Abstract
Plants have a lot of diversity in epigenetic modifications such as DNA methylation in their natural populations or cultivars. Although many studies observing the epigenetic diversity within and among species have been reported, the mechanisms how these variations are generated are still not clear. In addition to the de novo spontaneous epi-mutation, the intra- and inter-specific crossing can also cause a change of epigenetic modifications in their progenies. Here we report an example of diversification of DNA methylation by crossing and succeeding selfing. We traced the inheritance pattern of epigenetic modification during the crossing experiment between two natural strains Columbia (Col), and Landsberg electa (Ler) in model plant Arabidopsis thaliana to observe the inheritance of DNA methylation in two organellar DNA-like sequence regions in the nuclear genome. Because organellar DNA integration to the nuclear genome is common in flowering plants and these sequences are occasionally methylated, such DNA could be the novel source of plant genome evolution. The amplicon sequencing, using bisulfite-converted DNA and a next-generation auto-sequencer, was able to efficiently track the heredity of DNA methylation in F1 and F2 populations. One region showed hypomethylation in the F1 population and succeeding elevation of DNA methylation with large variance in the F2 population. The methylation level of Col and Ler alleles in F2 heterozygotes showed a significant positive correlation, implying the trans-chromosomal effect on DNA methylation. The results may suggest the possible mechanism causing the natural epigenetic diversity within plant populations.
1. Introduction
Recently, in addition to the genetic variations, a large amount of epigenetic diversity has been reported for plants [1,2,3,4]. Because it is easy to analyze the cytosine DNA methylation, many studies are focused on analyzing the status of DNA methylation. In the model plant Arabidopsis thaliana, DNA methylation is de novo established and/or maintained by several methyltransferases: DNA Methyltransferase 1 (MET1), Chromomethylase 3 & 2 (CMT3, CMT2), and Domains Rearranged Methyltransferase 2 (DRM2) [5,6]. DRM2 is involved in the RNA–directed DNA methylation (RdDM) with other factors such as Argonaute 4 (AGO4) and Dicer-like 2/3/4 (DCL2/3/4) [7]. In addition, there is a positive feedback of histone H3K9 methylation by SU(VAR)3-9 HOMOLOG 4/KRYPTONITE (SUVH4/KYP) and non–CpG (i.e., CpHpG and CpHpH, where H is A, T, or C) methylation by CMT3 and CMT2 [8]. The DNA methylation shows considerable within-species variations [1,4] and large divergences between different taxa [3], or even between close relatives [2]. For the analysis of DNA methylation, there are several methods that differ in their resolutions. While the approach of whole-genome methylome by next-generation sequencing (NGS) has the highest comprehensiveness and resolution for entire genomes (e.g., [9,10]), techniques such as methylation-sensitive amplified polymorphism are used as a cheaper way to analyze the whole-genome epigenetic status for multiple samples [11]. The Sanger sequencing of clones from bisulfite-converted DNA is also used for analyzing the DNA methylation status of a particular region in the genome.
Although there is a growing number of reports concerning diversity within plant genomes, the mechanism that causes epigenetic diversity within/among species is still not clear. The DNA methylation and histone modification patterns are associated with the genome architecture [12,13]. Epigenetic states induced by environmental stress can be reset and be prevented from transgenerational transmission [14]. A new spontaneous epi-mutation is brought about in the nuclear genome with relatively high forward and backward epi-mutation rates [15,16]. In addition to spontaneous epi-mutation on a single cytosine site, DNA methylation can be altered at the progeny of crossing between different strains of a species [17,18,19] or at the interspecific hybrid [20]. The ‘genome shock’ by these crossing and succeeding epigenetic and/or transcriptomic change in the progeny might be the cause of intriguing biological phenomena such as hybrid vigor, paramutations, and de novo epi-alleles. The examples of quantitative evaluation of epigenetic diversification brought about in the progenies’ population are, however, still poor, partly because of the methodology used to evaluate the level of variation.
