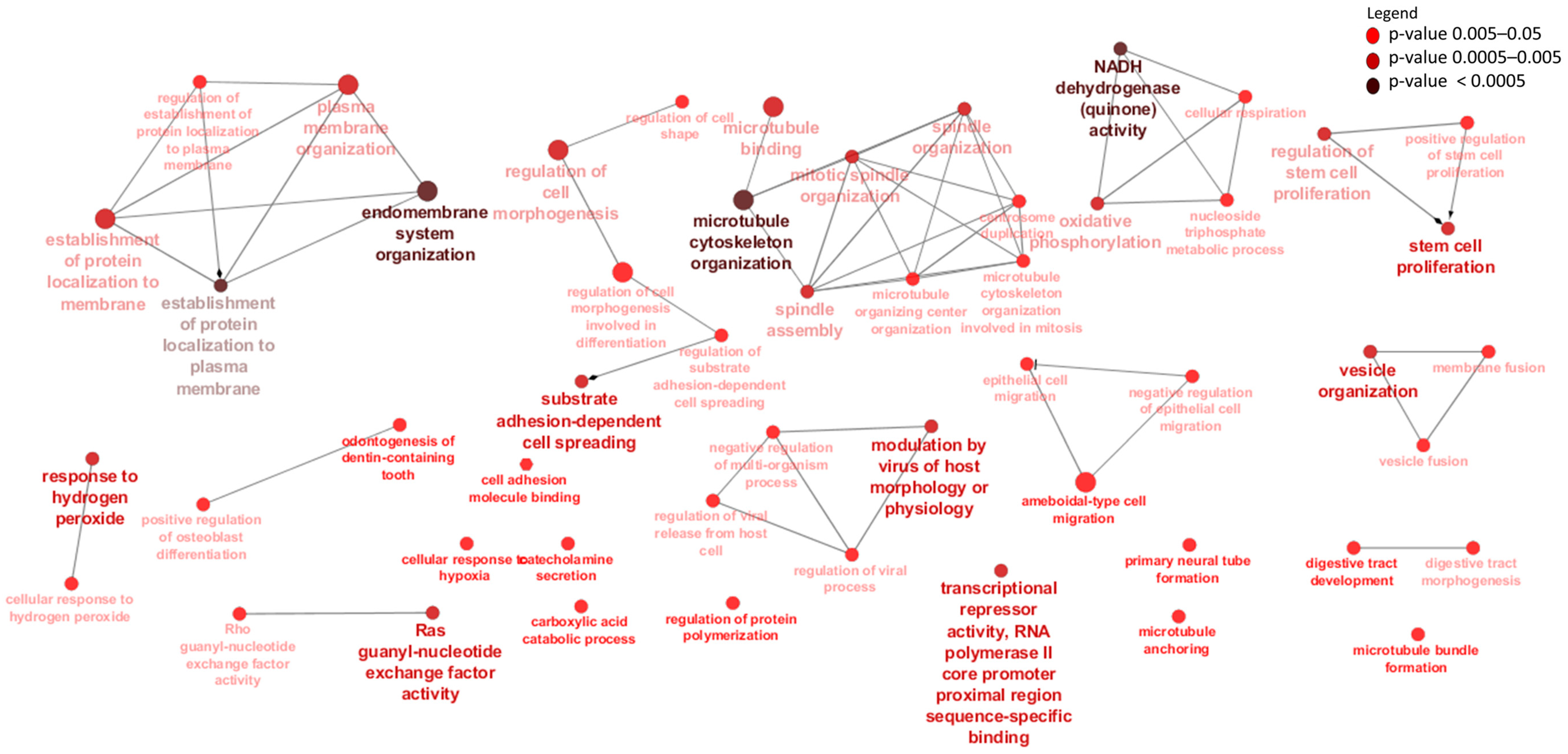
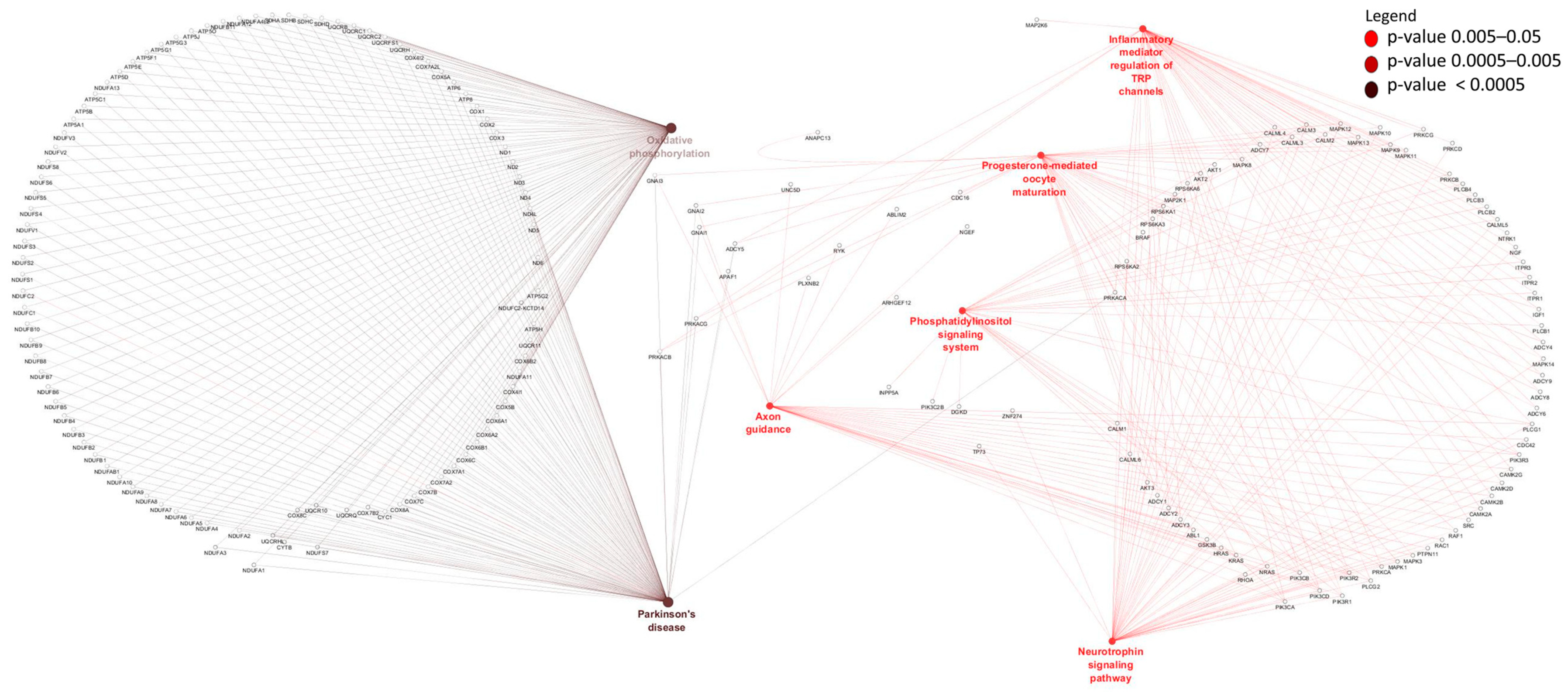
2.6. Gene Ontology and Pathways Enrichment of Differentially Expressed mRNAs
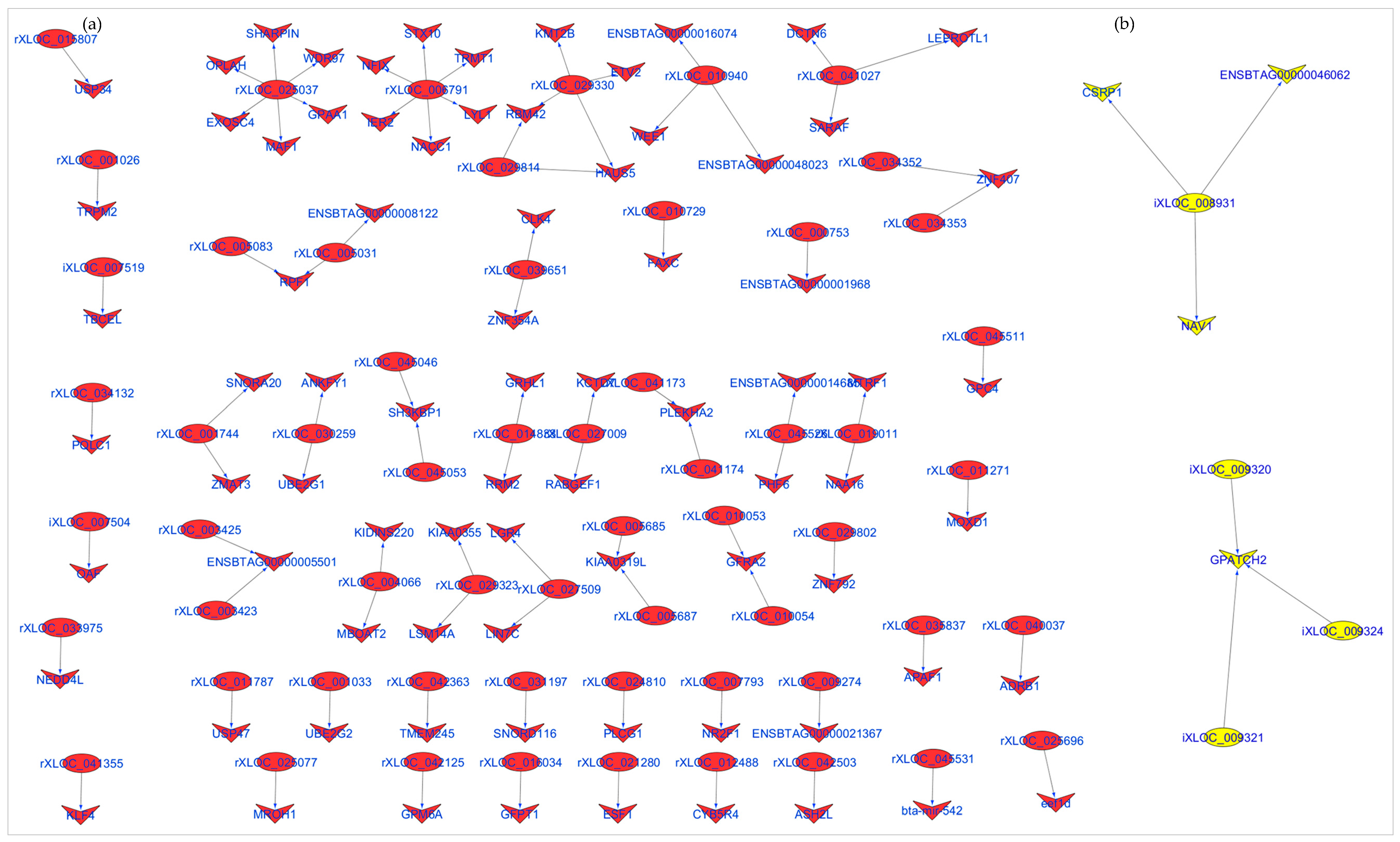
2.7. Identification of lncRNA cis Target Genes and cis Target Gene Enrichment
2.8. Real Time Quantitative Polymerase Chain Reaction
3. Results
3.1. RNA-Sequencing and Identification of Expressed mRNA and lncRNA Genes in the Gastrointestinal Tract of Calves
3.2. Genomic Features and Characteristics of Expressed lncRNAs
3.3. Differentially Expressed mRNAs and Enrichment Analyses
3.4. Identification and Enrichment Analyses of cis Target Genes of lncRNAs
3.5. Differentially Expressed lncRNAs in Ileum and Rumen Tissues
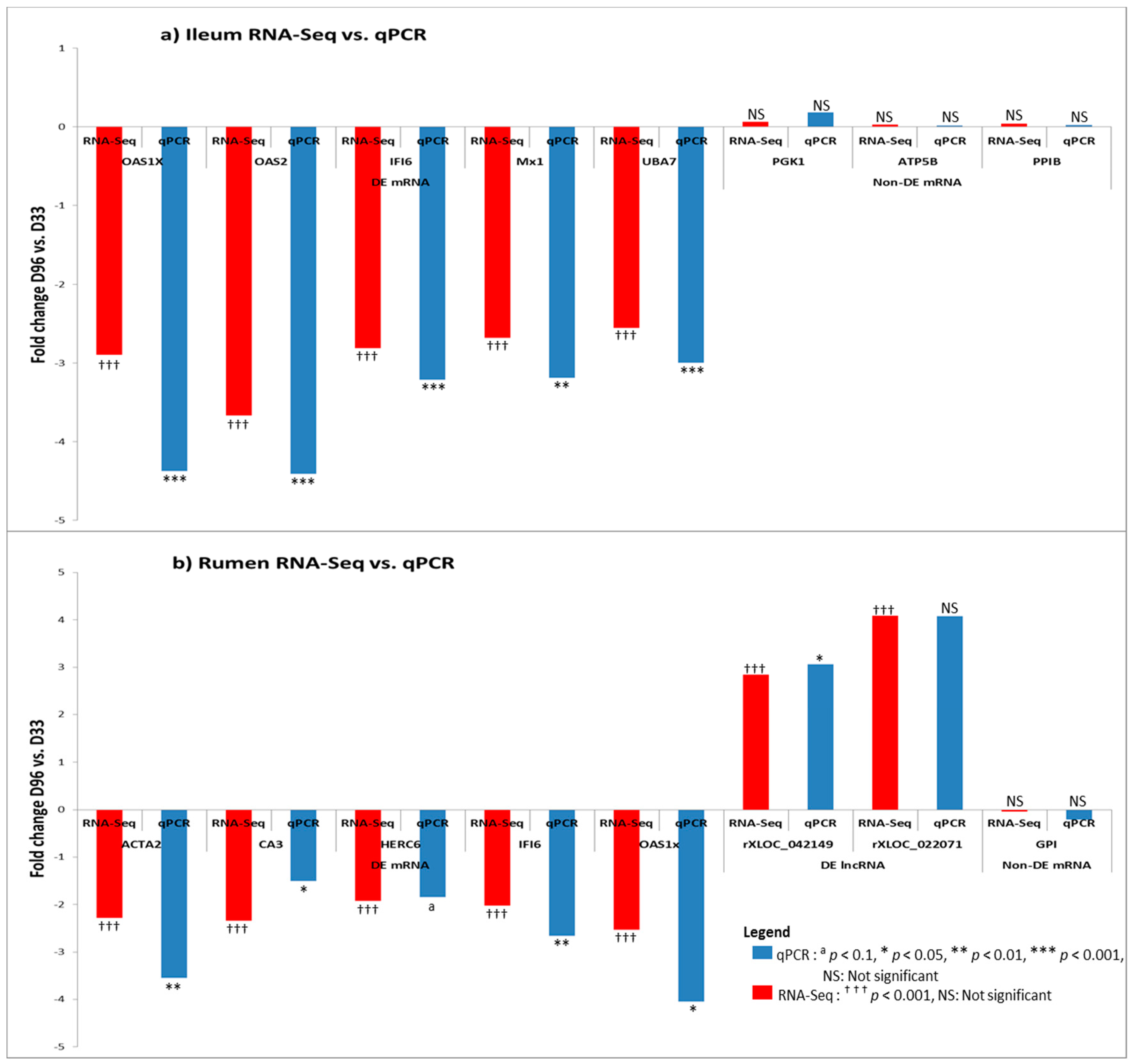
3.6. Real Time Quantitative PCR Confirmation of RNA-Seq Results
4. Discussion
4.1. Transcriptome mRNA Transition from Pre- to Post-Weaning Period in Rumen and Ileum Tissues
4.2. lncRNAs in Rumen and Ileum Tissues and Predicted Functions
5. Conclusions
Supplementary Materials
Availability of Sequence Data
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Guilloteau, P.; Zabielski, R.; Blum, J.W. Gastrointestinal tract and digestion in the young ruminant: Ontogenesis, adaptations, consequences and manipulations. J. Physiol. Pharmacol. 2009, 60, 37–46. [Google Scholar] [PubMed]
- Malmuthuge, N.; Griebel, P.J.; Guan, L.L. The gut microbiome and its potential role in the development and function of newborn calf gastrointestinal tract. Front. Vet. Sci. 2015, 2. [Google Scholar] [CrossRef] [PubMed]
- Turner, M.; Galloway, A.; Vigorito, E. Noncoding RNA and its associated proteins as regulatory elements of the immune system. Nat. Immunol. 2014, 15, 484–491. [Google Scholar] [CrossRef] [PubMed]
- Derrien, T.; Johnson, R.; Bussotti, G.; Tanzer, A.; Djebali, S.; Tilgner, H.; Guernec, G.; Martin, D.; Merkel, A.; Knowles, D.; et al. The gencode v7 catalog of human long noncoding RNAs: Analysis of their gene structure, evolution, and expression. Genome Res. 2012, 22, 1775–1789. [Google Scholar] [CrossRef] [PubMed]
- Mukherjee, N.; Calviello, L.; Hirsekorn, A.; de Pretis, S.; Pelizzola, M.; Ohler, U. Integrative classification of human coding and noncoding genes through RNA metabolism profiles. Nat. Struct. Mol. Biol. 2017, 24, 86–96. [Google Scholar] [CrossRef] [PubMed]
- Quinn, J.J.; Chang, H.Y. Unique features of long non-coding RNA biogenesis and function. Nat. Rev. Genet. 2016, 17, 47–62. [Google Scholar] [CrossRef] [PubMed]
- Melissari, M.T.; Grote, P. Roles for long non-coding RNAs in physiology and disease. Pflugers Arch.-Eur. J. Physiol. 2016, 468, 945–958. [Google Scholar] [CrossRef] [PubMed]
- Mercer, T.R.; Mattick, J.S. Structure and function of long noncoding RNAs in epigenetic regulation. Nat. Struct. Mol. Biol. 2013, 20, 300–307. [Google Scholar] [CrossRef] [PubMed]
- Djebali, S.; Davis, C.A.; Merkel, A.; Dobin, A.; Lassmann, T.; Mortazavi, A.; Tanzer, A.; Lagarde, J.; Lin, W.; Schlesinger, F.; et al. Landscape of transcription in human cells. Nature 2012, 489, 101–108. [Google Scholar] [CrossRef] [PubMed]
- Devaux, Y.; Zangrando, J.; Schroen, B.; Creemers, E.E.; Pedrazzini, T.; Chang, C.-P.; Dorn, G.W., II; Thum, T.; Heymans, S. Long noncoding RNAs in cardiac development and ageing. Nat. Rev. Cardiol. 2015, 12, 415–425. [Google Scholar] [PubMed]
