Homologous Recombination: To Fork and Beyond
Abstract
1. Introduction
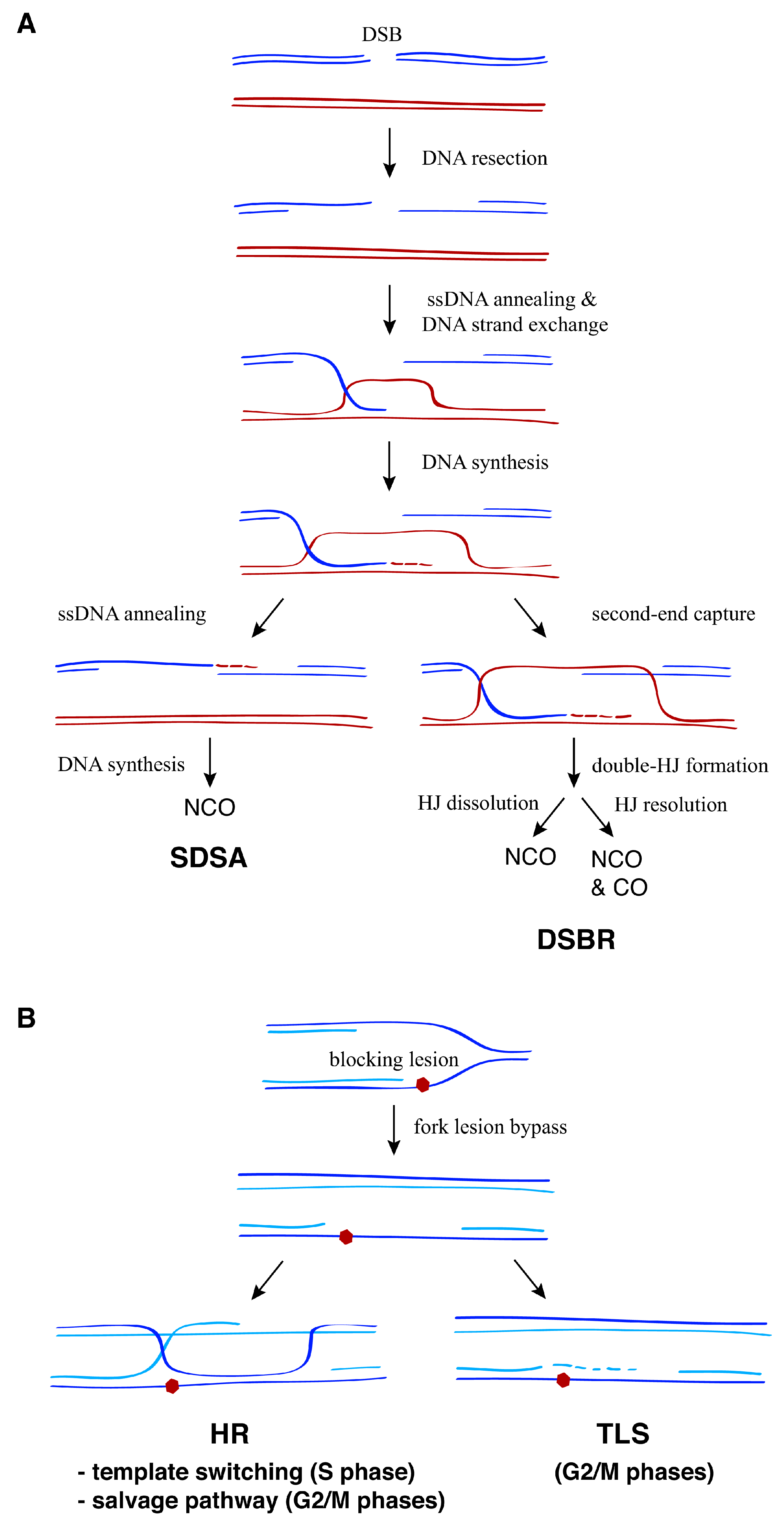
2. Two Different Scenarios for Recombination during DDT: ssDNA Gaps at and behind the Fork
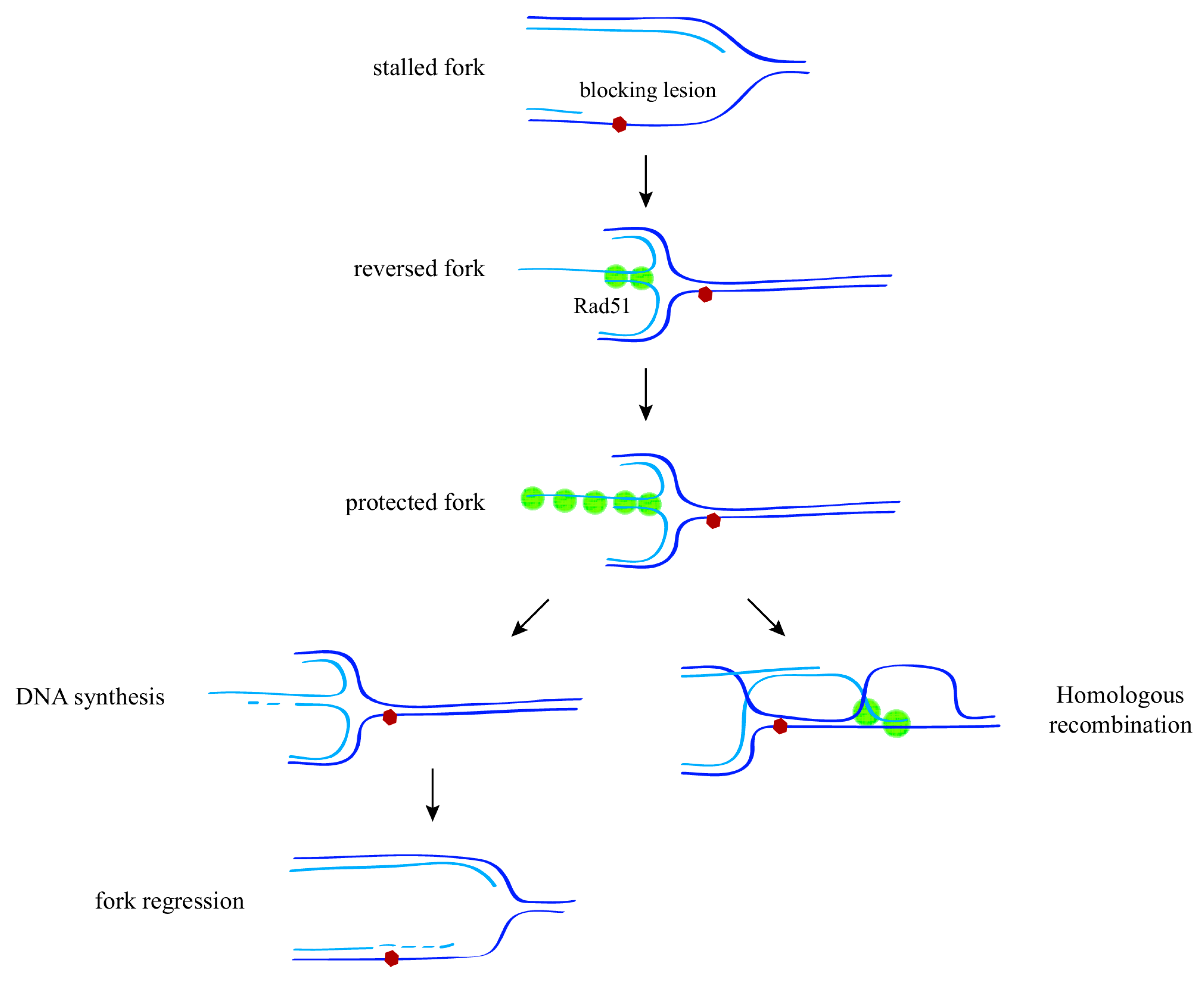
3. Recombination at the Fork
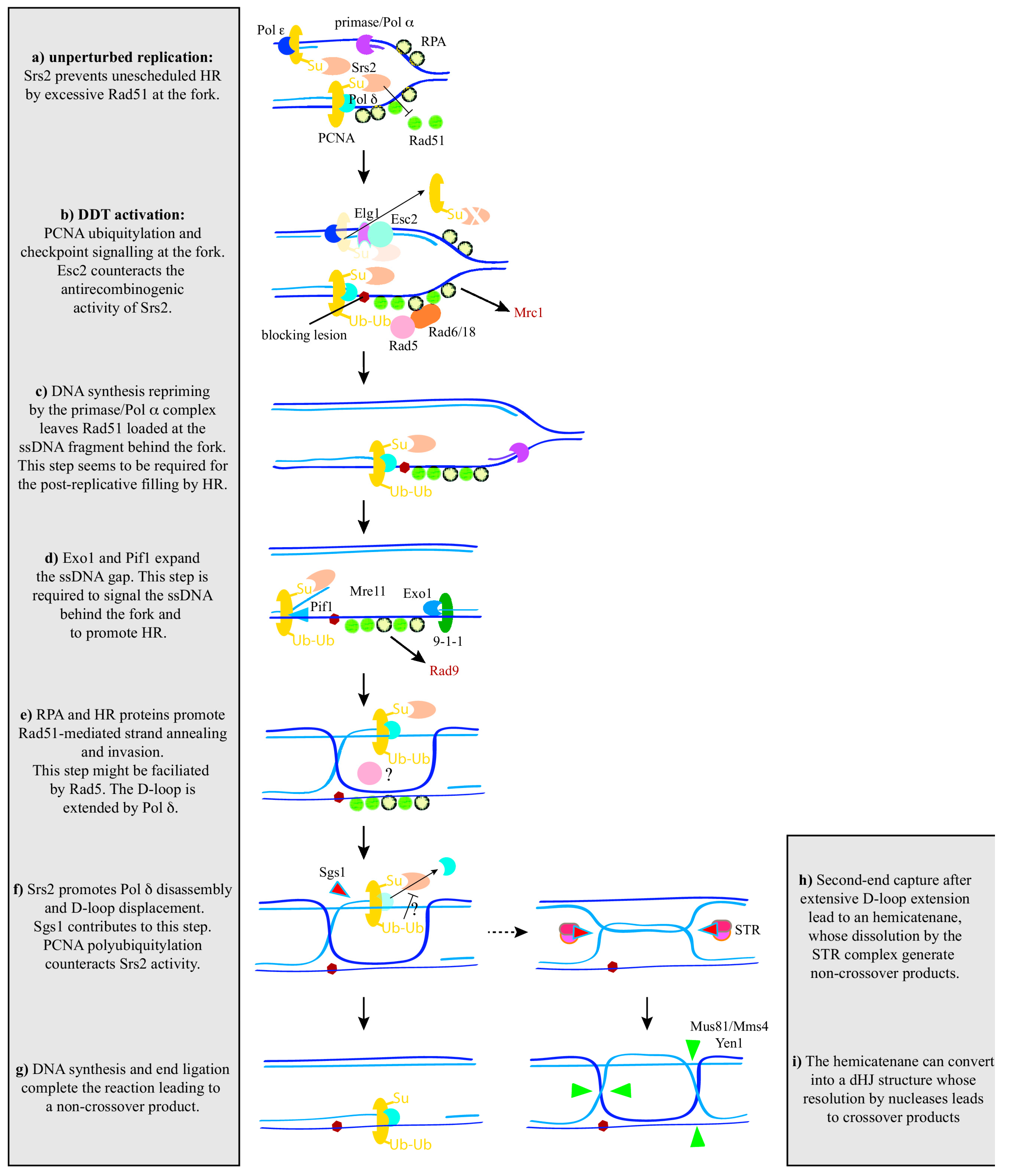
4. Controlling Recombination at the Fork
5. From the ssDNA Gap at the Fork to the ssDNA Gap behind the Fork
6. Recombination behind the Fork
7. Controlling Recombination behind the Fork
8. Are the Recombination Processes at and behind the Fork Mechanistically Connected?
9. Concluding Remarks
Funding
Acknowledgments
Conflicts of Interest
References
- Wright, W.D.; Shah, S.S.; Heyer, W.-D. Homologous recombination and the repair of DNA double-strand breaks. J. Biol. Chem. 2018, 293, 10524–10535. [Google Scholar] [CrossRef]
- Hunter, N. Meiotic recombination: The essence of heredity. Cold Spring Harb. Perspect. Biol. 2015, a016618. [Google Scholar] [CrossRef]
- Huertas, P. DNA resection in eukaryotes: Deciding how to fix the break. Nat. Struct. Mol. Biol. 2010, 17, 11–16. [Google Scholar] [CrossRef]
- Morrical, S.W. DNA-pairing and annealing processes in homologous recombination and homology-directed repair. Cold Spring Harb. Perspect. Biol. 2015, 7, a016444. [Google Scholar] [CrossRef]
- Mortensen, U.H.; Bendixen, C.; Sunjevaric, I.; Rothstein, R. DNA strand annealing is promoted by the yeast Rad52 protein. Proc. Natl. Acad. Sci. USA 1996, 93, 10729–10734. [Google Scholar] [CrossRef]
- Bi, B.; Rybalchenko, N.; Golub, E.I.; Radding, C.M. Human and yeast Rad52 proteins promote DNA strand exchange. Proc. Natl. Acad. Sci. USA 2004, 101, 9568–9572. [Google Scholar] [CrossRef]
- Sung, P. Catalysis of ATP-dependent homologous DNA pairing and strand exchange by yeast RAD51 protein. Science 1994, 265, 1241–1243. [Google Scholar] [CrossRef]
- Wu, Y.; Sugiyama, T.; Kowalczykowski, S.C. DNA annealing mediated by Rad52 and Rad59 proteins. J. Biol. Chem. 2006, 281, 15441–15449. [Google Scholar] [CrossRef]
- Burkovics, P.; Sebesta, M.; Balogh, D.; Haracska, L.; Krejci, L. Strand invasion by HLTF as a mechanism for template switch in fork rescue. Nucleic Acids Res. 2014, 42, 1711–1720. [Google Scholar] [CrossRef]
- Liu, J.; Heyer, W.D. Who’s who in human recombination: BRCA2 and RAD52. Proc. Natl. Acad. Sci. USA 2011, 108, 441–442. [Google Scholar] [CrossRef]
- Cejka, P.; Plank, J.L.; Bachrati, C.Z.; Hickson, I.D.; Kowalczykowski, S.C. Rmi1 stimulates decatenation of double Holliday junctions during dissolution by Sgs1-Top3. Nat. Struct. Mol. Biol. 2010, 17, 1377–1382. [Google Scholar] [CrossRef]
- Matos, J.; West, S.C. Holliday junction resolution: Regulation in space and time. DNA Repair 2014, 19, 176–181. [Google Scholar] [CrossRef]
- Zhao, L.; Washington, M. Translesion synthesis: Insights into the selection and switching of DNA polymerases. Genes 2017, 8, 24. [Google Scholar] [CrossRef]
- Branzei, D.; Szakal, B. DNA damage tolerance by recombination: Molecular pathways and DNA structures. DNA Repair 2016, 44, 68–75. [Google Scholar] [CrossRef]
- Prado, F. Homologous recombination maintenance of genome integrity during DNA damage tolerance. Mol. Cell. Oncol. 2014, 1, e957039. [Google Scholar] [CrossRef]
- Resnick, M.A.; Boyce, J.; Cox, B. Postreplication repair in Saccharomyces cerevisiae. J. Bacteriol. 1981, 146, 285–290. [Google Scholar]
- Jachymczyk, W.J.; Chlebowicz, E.; Swietlinska, Z.; Zuk, J. Alkaline sucrose sedimentation studies of MMS-induced DNA single-strand breakage and rejoining in the wild type and in UV-sensitive mutants of Saccharomyces cerevisiae. Mutat. Res. 1977, 43, 1–10. [Google Scholar] [CrossRef]
- Prakash, L. Characterization of postreplication repair in Saccharomyces cerevisiae and effects of rad6, rad18, rev3 and rad52 mutations. Mol. Gen. Genet. 1981, 184, 471–478. [Google Scholar] [CrossRef]