For evaluation of epigenetic change by crossing, endogenous and/or foreign DNAs in plant nuclear genomes such as transposable elements (TEs), retrovirus-derived DNAs, and organellar DNA-like sequences that are recently derived from organellar genomes can be a good target for analysis, because they are prone to be methylated and also bring new materials for genome evolution [21,22,23,24]. While it is often difficult to estimate the methylation level of particular TE regions because of their high copy number, organellar DNA-like elements, so-called nuclear plastid DNA (NUPT), and nuclear mitochondrial DNA (NUMT) are lower copy number in the genome and can also be methylated, possibly as a result of genome defense response [25,26,27]. In this study, we attempted to trace the epigenetic modification change at organellar DNA-like sequences by crossing two natural strains of A. thaliana (Columbia and Landsberg electa). Using a next-generation sequencer, amplicon sequencing (amplicon-seq) with bisulfite-converted genomic DNA (gDNA) was conducted for F1 and F2 populations to analyze the heritability of DNA methylations on organellar DNA-like sequences. We observed a contrasting pattern of transgenerational DNA methylation change: in one organellar DNA-like sequence, the level of DNA methylation was decreased in individuals of the F1 population, then diversified in the F2 population. The results may suggest a trans-chromosomal interaction that could diversify epigenetic modifications in succeeding generations.
2. Materials and Methods
2.1. Experimental Procedures
To design the primer sets for amplification of organellar DNA-like sequences, NUPTs and NUMTs in the A. thaliana reference genome analyzed in previous studies [28,29] were manually inspected. In this study, nuclear genome sequences with more than 80% homology to organellar genomes were considered as candidates for the analysis of DNA methylation. We sought to find differentially methylated regions (DMR) of NUPTs/NUMTs between two strains of A. thaliana, Columbia (Col) and Landsberg electa (Ler), to track the DNA methylation inheritance for Col and Ler alleles separately. Using 1001 project database ([4], available in: http://neomorph.salk.edu/1001_epigenomes.html), 49 NUPTs and 36 NUMTs were inspected for methylation level difference and Single Nucleotide Polymorphisms (SNPs) sites between Col and Ler strains. To avoid mis-priming of primers to TEs or the organellar genome during PCR, NUPTs/NUMTs located inside the TEs and those longer than 250 bp were filtered out. In total, 15 primer sets were tested for Bisulfite PCR and 2 primer sets that amplify NUMTs (region 1; Chr3: 16574335..16574467, region 2; Chr4: 6341088..6341288) were used in this study. The two regions are homologous to mitochondrial genome sequences (region 1; ChM: 15459..15600, region 2; ChM: 236742..236943). The level of DNA methylation in these regions is shown in Figure S1. Because there is no DNA methylation in Ler allele of region 1 by the inspection of 1001 epigenome data, we checked the DNA methylation level by bisulfite Sanger sequencing to confirm the DNA methylation in the Ler allele (Figure 1a).
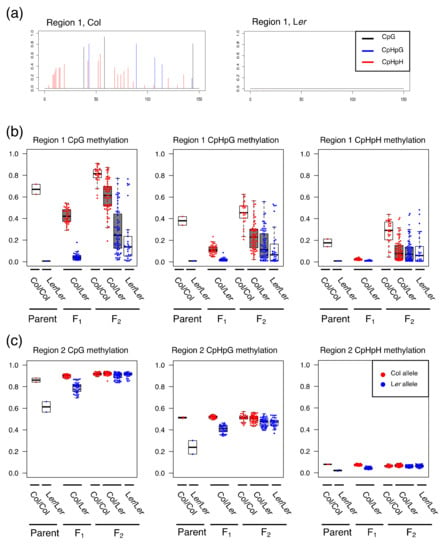
Figure 1.
Level of DNA methylations in parents, F1, and F2. (a) Bar plots of DNA methylation level of Col and Ler alleles in region 1; (b,c) The DNA methylation level of each individual for CpG, CpHpG, and CpHpH is plotted as boxplots and dots for region 1 (b) and region 2 (c). In each box, the thick line represents the median. Red and blue dots represent the methylation level of the Col and Ler alleles, respectively.