- Fatica, A.; Bozzoni, I. Long non-coding RNAs: New players in cell differentiation and development. Nat. Rev. Genet. 2014, 15, 7–21. [Google Scholar] [CrossRef] [PubMed]
- Heward, J.A.; Lindsay, M.A. Long non-coding RNAs in the regulation of the immune response. Trends Immunol. 2014, 35, 408–419. [Google Scholar] [CrossRef] [PubMed]
- Yang, Z.G.; Gao, L.; Guo, X.B.; Shi, Y.L. Roles of long non-coding RNAs in gastric cancer metastasis. World J. Gastroenterol. 2015, 21, 5220–5230. [Google Scholar] [CrossRef] [PubMed]
- Batista, Pedro J.; Chang, Howard Y. Long noncoding RNAs: Cellular address codes in development and disease. Cell 2013, 152, 1298–1307. [Google Scholar]
- Guttman, M.; Donaghey, J.; Carey, B.W.; Garber, M.; Grenier, J.K.; Munson, G.; Young, G.; Lucas, A.B.; Ach, R.; Bruhn, L.; et al. LincRNAs act in the circuitry controlling pluripotency and differentiation. Nature 2011, 477, 295–300. [Google Scholar] [CrossRef] [PubMed]
- Dinger, M.E.; Amaral, P.P.; Mercer, T.R.; Pang, K.C.; Bruce, S.J.; Gardiner, B.B.; Askarian-Amiri, M.E.; Ru, K.; Solda, G.; Simons, C.; et al. Long noncoding RNAs in mouse embryonic stem cell pluripotency and differentiation. Genome Res. 2008, 18, 1433–1445. [Google Scholar] [CrossRef] [PubMed]
- Chen, B.; Zhang, Y.; Zhang, X.; Jia, S.; Chen, S.; Kang, L. Genome-wide identification and developmental expression profiling of long noncoding RNAs during Drosophila metamorphosis. Sci. Rep. 2016, 6, 23330. [Google Scholar] [CrossRef] [PubMed]
- Caballero, J.; Gilbert, I.; Fournier, E.; Gagne, D.; Scantland, S.; Macaulay, A.; Robert, C. Exploring the function of long non-coding RNA in the development of bovine early embryos. Reprod. Fertil. Dev. 2014, 27, 40–52. [Google Scholar] [CrossRef] [PubMed]
- Sauvageau, M.; Goff, L.A.; Lodato, S.; Bonev, B.; Groff, A.F.; Gerhardinger, C.; Sanchez-Gomez, D.B.; Hacisuleyman, E.; Li, E.; Spence, M.; et al. Multiple knockout mouse models reveal lincRNAs are required for life and brain development. eLife 2013, 2, e01749. [Google Scholar] [CrossRef] [PubMed]
- Cabili, M.N.; Trapnell, C.; Goff, L.; Koziol, M.; Tazon-Vega, B.; Regev, A.; Rinn, J.L. Integrative annotation of human large intergenic noncoding RNAs reveals global properties and specific subclasses. Genes Dev. 2011, 25, 1915–1927. [Google Scholar] [CrossRef] [PubMed]
- Koufariotis, L.T.; Chen, Y.-P.P.; Chamberlain, A.; Vander Jagt, C.; Hayes, B.J. A catalogue of novel bovine long noncoding RNA across 18 tissues. PLoS ONE 2015, 10, e0141225. [Google Scholar] [CrossRef] [PubMed]
- Mirza, A.H.; Berthelsen, C.H.; Seemann, S.E.; Pan, X.; Frederiksen, K.S.; Vilien, M.; Gorodkin, J.; Pociot, F. Transcriptomic landscape of lncRNAs in inflammatory bowel disease. Genome Med. 2015, 7, 39. [Google Scholar] [CrossRef] [PubMed]
- Liang, L.; Ai, L.; Qian, J.; Fang, J.-Y.; Xu, J. Long noncoding RNA expression profiles in gut tissues constitute molecular signatures that reflect the types of microbes. Sci. Rep. 2015, 5, 11763. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.Y.; Xiao, L.; Wang, J.Y. Post-transcriptional regulation of intestinal epithelial integrity by noncoding RNAs. Wiley Interdiscip. Rev. RNA 2017, 8. [Google Scholar] [CrossRef] [PubMed]
- McHugh, C.A.; Chen, C.-K.; Chow, A.; Surka, C.F.; Tran, C.; McDonel, P.; Pandya-Jones, A.; Blanco, M.; Burghard, C.; Moradian, A.; et al. The Xist lncRNA interacts directly with sharp to silence transcription through HDAC3. Nature 2015, 521, 232–236. [Google Scholar] [CrossRef] [PubMed]