- Gangavarapu, V.; Prakash, S.; Prakash, L. Requirement of RAD52 group genes for postreplication repair of UV-damaged DNA in Saccharomyces cerevisiae. Mol. Cell. Biol. 2007, 27, 7758–7764. [Google Scholar] [CrossRef]
- Hashimoto, Y.; Ray Chaudhuri, A.; Lopes, M.; Costanzo, V. Rad51 protects nascent DNA from Mre11-dependent degradation and promotes continuous DNA synthesis. Nat. Struct. Mol. Biol. 2010, 17, 1305–1311. [Google Scholar] [CrossRef]
- Lopes, M.; Foiani, M.; Sogo, J.M. Multiple mechanisms control chromosome integrity after replication fork uncoupling and restart at irreparable UV lesions. Mol. Cell 2006, 21, 15–27. [Google Scholar] [CrossRef]
- Petermann, E.; Orta, M.L.; Issaeva, N.; Schultz, N.; Helleday, T. Hydroxyurea-stalled replication forks become progressively inactivated and require two different RAD51-mediated pathways for restart and repair. Mol. Cell 2010, 37, 492–502. [Google Scholar] [CrossRef]
- González-Prieto, R.; Muñoz-Cabello, A.M.; Cabello-Lobato, M.J.; Prado, F. Rad51 replication fork recruitment is required for DNA damage tolerance. EMBO J. 2013, 32, 1307–1321. [Google Scholar] [CrossRef]
- Meister, P.; Taddei, A.; Vernis, L.; Poidevin, M.; Gasser, S.M.; Baldacci, G. Temporal separation of replication and recombination requires the intra-S checkpoint. J. Cell Biol. 2005, 168, 537–544. [Google Scholar] [CrossRef]
- Alabert, C.; Bianco, J.N.; Pasero, P. Differential regulation of homologous recombination at DNA breaks and replication forks by the Mrc1 branch of the S-phase checkpoint. EMBO J. 2009, 28, 1131–1141. [Google Scholar] [CrossRef]
- Barlow, J.H.; Rothstein, R. Rad52 recruitment is DNA replication independent and regulated by Cdc28 and the Mec1 kinase. EMBO J. 2009, 28, 1121–1130. [Google Scholar] [CrossRef]
- Jossen, R.; Bermejo, R. The DNA damage checkpoint response to replication stress: A Game of Forks. Front. Genet. 2013, 4, 26. [Google Scholar] [CrossRef]
- Galanti, L.; Pfander, B. Right time, right place—DNA damage and DNA replication checkpoints collectively safeguard S phase. EMBO J. 2018, 37, e100681. [Google Scholar] [CrossRef]
- Tercero, J.A.; Longhese, M.P.; Diffley, J.F.X. A central role for DNA replication forks in checkpoint activation and response. Mol. Cell 2003, 11, 1323–1336. [Google Scholar] [CrossRef]
- Osborn, A.J.; Elledge, S.J. Mrc1 is a replication fork component whose phosphorylation in response to DNA replication stress activates Rad53. Genes Dev. 2003, 17, 1755–1767. [Google Scholar] [CrossRef]
- Prado, F. Genetic instability is prevented by Mrc1-dependent spatio-temporal separation of replicative and repair activities of homologous recombination: Homologous recombination tolerates replicative stress by Mrc1-regulated replication and repair activities operating at S and G2 in distinct subnuclear compartments. Bioessays 2014, 36, 451–462. [Google Scholar]
- García Rodríguez, N.; Morawska, M.; Wong, R.P.; Daigaku, Y.; Ulrich, H.D. Spatial separation between replisome- and template-induced replication stress signaling. EMBO J. 2018, 37, e98369. [Google Scholar] [CrossRef]
- Bacal, J.; Moriel-Carretero, M.; Pardo, B.; Barthe, A.; Sharma, S.; Chabes, A.; Lengronne, A.; Pasero, P. Mrc1 and Rad9 cooperate to regulate initiation and elongation of DNA replication in response to DNA damage. EMBO J. 2018, 37, e99319. [Google Scholar] [CrossRef]
- Zellweger, R.; Dalcher, D.; Mutreja, K.; Berti, M.; Schmid, J.A.; Herrador, R.; Vindigni, A.; LOPES, M. Rad51-mediated replication fork reversal is a global response to genotoxic treatments in human cells. J. Cell Biol. 2015, 208, 563–579. [Google Scholar] [CrossRef]
- Alabert, C.; Bukowski-Wills, J.-C.; Lee, S.-B.; Kustatscher, G.; Nakamura, K.; de Lima Alves, F.; Menard, P.; Mejlvang, J.; Rappsilber, J.; Groth, A. Nascent chromatin capture proteomics determines chromatin dynamics during DNA replication and identifies unknown fork components. Nat. Cell. Biol. 2014, 16, 281–293. [Google Scholar] [CrossRef]