Seeds of Col and Ler were sown in Murashige and Skoog medium (MS) and grown in a plant growth chamber with the conditions of 23 °C and 16 h daylight. Reciprocal crosses were conducted and F1 seeds were collected from both crosses. F1 seeds were sown with the same conditions and fresh leaves of 18 F1 (Col × Ler) and 19 F1 (Ler × Col) individuals were collected. Two individuals from both F1 populations were selected to collect seeds by self-pollination. For each, 28 to 30 individuals were selected as F2 populations (F2 (Col × Ler ind.2 Self), F2 (Col × Ler ind.4 Self), F2 (Ler × Col ind.3 Self), and F2 (Ler × Col ind.4 Self)). The number of collected samples is shown in Table 1. Genomic DNA was extracted from fresh leaves of F1 and F2 individuals using a DNeasy Plant Mini Kit (QIAGEN K.K., Tokyo, Japan). Extracted gDNAs were bisulfite-converted by a MethylCode Bisulfite Conversion Kit (Thermo Fisher Scientific K.K., Tokyo, Japan). Bisulfite-converged DNAs were used for PCR to construct the sequencing library for MiSeq autosequencer (Illumina K.K., Tokyo, Japan). We modified the protocol of 16S Metagenomic Sequencing Library Preparation (available from http://support.illumina.com) for amplicon-seq of two regions in the nuclear genome of A. thaliana. In brief, PCR primers with overhang adapters were designed for target regions (Table S1). Then, eight cycles of PCR were conducted to attach indices (Nextera XT Index Kit, Illumina) that enable us to distinguish each individual. Products with indices were normalized and pooled in the sequencing library. To improve the sequencing quality, PhiX Control (30% of sequencing library volume) was added by following the instructions from Illumina. The sequencing library was sequenced by a MiSeq autosequencer (Illumina) with 300 cycles (150 × 2). Reads were classified into individuals by attached indices.

Table 1.
The number of samples used for amplicon-Seq.
2.2. Data Analysis
Quality of sequenced reads was inspected by program FastQC v. 0.11.5 (available from https://www.bioinformatics.babraham.ac.uk/). Reads with low quality were discarded by program Trimmomatic ver. 3 ([30], available in: http://www.usadellab.org/). Sequences of amplified regions (Supplementary File 1) were used for preparing reference sequence by script bismark_genome_preparatopm in Bismark suite v. 0.14.5 ([31], available from https://www.bioinformatics.babraham.ac.uk/). Then, treated reads were mapped to the reference sequences by program bismark. For F1 and F2 individuals, reads from Col and Ler alleles were distinguished by four and two SNP sites for region 1 and 2. The level of DNA methylation in cytosine sites was extracted by script bismark_methylation_extractor in Bismark suite. The commands used in data analysis are described in Supplementary File 2. For analysis of DNA methylations, cytosine sites shared by both Col and Ler alleles were selected and used for subsequent analyses of DNA methylations (region 1: two sites with CpG, five sites with CpHpG, and 23 sites with CpHpH contexts, region 2: 10 sites with CpG, six sites with CpHpG, and 33 sites with CpHpH contexts, Table S2). From the extracted methylation data, the weighted methylation level [32] was calculated for each allele of each individual and each cytosine motif by the following equation:
in which Ci and Ti represent methylated (Ci) and unmethylated (Ti) cytosine counts of the ith cytosine site with CpG/CpHpG/CpHpH context.
To estimate the epigenetic regulation genes possibly involved in DNA methylation in the regions, genome-wide methylome data from mutant lines of epigenetic regulation genes [33] were analyzed. The data of single base-pair methylation level were downloaded from Gene Expression Omnibus (GEO, available in: https://www.ncbi.nlm.nih.gov/geo/) for mutants associated with epigenetic regulations (GSE39901). The fractional methylation level [32] was calculated for each region to compare the DNA methylation level between the wild type and mutants.