- Gupta, R.A.; Shah, N.; Wang, K.C.; Kim, J.; Horlings, H.M.; Wong, D.J.; Tsai, M.C.; Hung, T.; Argani, P.; Rinn, J.L.; et al. Long non-coding RNA HOTAIR reprograms chromatin state to promote cancer metastasis. Nature 2010, 464, 1071–1076. [Google Scholar] [CrossRef] [PubMed]
- Gutschner, T.; Hammerle, M.; Eissmann, M.; Hsu, J.; Kim, Y.; Hung, G.; Revenko, A.; Arun, G.; Stentrup, M.; Gross, M.; et al. The noncoding RNA MALAT1 is a critical regulator of the metastasis phenotype of lung cancer cells. Cancer Res. 2013, 73, 1180–1189. [Google Scholar] [CrossRef] [PubMed]
- Qu, Z.; Adelson, D.L. Bovine ncRNAs are abundant, primarily intergenic, conserved and associated with regulatory genes. PLoS ONE 2012, 7, e42638. [Google Scholar] [CrossRef] [PubMed]
- Weikard, R.; Hadlich, F.; Kuehn, C. Identification of novel transcripts and noncoding RNAs in bovine skin by deep next generation sequencing. BMC Genom. 2013, 14, 789. [Google Scholar] [CrossRef] [PubMed]
- Billerey, C.; Boussaha, M.; Esquerré, D.; Rebours, E.; Djari, A.; Meersseman, C.; Klopp, C.; Gautheret, D.; Rocha, D. Identification of large intergenic non-coding RNAs in bovine muscle using next-generation transcriptomic sequencing. BMC Genom. 2014, 15, 499. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Ding, X.; Li, X.; Jin, C.; Yue, Y.; Li, G.; Guo, H. An atlas and analysis of bovine skeletal muscle long noncoding RNAs. Anim. Genet. 2017, 48, 278–286. [Google Scholar] [CrossRef] [PubMed]
- Do, D.N.; Ibeagha-Awemu, E.M. Non-coding RNA roles in ruminant mammary gland development and lactation. In Current Topics in Lactation; InTech: London, UK, 2017. [Google Scholar]
- Tong, C.; Chen, Q.; Zhao, L.; Ma, J.; Ibeagha-Awemu, E.M.; Zhao, X. Identification and characterization of long intergenic noncoding RNAs in bovine mammary glands. BMC Genom. 2017, 18, 468. [Google Scholar] [CrossRef] [PubMed]
- Andersson, L.; Archibald, A.L.; Bottema, C.D.; Brauning, R.; Burgess, S.C.; Burt, D.W.; Casas, E.; Cheng, H.H.; Clarke, L.; Couldrey, C. Coordinated international action to accelerate genome-to-phenome with FAANG, the Functional Annotation of Animal Genomes Project. Genome Biol. 2015, 16, 57. [Google Scholar] [CrossRef] [PubMed]
- Tuggle, C.K.; Giuffra, E.; White, S.N.; Clarke, L.; Zhou, H.; Ross, P.J.; Acloque, H.; Reecy, J.M.; Archibald, A.; Bellone, R.R. Go-FAANG meeting: A Gathering on Functional Annotation of Animal Genomes. Anim. Genet. 2016, 47, 528–533. [Google Scholar] [CrossRef] [PubMed]
- CCAC. CCAC guidelines on: The care and use of farm animals in research, teaching and testing. Canadian Council on Animal Care, 2009. Available online: https://www.ccac.ca/Documents/Standards/Guidelines/Farm_Animals.pdf (accessed on 16 January 2013).
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A flexible trimmer for illumina sequence data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef] [PubMed]
- Dobin, A.; Davis, C.A.; Schlesinger, F.; Drenkow, J.; Zaleski, C.; Jha, S.; Batut, P.; Chaisson, M.; Gingeras, T.R. Star: Ultrafast universal RNA-seq aligner. Bioinformatics 2013, 29, 15–21. [Google Scholar] [CrossRef] [PubMed]
- Picard tools. Available online: https://broadinstitute.github.io/picard/ (accessed on 17 June 2016).