- López-Contreras, A.J.; Ruppen, I.; Nieto-Soler, M.; Murga, M.; Rodriguez-Acebes, S.; Remeseiro, S.; Rodrigo-Perez, S.; Rojas, A.M.; Méndez, J.; Muñoz, J.; et al. A proteomic characterization of factors enriched at nascent DNA molecules. Cell Rep. 2013, 3, 1105–1116. [Google Scholar] [CrossRef]
- Sirbu, B.M.; Couch, F.B.; Feigerle, J.T.; Bhaskara, S.; Hiebert, S.W.; Cortez, D. Analysis of protein dynamics at active, stalled, and collapsed replication forks. Genes Dev. 2011, 25, 1320–1327. [Google Scholar] [CrossRef]
- Henry-Mowatt, J.; Jackson, D.; Masson, J.-Y.; Johnson, P.A.; Clements, P.M.; Benson, F.E.; Thompson, L.H.; Takeda, S.; West, S.C.; Caldecott, K.W. XRCC3 and Rad51 modulate replication fork progression on damaged vertebrate chromosomes. Mol. Cell 2003, 11, 1109–1117. [Google Scholar] [CrossRef]
- Vázquez, M.V.; Rojas, V.; Tercero, J.A. Multiple pathways cooperate to facilitate DNA replication fork progression through alkylated DNA. DNA Repair 2008, 7, 1693–1704. [Google Scholar] [CrossRef]
- Sonoda, E.; Sasaki, M.S.; Buerstedde, J.M.; Bezzubova, O.; Shinohara, A.; Ogawa, H.; Takata, M.; Yamaguchi-Iwai, Y.; Takeda, S. Rad51-deficient vertebrate cells accumulate chromosomal breaks prior to cell death. EMBO J. 1998, 17, 598–608. [Google Scholar] [CrossRef]
- Daboussi, F.; Courbet, S.; Benhamou, S.; Kannouche, P.; Zdzienicka, M.Z.; Debatisse, M.; Lopez, B.S. A homologous recombination defect affects replication-fork progression in mammalian cells. J. Cell Sci. 2008, 121, 162–166. [Google Scholar] [CrossRef]
- Higgins, N.P.; Kato, K.; Strauss, B. A model for replication repair in mammalian cells. J. Mol. Biol. 1976, 101, 417–425. [Google Scholar] [CrossRef]
- Bugreev, D.V.; Rossi, M.J.; Mazin, A.V. Cooperation of RAD51 and RAD54 in regression of a model replication fork. Nucleic Acids Res. 2011, 39, 2153–2164. [Google Scholar] [CrossRef]
- Bhat, K.P.; Cortez, D. RPA and RAD51: Fork reversal, fork protection, and genome stability. Nat. Struct. Mol. Biol. 2018, 25, 446–453. [Google Scholar] [CrossRef]
- Neelsen, K.J.; Lopes, M. Replication fork reversal in eukaryotes: From dead end to dynamic response. Nature 2015, 16, 207–220. [Google Scholar] [CrossRef]
- Schlacher, K.; Christ, N.; Siaud, N.; Egashira, A.; Wu, H.; Jasin, M. Double-strand break repair-independent role for BRCA2 in blocking stalled replication fork degradation by MRE11. Cell 2011, 145, 529–542. [Google Scholar] [CrossRef]
- Vallerga, M.B.; Mansilla, S.F.; Federico, M.B.; Bertolin, A.P.; Gottifredi, V. Rad51 recombinase prevents Mre11 nuclease-dependent degradation and excessive PrimPol-mediated elongation of nascent DNA after UV irradiation. Proc. Natl. Acad. Sci. USA 2015, 112, E6624–E6633. [Google Scholar] [CrossRef]
- Mijic, S.; Zellweger, R.; Chappidi, N.; Berti, M.; Jacobs, K.; Mutreja, K.; Ursich, S.; Ray Chaudhuri, A.; Nussenzweig, A.; Janscak, P.; et al. Replication fork reversal triggers fork degradation in BRCA2-defective cells. Nat. Commun. 2017, 8, 859. [Google Scholar] [CrossRef]
- Wang, A.T.; Kim, T.; Wagner, J.E.; Conti, B.A.; Lach, F.P.; Huang, A.L.; Molina, H.; Sanborn, E.M.; Zierhut, H.; Cornes, B.K.; et al. A dominant mutation in human RAD51 reveals its function in DNA interstrand crosslink repair independent of homologous recombination. Mol. Cell 2015, 59, 478–490. [Google Scholar] [CrossRef]
- Iannascoli, C.; Palermo, V.; Murfuni, I.; Franchitto, A.; Pichierri, P. The WRN exonuclease domain protects nascent strands from pathological MRE11/EXO1-dependent degradation. Nucleic Acids Res. 2015, 43, 9788–9803. [Google Scholar] [CrossRef]
- Higgs, M.R.; Reynolds, J.J.; Winczura, A.; Blackford, A.N.; Borel, V.; Miller, E.S.; Zlatanou, A.; Nieminuszczy, J.; Ryan, E.L.; Davies, N.J.; et al. BOD1L is required to suppress deleterious resection of stressed replication forks. Mol. Cell 2015, 59, 462–477. [Google Scholar] [CrossRef]