3. Results
3.1. Summary of Amplicon Sequencing
For individuals, over 40,000 reads were mapped (Table S3, minimum mapped reads; 43,686, maximum mapped reads; 507,516). The number of reads mapped to each allele was compared with each other. If the number of reads mapped to one allele was smaller than that mapped to the other allele (less than 5% of total mapped reads), the reads were regarded as mapping error and not used in the following analysis. The rate of probable mapping error was small enough (median = 0.8%). For individuals in F1 populations, nearly equal amounts of reads were mapped for Col and Ler alleles, suggesting the mapping/distinguishing strategy of Col and Ler reads by SNPs works well and mapping errors at heterozygotes can be ignored. The genotype of individuals in F2 (Col/Col; CC, Col/Ler; CL, Ler/Ler; LL) was detected from mapped reads and the results are summarized in Table S4. F2 populations showed no deviation from the expected segregation ratio (CC = 0.25, CL = 0.5, LL = 0.25), except for region 2 of the F2(Col × Ler ind.2 Self) population showing a weak significance (Chi-square = 7.9, p = 0.02). These results indicate that the process of sequencing, mapping, and distinguishing Col/Ler reads is appropriate and there is no distortion of segregation in regions used. For Col and/or Ler alleles of each individual, the DNA methylation levels with different sequence motifs (CpG, CpHpG, CpHpH) were estimated (Table S5).
3.2. Fluctuation of DNA Methylation Level in F1 and F2 Populations in Region 1
The DNA methylation could be differently reset and re-initiated in maternal and paternal gametogenesis, resulting in a different level of DNA methylation in each Col and Ler allele between reciprocal crosses. For each allele in an F1 individual, we inspected the distribution of methylation level and tested the difference of distributions between reciprocal crosses (Figures S2 and S3). There were no strong significant differences in distribution (p < 0.001). Thus, in the following analyses, we merged data of reciprocal crosses in F1 and F2 populations. Although there might be a difference of inheritance between male/female gametes for the analyzed regions, the effects could be relatively small compared to other factors affecting the methylation levels in these regions.
In region 1, we observed a drastic change of DNA methylation pattern from parents through F2 offspring. The medians of DNA methylation level of parent Col and Ler in region 1 were 0.67/0.01 for CpG, 0.38/0.01 for CpHpG, and 0.18/0.01 for CpHpH motifs, respectively (Figure 1, Table 2). In the F1 population, a decrease of DNA methylation was observed in region 1 for all motifs (CpG, CPHpG, and CpHpH). In the F1 population, the values in the Col allele were 0.42 for CpG, 0.11 for CpHpG, and 0.02 for CpHpH, while the values in the Ler allele remained largely unchanged (0.04 for CpG, 0.02 for CpHpG, 0.01 for CpHpH). In the F2 population, in contrast to the F1 population, the DNA methylation level of both Col and Ler alleles was increased and largely diversified for all three motifs. The standard deviations of Col and Ler alleles in F2 were larger than those in F1 for all motifs (Table 2). In particular, the difference was prominent in Ler alleles, because their methylation levels were small in the F1 population. In contrast to the change of DNA methylation in region 1, the DNA methylation level of both Col and Ler alleles in region 2 showed convergence to a certain level of DNA methylation for all motifs. The medians of parent Col and Ler were 0.86/0.61 for CpG, 0.51/0.24 for CpHpG, and 0.08/0.02 for CpHpH, respectively. The methylation level of Ler alleles was then increased in succeeding generations (Table 2). Compared to region 1, region 2 showed a relatively small difference of dispersion for DNA methylation level between the F1 and F2 populations.

Table 2.
Diversification of DNA methylation level.