- DeLuca, D.S.; Levin, J.Z.; Sivachenko, A.; Fennell, T.; Nazaire, M.-D.; Williams, C.; Reich, M.; Winckler, W.; Getz, G. RNA-SeQC: RNA-seq metrics for quality control and process optimization. Bioinformatics 2012, 28, 1530–1532. [Google Scholar] [CrossRef] [PubMed]
- Trapnell, C.; Roberts, A.; Goff, L.; Pertea, G.; Kim, D.; Kelley, D.R.; Pimentel, H.; Salzberg, S.L.; Rinn, J.L.; Pachter, L. Differential gene and transcript expression analysis of rna-seq experiments with tophat and cufflinks. Nat. Protoc 2012, 7, 562. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Handsaker, B.; Wysoker, A.; Fennell, T.; Ruan, J.; Homer, N.; Marth, G.; Abecasis, G.; Durbin, R. The sequence alignment/map format and samtools. Bioinformatics 2009, 25, 2078–2079. [Google Scholar] [CrossRef] [PubMed]
- Sun, L.; Luo, H.; Bu, D.; Zhao, G.; Yu, K.; Zhang, C.; Liu, Y.; Chen, R.; Zhao, Y. Utilizing sequence intrinsic composition to classify protein-coding and long non-coding transcripts. Nucleic Acids Res. 2013, 41, e166. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Park, H.J.; Dasari, S.; Wang, S.; Kocher, J.-P.; Li, W. CPAT: Coding-potential assessment tool using an alignment-free logistic regression model. Nucleic Acids Res. 2013, 41, e74. [Google Scholar] [CrossRef] [PubMed]
- Edgar, R.C. Search and clustering orders of magnitude faster than BLAST. Bioinformatics 2010, 26, 2460–2461. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Y.; Li, H.; Fang, S.; Kang, Y.; Hao, Y.; Li, Z.; Bu, D.; Sun, N.; Zhang, M.Q.; Chen, R. NONCODE 2016: An informative and valuable data source of long non-coding RNAs. Nucleic Acids Res. 2016, 44, D203–D208. [Google Scholar] [CrossRef] [PubMed]
- Anders, S.; Pyl, P.T.; Huber, W. HTSeq—A python framework to work with high-throughput sequencing data. Bioinformatics 2015, 31, 166–169. [Google Scholar] [CrossRef] [PubMed]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef] [PubMed]
- R Development Core Team. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2013; ISBN 3-900051-07-0. [Google Scholar]
- Benjamini, Y.; Hochberg, Y. Controlling the false discovery rate: A practical and powerful approach to multiple testing. J. R. Stat. Soc. Ser. B (Methodol.) 1995, 57, 289–300. [Google Scholar]
- Bindea, G.; Mlecnik, B.; Hackl, H.; Charoentong, P.; Tosolini, M.; Kirilovsky, A.; Fridman, W.H.; Pages, F.; Trajanoski, Z.; Galon, J. ClueGO: A cytoscape plug-in to decipher functionally grouped gene ontology and pathway annotation networks. Bioinformatics 2009, 25, 1091–1093. [Google Scholar] [CrossRef] [PubMed]
- Shannon, P.; Markiel, A.; Ozier, O.; Baliga, N.S.; Wang, J.T.; Ramage, D.; Amin, N.; Schwikowski, B.; Ideker, T. Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Res. 2003, 13, 2498–2504. [Google Scholar] [CrossRef] [PubMed]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]
- Chao, T.; Wang, G.; Ji, Z.; Liu, Z.; Hou, L.; Wang, J.; Wang, J. Transcriptome analysis of three sheep intestinal regions reveals key pathways and hub regulatory genes of large intestinal lipid metabolism. Sci. Rep. 2017, 7, 5345. [Google Scholar] [CrossRef] [PubMed]
- Liang, G.; Malmuthuge, N.; Bao, H.; Stothard, P.; Griebel, P.J.; Guan, L.L. Transcriptome analysis reveals regional and temporal differences in mucosal immune system development in the small intestine of neonatal calves. BMC Genom. 2016, 17, 602. [Google Scholar] [CrossRef] [PubMed]
- Meale, S.; Li, S.; Azevedo, P.; Derakhshani, H.; DeVries, T.; Plaizier, J.; Steele, M.; Khafipour, E. Weaning age influences the severity of gastrointestinal microbiome shifts in dairy calves. Sci. Rep. 2017, 7, 198. [Google Scholar] [CrossRef] [PubMed]
- Inlay, M.A.; Bhattacharya, D.; Sahoo, D.; Serwold, T.; Seita, J.; Karsunky, H.; Plevritis, S.K.; Dill, D.L.; Weissman, I.L. Ly6d marks the earliest stage of B-cell specification and identifies the branchpoint between B-cell and T-cell development. Genes Dev. 2009, 23, 2376–2381. [Google Scholar] [CrossRef] [PubMed]
- Moncada, D.M.; Kammanadiminti, S.J.; Chadee, K. Mucin and Toll-like receptors in host defense against intestinal parasites. Trends Parasitol. 2003, 19, 305–311. [Google Scholar] [CrossRef]
- Chao, F.; Zhang, J.; Zhang, Y.; Liu, H.; Yang, C.; Wang, J.; Guo, Y.; Wen, X.; Zhang, K.; Huang, B.; et al. Embigin, regulated by HOXC8, plays a suppressive role in breast tumorigenesis. Oncotarget 2015, 6, 23496–23509. [Google Scholar] [CrossRef] [PubMed]
- Krueger, A.B.; Drasin, D.J.; Lea, W.A.; Patrick, A.N.; Patnaik, S.; Backos, D.S.; Matheson, C.J.; Hu, X.; Barnaeva, E.; Holliday, M.J.; et al. Allosteric inhibitors of the Eya2 phosphatase are selective and inhibit Eya2-mediated cell migration. J. Biol. Chem. 2014, 289, 16349–16361. [Google Scholar] [CrossRef] [PubMed]
- Tuteja, N. Signaling through g protein coupled receptors. Plant Signal. Behav. 2009, 4, 942–947. [Google Scholar] [CrossRef] [PubMed]
- Todini, L. Thyroid hormones in small ruminants: Effects of endogenous, environmental and nutritional factors. Animal 2007, 1, 997–1008. [Google Scholar] [CrossRef] [PubMed]
- Chin, K.-C.; Cresswell, P. Viperin (cig5), an IFN-inducible antiviral protein directly induced by human cytomegalovirus. Proc. Natl. Acad. Sci. USA 2001, 98, 15125–15130. [Google Scholar] [CrossRef] [PubMed]
- Pulendran, B. Learning immunology from the yellow fever vaccine: Innate immunity to systems vaccinology. Nat. Rev. Immunol. 2009, 9, 741–747. [Google Scholar] [CrossRef] [PubMed]
- Hovanessian, A.G.; Svab, J.; Marie, I.; Robert, N.; Chamaret, S.; Laurent, A.G. Characterization of 69- and 100-kDa forms of 2-5A-synthetase from interferon-treated human cells. J. Biol. Chem. 1988, 263, 4945–4949. [Google Scholar] [PubMed]
- Malmuthuge, N.; Li, M.; Fries, P.; Griebel, P.J.; Guan, L.L. Regional and age dependent changes in gene expression of Toll-like receptors and key antimicrobial defence molecules throughout the gastrointestinal tract of dairy calves. Vet. Immunol. Immunopathol. 2012, 146, 18–26. [Google Scholar] [CrossRef] [PubMed]
- Meale, S.J.; Li, S.; Azevedo, P.; Derakhshani, H.; Plaizier, J.C.; Khafipour, E.; Steele, M.A. Development of ruminal and fecal microbiomes are affected by weaning but not weaning strategy in dairy calves. Front. Microbiol. 2016, 7, 582. [Google Scholar] [CrossRef] [PubMed]
- Kotake, Y.; Nakagawa, T.; Kitagawa, K.; Suzuki, S.; Liu, N.; Kitagawa, M.; Xiong, Y. Long non-coding RNA ANRIL is required for the PRC2 recruitment to and silencing of p15INK4B tumor suppressor gene. Oncogene 2011, 30, 1956–1962. [Google Scholar] [CrossRef] [PubMed]
- Maamar, H.; Cabili, M.N.; Rinn, J.; Raj, A. Linc-HOXA1 is a noncoding RNA that represses HOXA1 transcription in cis. Genes Dev. 2013, 27, 1260–1271. [Google Scholar] [CrossRef] [PubMed]
- Ponjavic, J.; Oliver, P.L.; Lunter, G.; Ponting, C.P. Genomic and transcriptional co-localization of protein-coding and long non-coding RNA pairs in the developing brain. PLoS Genet. 2009, 5, e1000617. [Google Scholar] [CrossRef] [PubMed]
- Rinn, J.L.; Kertesz, M.; Wang, J.K.; Squazzo, S.L.; Xu, X.; Brugmann, S.A.; Goodnough, L.H.; Helms, J.A.; Farnham, P.J.; Segal, E.; et al. Functional demarcation of active and silent chromatin domains in human HOX loci by noncoding RNAs. Cell 2007, 129, 1311–1323. [Google Scholar] [CrossRef] [PubMed]
- Wang, K.C.; Yang, Y.W.; Liu, B.; Sanyal, A.; Corces-Zimmerman, R.; Chen, Y.; Lajoie, B.R.; Protacio, A.; Flynn, R.A.; Gupta, R.A.; et al. A long noncoding RNA maintains active chromatin to coordinate homeotic gene expression. Nature 2011, 472, 120–124. [Google Scholar] [CrossRef] [PubMed]
- Ren, H.; Wang, G.; Chen, L.; Jiang, J.; Liu, L.; Li, N.; Zhao, J.; Sun, X.; Zhou, P. Genome-wide analysis of long non-coding RNAs at early stage of skin pigmentation in goats (Capra hircus). BMC Genom. 2016, 17, 67. [Google Scholar] [CrossRef] [PubMed]
- Pauli, A.; Valen, E.; Lin, M.F.; Garber, M.; Vastenhouw, N.L.; Levin, J.Z.; Fan, L.; Sandelin, A.; Rinn, J.L.; Regev, A. Systematic identification of long noncoding RNAs expressed during zebrafish embryogenesis. Genome Res. 2012, 22, 577–591. [Google Scholar] [CrossRef] [PubMed]
- Sun, M.; Gadad, S.S.; Kim, D.-S.; Kraus, W.L. Discovery, annotation, and functional analysis of long noncoding RNAs controlling cell-cycle gene expression and proliferation in breast cancer cells. Mol. Cell 2015, 59, 698–711. [Google Scholar] [CrossRef] [PubMed]
- Do, D.N.; Dudemaine, P.-L.; Fomenky, B.; Ibeagha-Awemu, E.M. Transcriptome analysis of non-coding RNAs in livestock species: Elucidating the ambiguity. In Applications of RNA-seq and Omics Strategies-from Microorganisms to Human Health; InTech: London, UK, 2017. [Google Scholar]
- Aqeilan, R.I.; Trapasso, F.; Hussain, S.; Costinean, S.; Marshall, D.; Pekarsky, Y.; Hagan, J.P.; Zanesi, N.; Kaou, M.; Stein, G.S. Targeted deletion of Wwox reveals a tumor suppressor function. Proc. Natl. Acad. Sci. USA 2007, 104, 3949–3954. [Google Scholar] [CrossRef] [PubMed]
- Tong, W.; Yang, L.; Yu, Q.; Yao, J.; He, A. A new tumor suppressor lncRNA RP11-190D6. 2 inhibits the proliferation, migration, and invasion of epithelial ovarian cancer cells. Onco Targets Ther. 2017, 10, 1227–1235. [Google Scholar] [CrossRef] [PubMed]
- Zhou, X.-P.; Woodford-Richens, K.; Lehtonen, R.; Kurose, K.; Aldred, M.; Hampel, H.; Launonen, V.; Virta, S.; Pilarski, R.; Salovaara, R. Germline mutations in BMPR1A/ALK3 cause a subset of cases of juvenile polyposis syndrome and of cowden and bannayan-riley-ruvalcaba syndromes. Am. J. Hum. Genet. 2001, 69, 704–711. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Van Hattem, W.A.; Brosens, L.A.; de Leng, W.W.; Morsink, F.H.; Lens, S.; Carvalho, R.; Giardiello, F.M.; Offerhaus, G.J.A. Large genomic deletions of SMAD4, BMPR1A and PTEN in juvenile polyposis. Gut 2008. [Google Scholar] [CrossRef] [PubMed]
- Jalali, S.; Bhartiya, D.; Lalwani, M.K.; Sivasubbu, S.; Scaria, V. Systematic transcriptome wide analysis of lncRNA-miRNA interactions. PLoS ONE 2013, 8, e53823. [Google Scholar] [CrossRef] [PubMed]
- Wan, X.; Ding, X.; Chen, S.; Song, H.; Jiang, H.; Fang, Y.; Li, P.; Guo, J. The functional sites of miRNAs and lncRNAs in gastric carcinogenesis. Tumor Biol. 2015, 36, 521–532. [Google Scholar] [CrossRef] [PubMed]
- Naeem, A.; Drackley, J.; Stamey, J.; Loor, J. Role of metabolic and cellular proliferation genes in ruminal development in response to enhanced plane of nutrition in neonatal holstein calves. J. Dairy Sci. 2012, 95, 1807–1820. [Google Scholar] [CrossRef] [PubMed]
- Laarman, A.H.; Dionissopoulos, L.; AlZahal, O.; Steele, M.A.; Greenwood, S.L.; Matthews, J.C.; McBride, B.W. Butyrate supplementation affects mRNA abundance of genes involved in glycolysis, oxidative phosphorylation and lipogenesis in the rumen epithelium of holstein dairy cows. Am. J. Anim. Vet. Sci. 2013, 8, 239–245. [Google Scholar] [CrossRef][Green Version]
- Baldwin, R.; McLeod, K.; Klotz, J.; Heitmann, R. Rumen development, intestinal growth and hepatic metabolism in the pre- and postweaning ruminant. J. Dairy Sci. 2004, 87, E55–E65. [Google Scholar] [CrossRef]
- EXOSC4 Gene – GeneCards | EXOS4 Protein | EXOS4 Antibody. Available online: http://www.genecards.org/cgi-bin/carddisp.pl?gene=EXOSC4 (accsessed on 18 October 2017).