- Lemaçon, D.; Jackson, J.; Quinet, A.; Brickner, J.R.; Li, S.; Yazinski, S.; You, Z.; Ira, G.; Zou, L.; Mosammaparast, N.; et al. MRE11 and EXO1 nucleases degrade reversed forks and elicit MUS81-dependent fork rescue in BRCA2- deficient cells. Nat. Commun. 2017, 8, 860. [Google Scholar] [CrossRef]
- Michl, J.; Zimmer, J.; Tarsounas, M. Interplay between Fanconi anemia and homologous recombination pathways in genome integrity. EMBO J. 2016, 35, 909–923. [Google Scholar] [CrossRef]
- Toledo, L.I.; Altmeyer, M.; Rask, M.-B.; Lukas, C.; Larsen, D.H.; Povlsen, L.K.; Bekker-Jensen, S.; Mailand, N.; Bartek, J.; Lukas, J. ATR prohibits replication catastrophe by preventing global exhaustion of RPA. Cell 2013, 155, 1088–1103. [Google Scholar] [CrossRef]
- Chaudhuri, A.R.; Hashimoto, Y.; Herrador, R.; Neelsen, K.J.; Fachinetti, D.; Bermejo, R.; Cocito, A.; Costanzo, V.; Lopes, M. Topoisomerase I poisoning results in PARP-mediated replication fork reversal. Nat. Struct. Mol. Biol. 2012, 19, 417–423. [Google Scholar] [CrossRef]
- Giannattasio, M.; Zwicky, K.; Follonier, C.; Foiani, M.; Lopes, M.; Branzei, D. Visualization of recombination-mediated damage bypass by template switching. Nat. Struct. Mol. Biol. 2014, 21, 884–892. [Google Scholar] [CrossRef]
- Sogo, J.M.; Lopes, M.; Foiani, M. Fork reversal and ssDNA accumulation at stalled replication forks owing to checkpoint defects. Science 2002, 297, 599–602. [Google Scholar] [CrossRef]
- Cotta-Ramusino, C.; Fachinetti, D.; Lucca, C.; Doksani, Y.; Lopes, M.; Sogo, J.; Foiani, M. Exo1 processes stalled replication forks and counteracts fork reversal in checkpoint-defective cells. Mol. Cell 2005, 17, 153–159. [Google Scholar] [CrossRef]
- Fumasoni, M.; Zwicky, K.; Vanoli, F.; Lopes, M.; Branzei, D. Error-free DNA damage tolerance and sister chromatid proximity during DNA replication rely on the Polα/Primase/Ctf4 Complex. Mol. Cell 2015, 57, 812–823. [Google Scholar] [CrossRef]
- Vos, S.M.; Tretter, E.M.; Schmidt, B.H.; Berger, J.M. All tangled up: How cells direct, manage and exploit topoisomerase function. Nature 2011, 12, 827–841. [Google Scholar] [CrossRef]
- Chaudhury, I.; Koepp, D.M. Degradation of Mrc1 promotes recombination-mediated restart of stalled replication forks. Nucleic Acids Res. 2016, 45, 2558–2570. [Google Scholar] [CrossRef]
- Minca, E.C.; Kowalski, D. Multiple Rad5 activities mediate sister chromatid recombination to bypass DNA damage at stalled replication forks. Mol. Cell 2010, 38, 649–661. [Google Scholar] [CrossRef]
- Bailis, J.M.; Luche, D.D.; Hunter, T.; Forsburg, S.L. Minichromosome maintenance proteins interact with checkpoint and recombination proteins to promote S-phase genome stability. Mol. Cell. Biol. 2008, 28, 1724–1738. [Google Scholar] [CrossRef]
- Shukla, A.; Navadgi, V.M.; Mallikarjuna, K.; Rao, B.J. Interaction of hRad51 and hRad52 with MCM complex: A cross-talk between recombination and replication proteins. Biochem. Biophys. Res. Commun. 2005, 329, 1240–1245. [Google Scholar] [CrossRef]
- Klein, H.L. The consequences of Rad51 overexpression for normal and tumor cells. DNA Repair 2008, 7, 686–693. [Google Scholar] [CrossRef]
- Dungrawala, H.; Bhat, K.P.; Le Meur, R.; Chazin, W.J.; Ding, X.; Sharan, S.K.; Wessel, S.R.; Sathe, A.A.; Zhao, R.; Cortez, D. RADX promotes genome stability and modulates chemosensitivity by regulating RAD51 at replication forks. Mol. Cell 2017, 67, 374–386. [Google Scholar] [CrossRef]
- Bhat, K.P.; Krishnamoorthy, A.; Dungrawala, H.; Garcin, E.B.; Modesti, M.; Cortez, D. RADX modulates RAD51 activity to control replication fork protection. Cell Rep. 2018, 24, 538–545. [Google Scholar] [CrossRef]
- Hu, Y.; Raynard, S.; Sehorn, M.G.; Lu, X.; Bussen, W.; Zheng, L.; Stark, J.M.; Barnes, E.L.; Chi, P.; Janscak, P.; et al. RECQL5/Recql5 helicase regulates homologous recombination and suppresses tumor formation via disruption of Rad51 presynaptic filaments. Genes Dev. 2007, 21, 3073–3084. [Google Scholar] [CrossRef]