- Picco, G.; Julien, S.; Brockhausen, I.; Beatson, R.; Antonopoulos, A.; Haslam, S.; Mandel, U.; Dell, A.; Pinder, S.; Taylor-Papadimitriou, J.; et al. Over-expression of ST3Gal-I promotes mammary tumorigenesis. Glycobiology 2010, 20, 1241–1250. [Google Scholar] [CrossRef] [PubMed]
- Ramsey, I.S.; Delling, M.; Clapham, D.E. An introduction to TRP channels. Annu. Rev. Physiol. 2006, 68, 619–647. [Google Scholar] [CrossRef] [PubMed]
- Boesmans, W.; Owsianik, G.; Tack, J.; Voets, T.; Berghe, P.V. TRP channels in neurogastroenterology: Opportunities for therapeutic intervention. Br. J. Pharmacol. 2011, 162, 18–37. [Google Scholar] [CrossRef] [PubMed]
- Hu, F.; Gou, L.; Liu, Q.; Zhang, W.; Luo, M.; Zhang, X. G-patch domain containing 2, a gene highly expressed in testes, inhibits nuclear factor-κB and cell proliferation. Mol. Med. Rep. 2015, 11, 1252–1257. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Lin, M.L.; Fukukawa, C.; Park, J.H.; Naito, K.; Kijima, K.; Shimo, A.; Ajiro, M.; Nishidate, T.; Nakamura, Y.; Katagiri, T. Involvement of G-patch domain containing 2 overexpression in breast carcinogenesis. Cancer Sci. 2009, 100, 1443–1450. [Google Scholar] [CrossRef] [PubMed]
- He, H.; Liu, X.-l.; Zhang, H.-l.; Yang, J.; Niu, F.-b.; Li, Z.-x.; Liu, Y.; Chen, L. Snv and haplotype analysis reveals new csrp1 variants associated with growth and carcass traits. Gene 2013, 522, 206–213. [Google Scholar] [CrossRef] [PubMed]
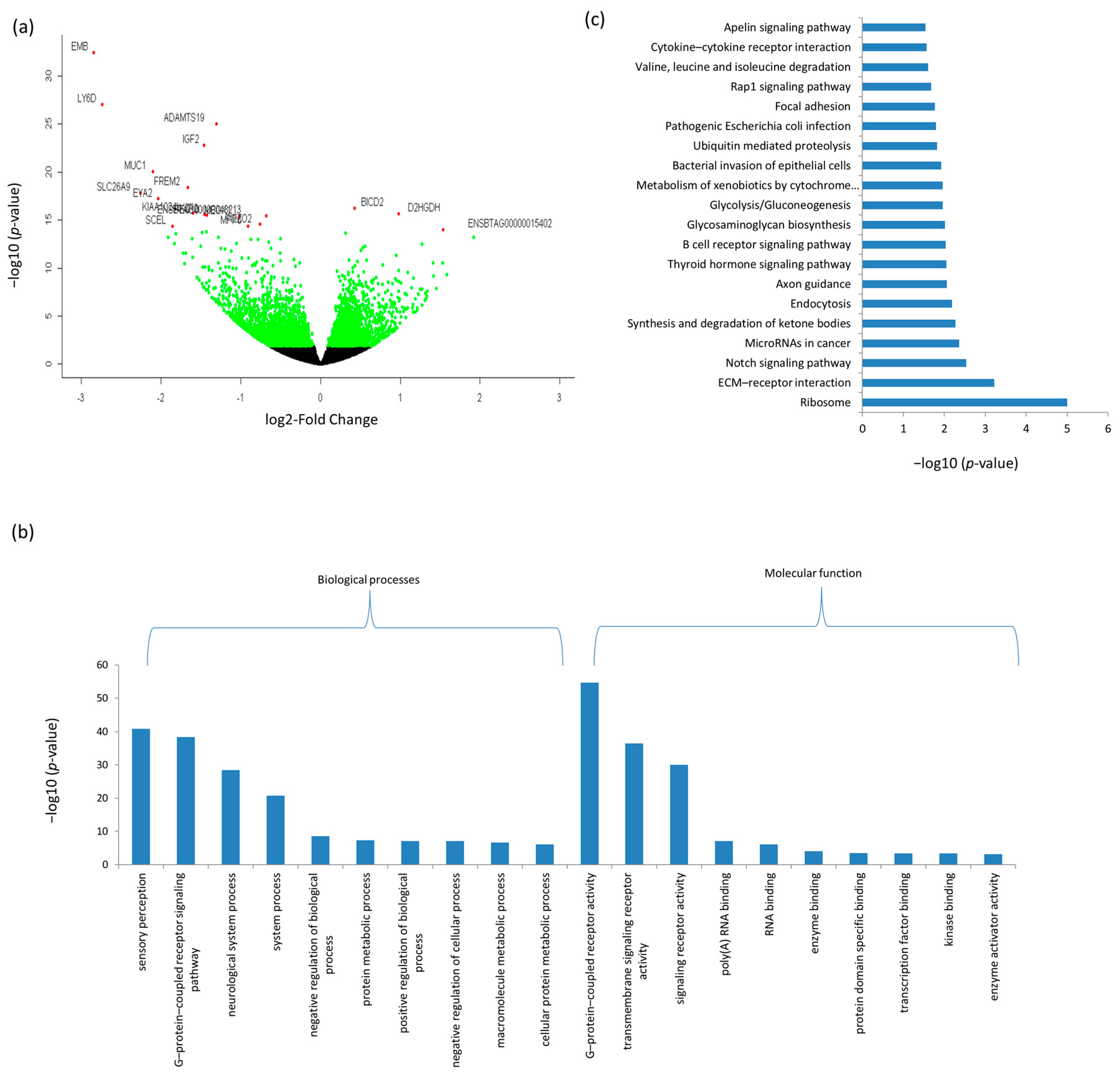
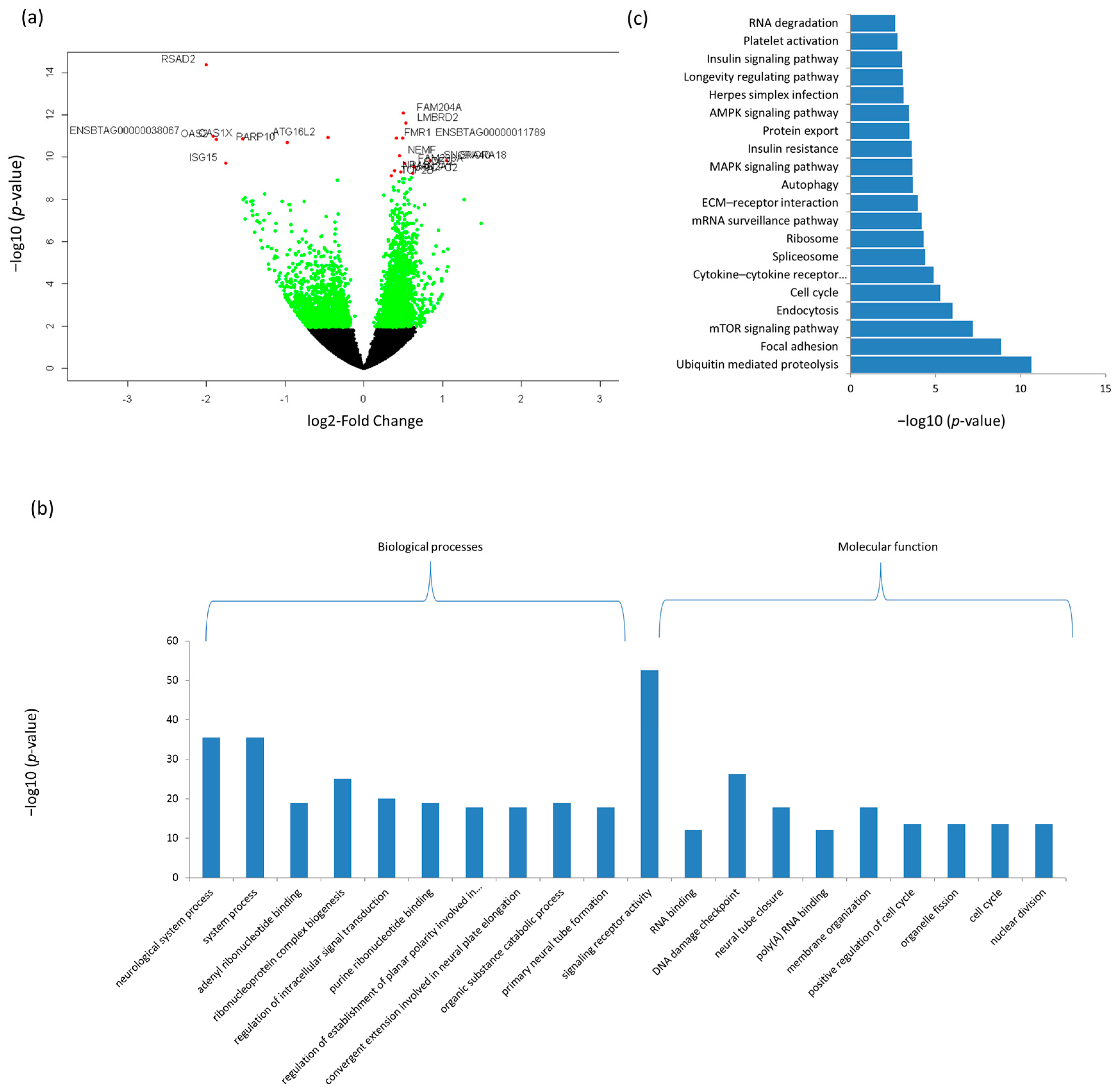
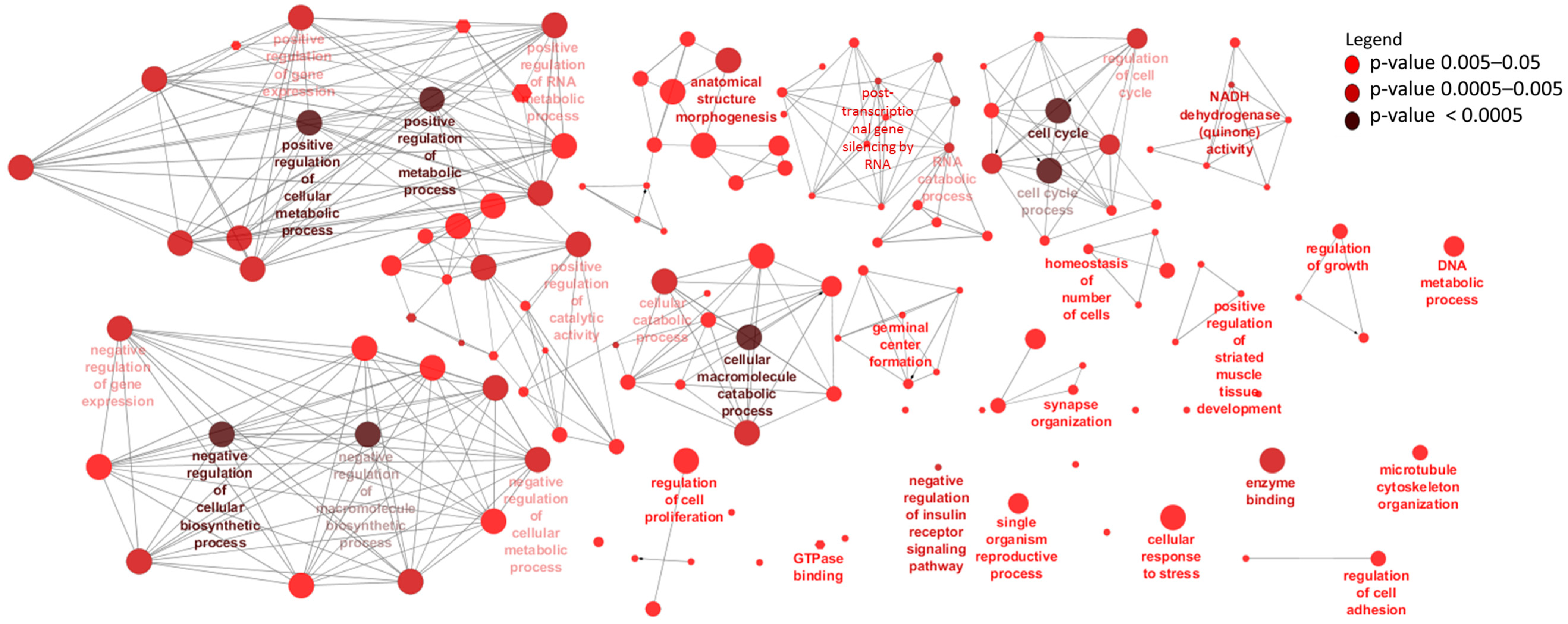
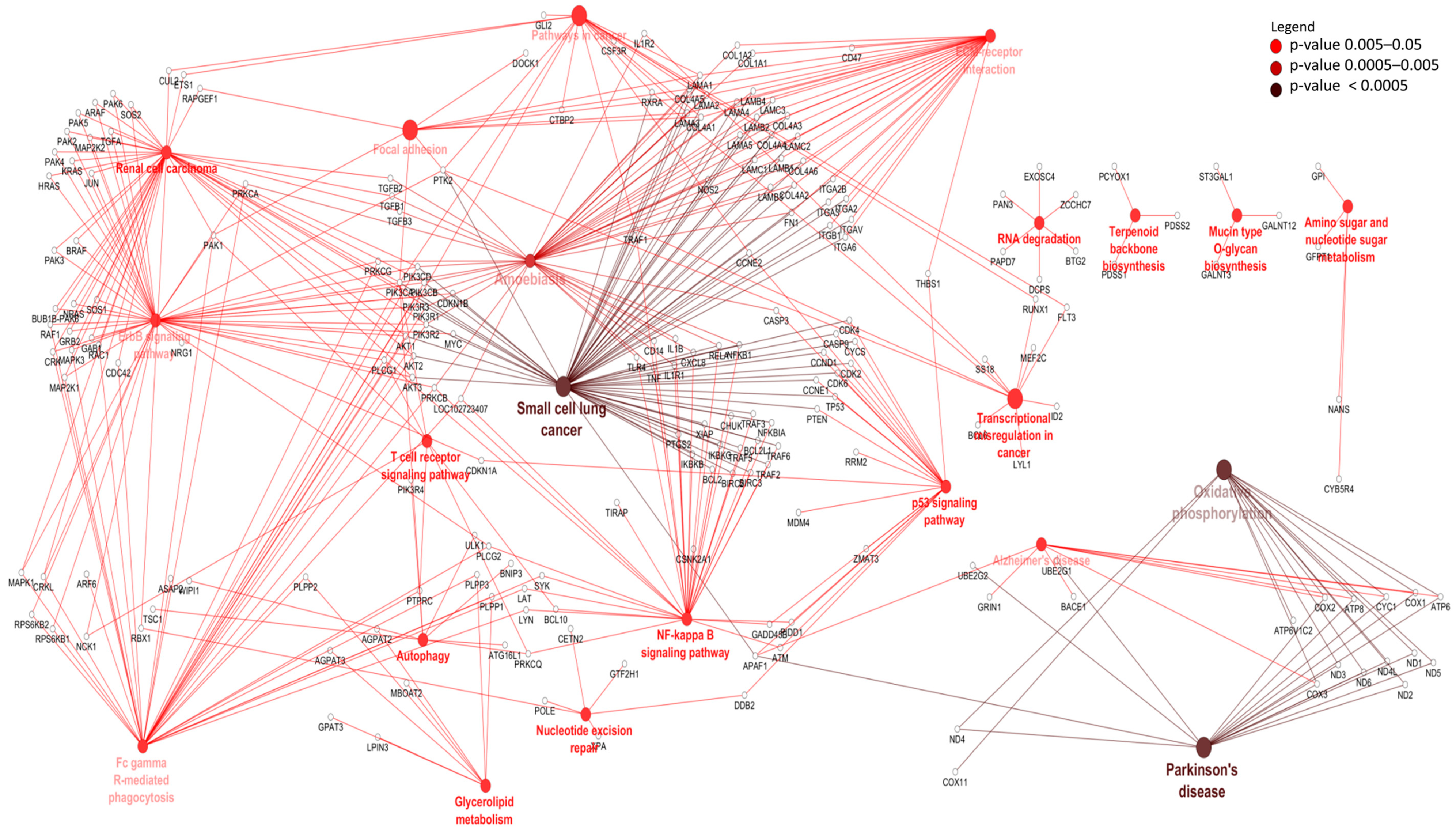
| Ensemble Name | Gene Name | Base Mean | Log2FC 1 | p-Value | p.BH 2 |
|---|---|---|---|---|---|
| Rumen | |||||
| ENSBTAG00000044010 | EMB | 45.41 | −2.85 | 3.43 × 10−33 | 2.68 × 10−29 |
| ENSBTAG00000034498 | LY6D | 45.30 | −2.74 | 9.13 × 10−28 | 4.76 × 10−24 |
| ENSBTAG00000016145 | ADAMTS19 | 281.43 | −1.30 | 9.85 × 10−26 | 3.85 × 10−22 |
| ENSBTAG00000013066 | IGF2 | 2769.08 | −1.46 | 1.57 × 10−23 | 4.91 × 10−20 |
| ENSBTAG00000017104 | MUC1 | 18.85 | −2.10 | 8.69 × 10−21 | 2.26 × 10−17 |
| ENSBTAG00000017032 | FREM2 | 344.45 | −1.67 | 3.78 × 10−19 | 8.45 × 10−16 |
| ENSBTAG00000001382 | SLC26A9 | 26.28 | −2.26 | 1.52 × 10−18 | 2.97 × 10−15 |
| ENSBTAG00000013336 | EYA2 | 5.79 | −2.04 | 6.12 × 10−18 | 1.06 × 10−14 |
| ENSBTAG00000046549 | BICD2 | 785.41 | 0.42 | 5.94 × 10−17 | 9.28 × 10−14 |
| ENSBTAG00000048309 | KIAA1024L | 12.77 | −1.60 | 1.73 × 10−16 | 2.46 × 10−13 |
| ENSBTAG00000002847 | D2HGDH | 305.50 | 0.98 | 2.26 × 10−16 | 2.94 × 10−13 |
| ENSBTAG00000037804 | IKZF2 | 73.24 | −1.46 | 2.73 × 10−16 | 3.28 × 10−13 |
| ENSBTAG00000038093 | PEG10 | 91.51 | −1.43 | 3.29 × 10−16 | 3.68 × 10−13 |
| ENSBTAG00000048213 | ENSBTAG00000048213 | 87.58 | −0.68 | 3.71 × 10−16 | 3.86 × 10−13 |
| ENSBTAG00000015751 | MEOX1 | 47.50 | −1.02 | 4.22 × 10−16 | 4.13 × 10−13 |
| ENSBTAG00000011171 | PIEZO2 | 194.57 | −0.76 | 2.83 × 10−15 | 2.61 × 10−12 |
| ENSBTAG00000015303 | MPP6 | 35.74 | −0.91 | 4.26 × 10−15 | 3.70 × 10−12 |
| ENSBTAG00000032821 | SCEL | 95.16 | −1.85 | 4.50 × 10−15 | 3.70 × 10−12 |
| ENSBTAG00000015402 | ENSBTAG00000015402 | 17.68 | 1.54 | 9.55 × 10−15 | 7.46 × 10−12 |
| ENSBTAG00000021420 | EPHA7 | 45.08 | −1.43 | 1.76 × 10−14 | 1.31 × 10−11 |
| ENSBTAG00000018303 | PAPPA2 | 52.76 | −1.48 | 1.95 × 10−14 | 1.39 × 10−11 |
| Ileum | |||||
| ENSBTAG00000016061 | RSAD2 | 562.08 | −2.00 | 4.13 × 10−15 | 6.05 × 10−11 |
| ENSBTAG00000005057 | FAM204A | 296.54 | 0.50 | 8.36 × 10−13 | 6.13 × 10−09 |
| ENSBTAG00000020142 | LMBRD2 | 296.05 | 0.53 | 2.39 × 10−12 | 1.17 × 10−08 |
| ENSBTAG00000011789 | ENSBTAG00000011789 | 410.62 | 0.50 | 1.22 × 10−11 | 2.34 × 10−08 |
| ENSBTAG00000012552 | FMR1 | 676.18 | 0.42 | 1.23 × 10−11 | 2.34 × 10−08 |
| ENSBTAG00000014628 | OAS2 | 243.63 | −1.88 | 1.43 × 10−11 | 2.34 × 10−08 |
| ENSBTAG00000019059 | ATG16L2 | 319.11 | −0.45 | 1.14 × 10−11 | 2.34 × 10−08 |
| ENSBTAG00000037527 | OAS1X | 643.29 | −1.54 | 1.38 × 10−11 | 2.34 × 10−08 |
| ENSBTAG00000038067 | ENSBTAG00000038067 | 124.15 | −1.92 | 9.93 × 10−12 | 2.34 × 10−08 |
| ENSBTAG00000009677 | PARP10 | 704.04 | −0.98 | 2.05 × 10−11 | 3.01 × 10−08 |
| ENSBTAG00000004934 | NEMF | 958.94 | 0.46 | 8.49 × 10−11 | 1.13 × 10−07 |
| ENSBTAG00000042280 | SNORA40 | 55.73 | 0.85 | 1.46 × 10−10 | 1.65 × 10−07 |
| ENSBTAG00000043258 | SNORA18 | 33.13 | 1.05 | 1.44 × 10−10 | 1.65 × 10−07 |
| ENSBTAG00000014707 | ISG15 | 767.04 | −1.75 | 1.95 × 10−10 | 1.96 × 10−07 |
| ENSBTAG00000019748 | FAM208A | 2070.64 | 0.52 | 2.00 × 10−10 | 1.96 × 10−07 |
| ENSBTAG00000012907 | ODF2L | 283.56 | 0.64 | 2.85 × 10−10 | 2.62 × 10−07 |
| ENSBTAG00000046797 | NRAS | 1034.73 | 0.39 | 4.52 × 10−10 | 3.90 × 10−07 |
| ENSBTAG00000025029 | MAN2A1 | 1673.74 | 0.47 | 4.90 × 10−10 | 4.00 × 10−07 |
| ENSBTAG00000008048 | GCFC2 | 407.32 | 0.62 | 5.88 × 10−10 | 4.54 × 10−07 |
| ENSBTAG00000004593 | TOP2B | 4025.86 | 0.35 | 7.58 × 10−10 | 5.56 × 10−07 |
| LncRNA 1 | Bta 2 | Start | End | NONCODE Name 3 | log2FC 4 | FC 5 | p-Value | p.BH 6 |
|---|---|---|---|---|---|---|---|---|
| Rumen | ||||||||
| rXLOC_027852 | 17 | 59,152,438 | 59,152,902 | Novel | −2.36 | −5.13 | 9.59 × 10−15 | 4.07 × 10−11 |
| rXLOC_020809 | 12 | 49,167,421 | 49,168,996 | Novel | 2.41 | 5.33 | 3.35 × 10−14 | 7.11 × 10−11 |
| rXLOC_013111 | 9 | 70,095,274 | 70,095,655 | Novel | −2.82 | −7.06 | 1.61 × 10−12 | 2.27 × 10−09 |
| rXLOC_025879 | 15 | 23,812,355 | 23,812,636 | Novel | −0.83 | −1.78 | 3.29 × 10−12 | 3.49 × 10−09 |
| rXLOC_022247 | 13 | 18,885,057 | 18,885,409 | Novel | 2.61 | 6.11 | 9.49 × 10−12 | 8.05 × 10−09 |
| rXLOC_035178 | 24 | 59,070,221 | 59,070,559 | Novel | 1.36 | 2.57 | 2.79 × 10−11 | 1.97 × 10−08 |
| rXLOC_016360 | 11 | 80,849,366 | 80,849,978 | Novel | 1.79 | 3.45 | 1.02 × 10−10 | 6.12 × 10−08 |
| rXLOC_041953 | 27 | 44,295,463 | 44,296,339 | Novel | 1.61 | 3.05 | 1.15 × 10−10 | 6.12 × 10−08 |
| rXLOC_022213 | 13 | 18,879,759 | 18,880,268 | Novel | 2.16 | 4.47 | 2.45 × 10−10 | 1.05 × 10−07 |
| rXLOC_033684 | 3 | 78,439,929 | 78,440,225 | NONBTAG012391.2 | 1.05 | 2.07 | 2.47 × 10−10 | 1.05 × 10−07 |
| rXLOC_039125 | 26 | 33,565,524 | 33,566,229 | Novel | 1.59 | 3.01 | 3.29 × 10−10 | 1.27 × 10−07 |
| rXLOC_041968 | 27 | 44,323,639 | 44,323,953 | Novel | 1.39 | 2.63 | 3.86 × 10−10 | 1.36 × 10−07 |
| rXLOC_006593 | 4 | 97,107,954 | 97,108,721 | Novel | 2.64 | 6.22 | 4.86 × 10−10 | 1.58 × 10−07 |
| rXLOC_008000 | 7 | 95,774,878 | 95,775,405 | Novel | 1.26 | 2.40 | 1.68 × 10−09 | 4.95 × 10−07 |
| rXLOC_022071 | 21 | 1,659,256 | 1,663,385 | NONBTAG020155.1 | 2.03 | 4.09 | 1.85 × 10−09 | 4.95 × 10−07 |
| rXLOC_035173 | 24 | 59,051,654 | 59,052,324 | Novel | 1.52 | 2.87 | 1.87 × 10−09 | 4.95 × 10−07 |
| rXLOC_022254 | 13 | 18,885,611 | 18,886,350 | Novel | 2.49 | 5.61 | 2.47 × 10−09 | 6.17 × 10−07 |
| rXLOC_022211 | 13 | 18,876,739 | 18,879,073 | Novel | 2.26 | 4.80 | 2.65 × 10−09 | 6.25 × 10−07 |
| rXLOC_039103 | 26 | 33,517,784 | 33,518,636 | Novel | 1.37 | 2.59 | 2.90 × 10−09 | 6.47 × 10−07 |
| rXLOC_045518 | X | 17,862,231 | 17,862,527 | NONBTAG017530.2 | 1.73 | 3.31 | 3.18 × 10−09 | 6.74 × 10−07 |