- Bugreev, D.V.; Yu, X.; Egelman, E.H.; Mazin, A.V. Novel pro- and anti-recombination activities of the Bloom’s syndrome helicase. Genes Dev. 2007, 21, 3085–3094. [Google Scholar] [CrossRef]
- Simandlova, J.; Zagelbaum, J.; Payne, M.J.; Chu, W.K.; Shevelev, I.; Hanada, K.; Chatterjee, S.; Reid, D.A.; Liu, Y.; Janscak, P.; et al. FBH1 helicase disrupts RAD51 filaments in vitro and modulates homologous recombination in mammalian cells. J. Biol. Chem. 2013, 288, 34168–34180. [Google Scholar] [CrossRef]
- Krejci, L.; van Komen, S.; Li, Y.; Villemain, J.; Reddy, M.S.; Klein, H.; Ellenberger, T.; Sung, P. DNA helicase Srs2 disrupts the Rad51 presynaptic filament. Nature 2003, 423, 305–309. [Google Scholar] [CrossRef]
- Veaute, X.; Jeusset, J.; Soustelle, C.; Kowalczykowski, S.C.; Le Cam, E.; Fabre, F. The Srs2 helicase prevents recombination by disrupting Rad51 nucleoprotein filaments. Nature 2003, 423, 309–312. [Google Scholar] [CrossRef]
- Moldovan, G.-L.; Dejsuphong, D.; Petalcorin, M.I.R.; Hofmann, K.; Takeda, S.; Boulton, S.J.; D’Andrea, A.D. Inhibition of homologous recombination by the PCNA-interacting protein PARI. Mol. Cell 2012, 45, 75–86. [Google Scholar] [CrossRef]
- Hoege, C.; Pfander, B.; Moldovan, G.-L.; Pyrowolakis, G.; Jentsch, S. RAD6-dependent DNA repair is linked to modification of PCNA by ubiquitin and SUMO. Nature 2002, 419, 135–141. [Google Scholar] [CrossRef]
- Pfander, B.; Moldovan, G.-L.; Sacher, M.; Hoege, C.; Jentsch, S. SUMO-modified PCNA recruits Srs2 to prevent recombination during S phase. Nature 2005, 436, 428–433. [Google Scholar] [CrossRef]
- Papouli, E.; Chen, S.; Davies, A.A.; Huttner, D.; Krejci, L.; Sung, P.; Ulrich, H.D. Crosstalk between SUMO and ubiquitin on PCNA is mediated by recruitment of the helicase Srs2p. Mol. Cell 2005, 19, 123–133. [Google Scholar] [CrossRef]
- Urulangodi, M.; Sebesta, M.; Menolfi, D.; Szakal, B.; Sollier, J.; Sisakova, A.; Krejci, L.; Branzei, D. Local regulation of the Srs2 helicase by the SUMO-like domain protein Esc2 promotes recombination at sites of stalled replication. Genes Dev. 2015, 29, 2067–2080. [Google Scholar] [CrossRef]
- Burkovics, P.; Sebesta, M.; Sisakova, A.; Plault, N.; Szukacsov, V.; Robert, T.; Pintér, L.; Marini, V.; Kolesar, P.; Haracska, L.; et al. Srs2 mediates PCNA-SUMO-dependent inhibition of DNA repair synthesis. EMBO J. 2013, 32, 742–755. [Google Scholar] [CrossRef]
- McInerney, P.; O’Donnell, M. Functional uncoupling of twin polymerases: Mechanism of polymerase dissociation from a lagging-strand block. J. Biol. Chem. 2004, 279, 21543–21551. [Google Scholar] [CrossRef]
- Pagès, V.; Fuchs, R.P. Uncoupling of leading- and lagging-strand DNA replication during lesion bypass in vivo. Science 2003, 300, 1300–1303. [Google Scholar] [CrossRef]
- Higuchi, K.; Katayama, T.; Iwai, S.; Hidaka, M.; Horiuchi, T.; Maki, H. Fate of DNA replication fork encountering a single DNA lesion during oriC plasmid DNA replication in vitro. Genes Cells 2003, 8, 437–449. [Google Scholar] [CrossRef]
- Taylor, M.R.G.; Yeeles, J.T.P. The initial response of a eukaryotic replisome to DNA damage. Mol. Cell 2018, 70, 1067–1080. [Google Scholar] [CrossRef]
- Mourón, S.; Rodriguez-Acebes, S.; Martínez-Jiménez, M.I.; García-Gómez, S.; Chocrón, S.; Blanco, L.; Méndez, J. Repriming of DNA synthesis at stalled replication forks by human PrimPol. Nat. Struct. Mol. Biol. 2013, 20, 1383–1389. [Google Scholar] [CrossRef]
- García-Gómez, S.; Reyes, A.; Martínez-Jiménez, M.I.; Chocrón, E.S.; Mourón, S.; Terrados, G.; Powell, C.; Salido, E.; Méndez, J.; Holt, I.J.; et al. PrimPol, an archaic Primase/Polymerase operating in human cells. Mol. Cell 2013, 52, 541–553. [Google Scholar] [CrossRef]
- Jansen, J.G.; Tsaalbi-Shtylik, A.; Hendriks, G.; Verspuy, J.; Gali, H.; Haracska, L.; de Wind, N. Mammalian polymerase zeta is essential for post-replication repair of UV-induced DNA lesions. DNA Repair 2009, 8, 1444–1451. [Google Scholar] [CrossRef]
- Elvers, I.; Johansson, F.; Groth, P.; Erixon, K.; Helleday, T. UV stalled replication forks restart by re-priming in human fibroblasts. Nucleic Acids Res. 2011, 39, 7049–7057. [Google Scholar] [CrossRef]
- Barlow, T.; Eliasson, R.; Platz, A.; Reichard, P.; Sjöberg, B.M. Enzymic modification of a tyrosine residue to a stable free radical in ribonucleotide reductase. Proc. Natl. Acad. Sci. USA 1983, 80, 1492–1495. [Google Scholar] [CrossRef]
- Liberi, G.; Maffioletti, G.; Lucca, C.; Chiolo, I.; Baryshnikova, A.; Cotta-Ramusino, C.; Lopes, M.; Pellicioli, A.; Haber, J.E.; Foiani, M. Rad51-dependent DNA structures accumulate at damaged replication forks in sgs1 mutants defective in the yeast ortholog of BLM RecQ helicase. Genes Dev. 2005, 19, 339–350. [Google Scholar] [CrossRef]
- García Rodríguez, N.; Wong, R.P.; Ulrich, H.D. The helicase Pif1 functions in the template switching pathway of DNA damage bypass. Nucleic Acids Res. 2018, 46, 8347–8356. [Google Scholar] [CrossRef]
- Vanoli, F.; Fumasoni, M.; Szakal, B.; Maloisel, L.; Branzei, D. Replication and recombination factors contributing to recombination-dependent bypass of DNA lesions by template switch. PLoS Genet. 2010, 6, e1001205. [Google Scholar] [CrossRef]
- Karras, G.I.; Fumasoni, M.; Sienski, G.; Vanoli, F.; Branzei, D.; Jentsch, S. Noncanonical role of the 9-1-1 clamp in the error-free DNA damage tolerance pathway. Mol. Cell 2013, 49, 536–546. [Google Scholar] [CrossRef]
- Mankouri, H.W.; Ngo, H.-P.; Hickson, I.D. Shu proteins promote the formation of homologous recombination intermediates that are processed by Sgs1-Rmi1-Top3. Mol. Biol. Cell 2007, 18, 4062–4073. [Google Scholar] [CrossRef]
- Branzei, D.; Vanoli, F.; Foiani, M. SUMOylation regulates Rad18-mediated template switch. Nature 2008, 456, 915–920. [Google Scholar] [CrossRef]
- Shor, E.; Weinstein, J.; Rothstein, R. A genetic screen for top3 suppressors in Saccharomyces cerevisiae identifies SHU1, SHU2, PSY3 and CSM2: Four genes involved in error-free DNA repair. Genetics 2005, 169, 1275–1289. [Google Scholar] [CrossRef]
- Choi, K.; Szakal, B.; Chen, Y.-H.; Branzei, D.; Zhao, X. The Smc5/6 complex and Esc2 influence multiple replication-associated recombination processes in Saccharomyces cerevisiae. Mol. Biol. Cell 2010, 21, 2306–2314. [Google Scholar] [CrossRef]
- Gonzalez-Huici, V.; Szakal, B.; Urulangodi, M.; Psakhye, I.; Castellucci, F.; Menolfi, D.; Rajakumara, E.; Fumasoni, M.; Bermejo, R.; Jentsch, S.; et al. DNA bending facilitates the error-free DNA damage tolerance pathway and upholds genome integrity. EMBO J. 2014, 33, 327–340. [Google Scholar] [CrossRef]
- Branzei, D.; Sollier, J.; Liberi, G.; Zhao, X.; Maeda, D.; Seki, M.; Enomoto, T.; Ohta, K.; Foiani, M. Ubc9- and Mms21-mediated sumoylation counteracts recombinogenic events at damaged replication forks. Cell 2006, 127, 509–522. [Google Scholar] [CrossRef]
- Sollier, J.; Driscoll, R.; Castellucci, F.; Foiani, M.; Jackson, S.P.; Branzei, D. The Saccharomyces cerevisiae Esc2 and Smc5-6 proteins promote sister chromatid junction-mediated intra-S repair. Mol. Biol. Cell 2009, 20, 1671–1682. [Google Scholar] [CrossRef]
- Menolfi, D.; Delamarre, A.; Lengronne, A.; Pasero, P.; Branzei, D. Essential roles of the Smc5/6 complex in replication through natural pausing sites and endogenous DNA damage tolerance. Mol. Cell 2015, 60, 835–846. [Google Scholar] [CrossRef]
- Bonner, J.N.; Choi, K.; Xue, X.; Torres, N.P.; Szakal, B.; Wei, L.; Wan, B.; Arter, M.; Matos, J.; Sung, P.; et al. Smc5/6 mediated sumoylation of the Sgs1-Top3- Rmi1 complex promotes removal of recombination intermediates. Cell Rep. 2016, 16, 368–378. [Google Scholar] [CrossRef]