| Ileum | ||||||||
| iXLOC_002882 | 11 | 78,563,630 | 78,565,402 | Novel | 0.76 | 1.69 | 2.10 × 10−08 | 1.63 × 10−05 |
| iXLOC_009320 | 16 | 21,758,311 | 21,761,092 | Novel | 0.68 | 1.60 | 2.65 × 10−06 | 1.03 × 10−03 |
| iXLOC_007504 | 15 | 31,279,333 | 31,283,846 | Novel | −1.15 | −2.22 | 1.37 × 10−05 | 3.54 × 10−03 |
| iXLOC_007519 | 15 | 32,192,174 | 32,195,976 | Novel | 0.66 | 1.58 | 9.51 × 10−05 | 1.48 × 10−02 |
| iXLOC_033017 | 8 | 43,640,113 | 43,644,666 | Novel | 1.42 | 2.68 | 8.18 × 10−05 | 1.48 × 10−02 |
| iXLOC_009321 | 16 | 21,761,255 | 21,764,378 | Novel | 0.60 | 1.51 | 1.82 × 10−04 | 2.36 × 10−02 |
| iXLOC_008931 | 16 | 49,402,721 | 49,405,957 | Novel | −0.88 | −1.84 | 2.88 × 10−04 | 3.10 × 10−02 |
| iXLOC_009324 | 16 | 21,772,956 | 21,777,346 | Novel | 0.48 | 1.40 | 3.41 × 10−04 | 3.10 × 10−02 |
| iXLOC_031185 | 7 | 81,563,609 | 81,568,847 | Novel | 1.08 | 2.11 | 3.59 × 10−04 | 3.10 × 10−02 |
| iXLOC_013444 | 19 | 63,355,768 | 63,357,128 | Novel | −0.95 | −1.93 | 4.56 × 10−04 | 3.13 × 10−02 |
| iXLOC_013449 | 19 | 63,365,069 | 63,365,658 | Novel | −1.04 | −2.06 | 4.84 × 10−04 | 3.13 × 10−02 |
| iXLOC_015752 | 20 | 63,795,770 | 63,798,826 | Novel | −0.91 | −1.88 | 4.69 × 10−04 | 3.13 × 10−02 |
| iXLOC_013457 | 19 | 63,385,512 | 63,386,885 | Novel | −1.07 | −2.10 | 5.81 × 10−04 | 3.47 × 10−02 |
| iXLOC_009284 | 16 | 21,647,689 | 21,649,116 | Novel | 0.76 | 1.69 | 7.07 × 10−04 | 3.93 × 10−02 |
| LncRNA 1 | Bta 2 | Start | End | Ensembl Gene | Gene Symbol | Start | Stop | Cor 3 | p.BH 4 |
|---|---|---|---|---|---|---|---|---|---|
| Ileum | |||||||||
| iXLOC_007504 | 15 | 31,279,333 | 31,283,846 | ENSBTAG00000021338 | OAF | 31,312,383 | 31,330,638 | 0.97 | 2.30 × 10−10 |
| iXLOC_008931 | 16 | 49,402,721 | 49,405,957 | ENSBTAG00000016057 | CSRP1 | 49,332,770 | 49,353,517 | 0.83 | 7.72 × 10−05 |
| iXLOC_009320 | 16 | 21,758,311 | 21,761,092 | ENSBTAG00000001574 | GPATCH2 | 21,783,189 | 21,808,519 | 0.84 | 3.86 × 10−05 |
| iXLOC_009321 | 16 | 21,761,255 | 21,764,378 | ENSBTAG00000001574 | GPATCH2 | 21,783,189 | 21,808,519 | 0.75 | 9.20 × 10−04 |
| iXLOC_009324 | 16 | 21,772,956 | 21,777,346 | ENSBTAG00000001574 | GPATCH2 | 21,783,189 | 21,808,519 | 0.92 | 6.39 × 10−07 |
| Rumen | |||||||||
| rXLOC_001026 | 1 | 145,823,035 | 145,824,769 | ENSBTAG00000021901 | TRPM2 | 145,830,610 | 145,871,792 | 0.77 | 4.46 × 10−07 |
| rXLOC_001033 | 1 | 146,129,434 | 146,206,714 | ENSBTAG00000048090 | UBE2G2 | 146,114,659 | 146,120,826 | 0.45 | 1.02 × 10−02 |
| rXLOC_001744 | 1 | 88,589,659 | 88,590,085 | ENSBTAG00000012463 | ZMAT3 | 88,632,287 | 88,663,997 | 0.55 | 1.39 × 10−03 |
| rXLOC_004066 | 11 | 88,446,930 | 88,450,243 | ENSBTAG00000008160 | MBOAT2 | 88,390,471 | 88,444,776 | 0.85 | 1.72 × 10−09 |
| rXLOC_006791 | 7 | 13,559,789 | 13,561,916 | ENSBTAG00000016352 | STX10 | 13,548,241 | 13,551,708 | 0.78 | 2.51 × 10−07 |
| rXLOC_006791 | 7 | 13,559,789 | 13,561,916 | ENSBTAG00000018229 | NFIX | 13,596,367 | 13,658,112 | 0.58 | 5.62 × 10−04 |
| rXLOC_006791 | 7 | 13,559,789 | 13,561,916 | ENSBTAG00000019325 | TRMT1 | 13,572,878 | 13,581,965 | 0.76 | 7.57 × 10−07 |
| rXLOC_009274 | 8 | 63,568,396 | 63,573,554 | ENSBTAG00000021367 | ENSBTAG00000021367 | 63,617,041 | 63,630,082 | 0.74 | 2.49 × 10−06 |
| rXLOC_010940 | 15 | 43,689,413 | 43,689,680 | ENSBTAG00000016074 | ENSBTAG00000016074 | 43,703,911 | 43,770,016 | 0.56 | 1.13 × 10−03 |
| rXLOC_011271 | 9 | 71,006,423 | 71,217,097 | ENSBTAG00000019460 | MOXD1 | 71,245,109 | 71,341,930 | 0.57 | 7.51 × 10−04 |