- Bermúdez-López, M.; Villoria, M.T.; Esteras, M.; Jarmuz, A.; Torres-Rosell, J.; Clemente-Blanco, A.; Aragón, L. Sgs1’s roles in DNA end resection, HJ dissolution, and crossover suppression require a two-step SUMO regulation dependent on Smc5/6. Genes Dev. 2016, 30, 1339–1356. [Google Scholar] [CrossRef]
- Garg, P.; Burgers, P.M. Ubiquitinated proliferating cell nuclear antigen activates translesion DNA polymerases η and REV1. Proc. Natl. Acad. Sci. USA 2005, 102, 18361–18366. [Google Scholar] [CrossRef]
- Davies, A.A.; Huttner, D.; Daigaku, Y.; Chen, S.; Ulrich, H.D. Activation of ubiquitin-dependent DNA damage bypass is mediated by replication protein A. Mol. Cell 2008, 29, 625–636. [Google Scholar] [CrossRef]
- Karras, G.I.; Jentsch, S. The RAD6 DNA damage tolerance pathway operates uncoupled from the replication fork and is functional beyond S phase. Cell 2010, 141, 255–267. [Google Scholar] [CrossRef]
- Daigaku, Y.; Davies, A.A.; Ulrich, H.D. Ubiquitin-dependent DNA damage bypass is separable from genome replication. Nature 2010, 465, 951–955. [Google Scholar] [CrossRef]
- Ortiz-Bazán, M.Á.; Gallo-Fernández, M.; Saugar, I.; Jiménez-Martín, A.; Vázquez, M.V.; Tercero, J.A. Rad5 plays a major role in the cellular response to DNA damage during chromosome replication. Cell Rep. 2014, 9, 460–468. [Google Scholar] [CrossRef]
- Parker, J.L.; Bucceri, A.; Davies, A.A.; Heidrich, K.; Windecker, H.; Ulrich, H.D. SUMO modification of PCNA is controlled by DNA. EMBO J. 2008, 27, 2422–2431. [Google Scholar] [CrossRef]
- Parker, J.L.; Ulrich, H.D. A SUMO-interacting motif activates budding yeast ubiquitin ligase Rad18 towards SUMO-modified PCNA. Nucleic Acids Res. 2012, 40, 11380–11388. [Google Scholar] [CrossRef]
- Schiestl, R.H.; Prakash, S.; Prakash, L. The SRS2 suppressor of rad6 mutations of Saccharomyces cerevisiae acts by channeling DNA lesions into the RAD52 DNA repair pathway. Genetics 1990, 124, 817–831. [Google Scholar]
- Motegi, A.; Liaw, H.-J.; Lee, K.-Y.; Roest, H.P.; Maas, A.; Wu, X.; Moinova, H.; Markowitz, S.D.; Ding, H.; Hoeijmakers, J.H.J.; et al. Polyubiquitination of proliferating cell nuclear antigen by HLTF and SHPRH prevents genomic instability from stalled replication forks. Proc. Natl. Acad. Sci. USA 2008, 105, 12411–12416. [Google Scholar] [CrossRef]
- Waters, L.S.; Walker, G.C. The critical mutagenic translesion DNA polymerase Rev1 is highly expressed during G(2)/M phase rather than S phase. Proc. Natl. Acad. Sci. USA 2006, 103, 8971–8976. [Google Scholar] [CrossRef]
- Unk, I.; Hajdu, I.; Blastyák, A.; Haracska, L. Role of yeast Rad5 and its human orthologs, HLTF and SHPRH in DNA damage tolerance. DNA Repair 2010, 9, 257–267. [Google Scholar] [CrossRef]
- Prado, F.; Cortés-Ledesma, F.; Huertas, P.; Aguilera, A. Mitotic recombination in Saccharomyces cerevisiae. Curr. Genet. 2003, 42, 185–198. [Google Scholar]
- Matos, J.; Blanco, M.G.; Maslen, S.; Skehel, J.M.; West, S.C. Regulatory control of the resolution of DNA recombination intermediates during meiosis and mitosis. Cell 2011, 147, 158–172. [Google Scholar] [CrossRef]
- Saugar, I.; Vázquez, M.V.; Gallo-Fernández, M.; Ortiz-Bazán, M.Á.; Segurado, M.; Calzada, A.; Tercero, J.A. Temporal regulation of the Mus81-Mms4 endonuclease ensures cell survival under conditions of DNA damage. Nucleic Acids Res. 2013, 41, 8943–8958. [Google Scholar] [CrossRef]
- Szakal, B.; Branzei, D. Premature Cdk1/Cdc5/Mus81 pathway activation induces aberrant replication and deleterious crossover. EMBO J. 2013, 32, 1155–1167. [Google Scholar] [CrossRef]
- Mankouri, H.W.; Ashton, T.M.; Hickson, I.D. Holliday junction-containing DNA structures persist in cells lacking Sgs1 or Top3 following exposure to DNA damage. Proc. Natl. Acad. Sci. USA 2011, 108, 4944–4949. [Google Scholar] [CrossRef]
- Saugar, I.; Parker, J.L.; Zhao, S.; Ulrich, H.D. The genome maintenance factor Mgs1 is targeted to sites of replication stress by ubiquitylated PCNA. Nucleic Acids Res. 2011, 40, 245–257. [Google Scholar] [CrossRef]