| rXLOC_016034 | 11 | 67,744,515 | 67,747,453 | ENSBTAG00000017626 | GFPT1 | 67,684,390 | 67,728,959 | 0.65 | 8.63 × 10−05 |
| rXLOC_025037 | 14 | 1,907,631 | 1,909,665 | ENSBTAG00000000658 | WDR97 | 1,913,048 | 1,921,667 | 0.42 | 1.91 × 10−02 |
| rXLOC_025037 | 14 | 1,907,631 | 1,909,665 | ENSBTAG00000012242 | MAF1 | 1,921,784 | 1,924,818 | 0.64 | 1.17 × 10−04 |
| rXLOC_025037 | 14 | 1,907,631 | 1,909,665 | ENSBTAG00000014607 | EXOSC4 | 1,947,198 | 1,949,074 | 0.43 | 1.48 × 10−02 |
| rXLOC_025037 | 14 | 1,907,631 | 1,909,665 | ENSBTAG00000014610 | GPAA1 | 1,942,672 | 1,945,910 | 0.42 | 1.77 × 10−02 |
| rXLOC_025696 | 14 | 2,350,228 | 2,354,178 | ENSBTAG00000014643 | eef1d | 2,317,971 | 2,326,718 | 0.44 | 1.42 × 10−02 |
| rXLOC_029330 | 18 | 46,573,486 | 46,573,914 | ENSBTAG00000002763 | KMT2B | 46,620,836 | 46,640,904 | 0.63 | 1.31 × 10−04 |
| rXLOC_030259 | 19 | 25,389,242 | 25,389,587 | ENSBTAG00000021091 | ANKFY1 | 25,337,350 | 25,387,068 | 0.59 | 5.42 × 10−04 |
| rXLOC_041027 | 27 | 25,434,455 | 25,440,584 | ENSBTAG00000013581 | LEPROTL1 | 25,422,502 | 25,432,327 | 0.53 | 2.25 × 10−03 |
| rXLOC_041173 | 27 | 33,807,735 | 33,808,050 | ENSBTAG00000003509 | PLEKHA2 | 33,747,818 | 33,776,271 | 0.59 | 4.35 × 10−04 |
| rXLOC_041174 | 27 | 33,813,850 | 33,814,207 | ENSBTAG00000003509 | PLEKHA2 | 33,747,818 | 33,776,271 | 0.65 | 8.60 × 10−05 |
| rXLOC_042125 | 27 | 6,446,299 | 6,446,849 | ENSBTAG00000004231 | GPM6A | 6,267,201 | 6,417,208 | 0.40 | 2.47 × 10−02 |
| rXLOC_042503 | 27 | 32,987,255 | 32,989,352 | ENSBTAG00000000978 | ASH2L | 32,989,769 | 33,014,982 | 0.74 | 1.74 × 10−06 |
| rXLOC_045046 | X | 130,424,538 | 130,424,911 | ENSBTAG00000003727 | SH3KBP1 | 130,459,281 | 130,711,913 | 0.42 | 1.97 × 10−02 |
| rXLOC_045053 | X | 130,432,550 | 130,433,452 | ENSBTAG00000003727 | SH3KBP1 | 130,459,281 | 130,711,913 | 0.53 | 2.12 × 10−03 |
| rXLOC_045511 | X | 17,169,634 | 17,218,596 | ENSBTAG00000020644 | GPC4 | 17,087,447 | 17,122,426 | 0.73 | 2.75 × 10−06 |
| rXLOC_045531 | X | 18,185,751 | 18,188,438 | ENSBTAG00000030024 | ENSBTAG00000030024 | 18,180,133 | 18,180,229 | 0.59 | 4.97 × 10−04 |
© 2018 by Her Majesty the Queen in Right of Canada, as represented by the Minister of Agriculture and Agri-Food Canada; Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ibeagha-Awemu, E.M.; Do, D.N.; Dudemaine, P.-L.; Fomenky, B.E.; Bissonnette, N. Integration of lncRNA and mRNA Transcriptome Analyses Reveals Genes and Pathways Potentially Involved in Calf Intestinal Growth and Development during the Early Weeks of Life. Genes 2018, 9, 142. https://doi.org/10.3390/genes9030142
Ibeagha-Awemu EM, Do DN, Dudemaine P-L, Fomenky BE, Bissonnette N. Integration of lncRNA and mRNA Transcriptome Analyses Reveals Genes and Pathways Potentially Involved in Calf Intestinal Growth and Development during the Early Weeks of Life. Genes. 2018; 9(3):142. https://doi.org/10.3390/genes9030142
Chicago/Turabian StyleIbeagha-Awemu, Eveline M., Duy N. Do, Pier-Luc Dudemaine, Bridget E. Fomenky, and Nathalie Bissonnette. 2018. "Integration of lncRNA and mRNA Transcriptome Analyses Reveals Genes and Pathways Potentially Involved in Calf Intestinal Growth and Development during the Early Weeks of Life" Genes 9, no. 3: 142. https://doi.org/10.3390/genes9030142
APA StyleIbeagha-Awemu, E. M., Do, D. N., Dudemaine, P.-L., Fomenky, B. E., & Bissonnette, N. (2018). Integration of lncRNA and mRNA Transcriptome Analyses Reveals Genes and Pathways Potentially Involved in Calf Intestinal Growth and Development during the Early Weeks of Life. Genes, 9(3), 142. https://doi.org/10.3390/genes9030142