- Hishida, T.; Iwasaki, H.; Ohno, T.; Morishita, T.; Shinagawa, H. A yeast gene, MGS1, encoding a DNA-dependent AAA(+) ATPase is required to maintain genome stability. Proc. Natl. Acad. Sci. USA 2001, 98, 8283–8289. [Google Scholar] [CrossRef]
- Zeman, M.K.; Cimprich, K.A. Finally, polyubiquitinated PCNA gets recognized. Mol. Cell 2012, 47, 333–334. [Google Scholar] [CrossRef]
- Choi, K.; Batke, S.; Szakal, B.; Lowther, J.; Hao, F.; Sarangi, P.; Branzei, D.; Ulrich, H.D.; Zhao, X. Concerted and differential actions of two enzymatic domains underlie Rad5 contributions to DNA damage tolerance. Nucleic Acids Res. 2015, 43, 2666–2677. [Google Scholar] [CrossRef]
- Bi, X.; Yu, Q.; Siler, J.; Li, C.; Khan, A. Functions of Fun30 chromatin remodeler in regulating cellular resistance to genotoxic stress. PLoS ONE 2015, 10, e0121341. [Google Scholar] [CrossRef]
- Costelloe, T.; Louge, R.; Tomimatsu, N.; Mukherjee, B.; Martini, E.; Khadaroo, B.; Dubois, K.; Wiegant, W.W.; Thierry, A.; Burma, S.; et al. The yeast Fun30 and human SMARCAD1 chromatin remodellers promote DNA end resection. Nature 2012, 489, 581–584. [Google Scholar] [CrossRef]
- Chen, X.; Cui, D.; Papusha, A.; Zhang, X.; Chu, C.-D.; Tang, J.; Chen, K.; Pan, X.; Ira, G. The Fun30 nucleosome remodeller promotes resection of DNA double-strand break ends. Nature 2012, 489, 576–580. [Google Scholar] [CrossRef]
- Eapen, V.V.; Sugawara, N.; Tsabar, M.; Wu, W.-H.; Haber, J.E. The Saccharomyces cerevisiae chromatin remodeler Fun30 regulates DNA end resection and checkpoint deactivation. Mol. Cell. Biol. 2012, 32, 4727–4740. [Google Scholar] [CrossRef]
- Bantele, S.C.; Ferreira, P.; Gritenaite, D.; Boos, D.; Pfander, B. Targeting of the Fun30 nucleosome remodeller by the Dpb11 scaffold facilitates cell cycle-regulated DNA end resection. eLife 2017, 6, 836. [Google Scholar] [CrossRef]
- Pietrobon, V.; Fréon, K.; Hardy, J.; Costes, A.; Iraqui, I.; Ochsenbein, F.; Lambert, S.A.E. The chromatin assembly factor 1 promotes Rad51-dependent template switches at replication forks by counteracting D-Loop Disassembly by the RecQ-Type Helicase Rqh1. PLoS Biol. 2014, 12, e1001968. [Google Scholar] [CrossRef]
- Hung, S.-H.; Wong, R.P.; Ulrich, H.D.; Kao, C.-F. Monoubiquitylation of histone H2B contributes to the bypass of DNA damage during and after DNA replication. Proc. Natl. Acad. Sci. USA 2017, 114, E2205–E2214. [Google Scholar] [CrossRef]
- House, N.C.M.; Yang, J.H.; Walsh, S.C.; Moy, J.M.; Freudenreich, C.H. NuA4 initiates dynamic histone H4 acetylationto promote high-fidelity sister chromatid recombination at postreplication gaps. Mol. Cell 2014, 55, 818–828. [Google Scholar] [CrossRef]
- Trujillo, K.M.; Osley, M.A. A Role for H2B Ubiquitylation in DNA Replication. Mol. Cell 2012, 48, 734–746. [Google Scholar] [CrossRef]
- Litwin, I.; Bakowski, T.; Szakal, B.; Pilarczyk, E.; Maciaszczyk-Dziubinska, E.; Branzei, D.; Wysocki, R. Error-free DNA damage tolerance pathway is facilitated by the Irc5 translocase through cohesin. EMBO J. 2018, 37, e98732. [Google Scholar] [CrossRef]
- Prado, F.; Maya, D. Regulation of replication fork advance and stability by nucleosome assembly. Genes 2017, 8, 49. [Google Scholar] [CrossRef]
- Bishop, A. Role of homologous recombination in carcinogenesis. Exp. Mol. Pathol. 2003, 74, 94–105. [Google Scholar] [CrossRef]
© 2018 by the author. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Prado, F. Homologous Recombination: To Fork and Beyond. Genes 2018, 9, 603. https://doi.org/10.3390/genes9120603
Prado F. Homologous Recombination: To Fork and Beyond. Genes. 2018; 9(12):603. https://doi.org/10.3390/genes9120603
Chicago/Turabian StylePrado, Félix. 2018. "Homologous Recombination: To Fork and Beyond" Genes 9, no. 12: 603. https://doi.org/10.3390/genes9120603
APA StylePrado, F. (2018). Homologous Recombination: To Fork and Beyond. Genes, 9(12), 603. https://doi.org/10.3390/genes9120603