3.3. Correlation of Region 1 Methylation Level between Alleles in F2 Heterogyzotes
In region 1, the DNA methylation level of Col allele in F1 and F2 populations were higher than those of Ler alleles (Figure 1 and Figure S2). The increase of DNA methylation in Ler alleles in the F2 population could be caused by trans-chromosomal effect of Col allele. The DNA methylation levels in F2 heterozygotes showed significant correlation between Col and Ler alleles (CpG; ρ = 0.57, p = 8.50E-7, CpHpG; ρ = 0.75, p = 2.15E-12, CpHpH; ρ = 0.97, p < 2.20E-16). The DNA methylation level with each motif showed positive regression coefficients (0.800 for CpG, 0.751 for CpHpG, and 1.054 for CpHpH, Figure 2). The result may indicate a trans effect, possibly from a chromosome derived from Col to that from Ler, that influences the methylation level of region 1 in the F2 population. The trans-chromosomal effect can be caused by small interference RNA (siRNA) and the RNA-directed DNA methylation (RdDM) machinery. There are several reads of small RNA in both region 1 and region 2 (Figure S4). Whether siRNA will be expressed from other regions in the Col nuclear genome, the sequence having similarity to region 1 was confirmed by the program blastn implemented in NCBI BLAST ver.2.6.0 (https://blast.ncbi.nlm.nih.gov/) with default settings (Table S6). There was no significant hit more than 50 bp long, suggesting that small RNAs expressed only from this region can mainly affect the DNA methylation by trans-chromosome interaction.
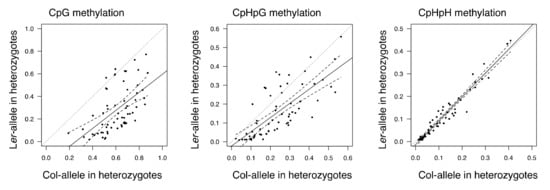
Figure 2.
Methylation level correlation between Col and Ler alleles in F2 heterozygotes. Plot of methylation level correlation between Col and Ler alleles in F2 heterogyzotes. Solid lines represent best-fit linear regression lines. Dashed lines represent 95% lower and upper bounds of regression.
3.4. DNA Methylation in Mutant Lines
In region 1, the ago4 and dcl2/3/4 mutants showed a decreased level of non-CpG methylation compared to that of wild type (WT) (Table 3, Figure S5). It may indicate that the DNA methylation in this region is in part dependent on the RdDM machinery. In addition, ddm1, suvh4/kyp, and met1 showed a more remarkable decrease of DNA methylation for both CpG and non–CpG contexts (Table 3, Figure S5). It suggests that maintenance methylation and chromatin remodeling are associated with the DNA methylation in this region. In region 2, there was no remarkable decrease of DNA methylation in mutant lines of ago4 and dcl2/3/4, while ddm1, suvh4/kyp, and met1 showed a decrease in DNA methylation (Table 3, Figure S6). This suggests that RdDM machinery is not involved in the DNA methylation of region 2.

Table 3.
Level of DNA methylation at mutants associated with epigenetic regulation.
4. Discussion
In previous studies, as well as genetic variation, a high epigenetic diversity is reported for natural population of A. thaliana [1,4]. Other plants also have epigenetic diversity in their populations (e.g., [34,35,36]). Some studies indicate the act of natural selection on the epigenetic variations for their relation to heat stress response [37,38] or for different environmental conditions [39]. If the epigenetic modification on DNA or histone is the target of selection, the diversity of such epigenetic modification should be given more attention as a source of variation. While there are some reports focused on the epigenetic variation, the mechanism by which these variations are newly generated, inherited, or maintained, and how they are erased, is still not clear. In addition to the epi-mutation at a single site of A. thaliana mutation accumulation lines [15], it is shown that both intra- and interspecific crossing can cause the difference of epigenetic modifications [17,18,19,20]. The insertion of TEs near genes can create TE- and repeat-associated epimutable alleles. During the cross between different strains, such epimutable alleles can be DNA methylated with a transchromosomal effect of other TEs on different chromosomes [16]. In this study, we observed two contrasting trajectories of DNA methylation inheritance and diversification of methylation of organellar DNA-like sequences in the F1 and F2 populations. While region 2 showed an almost even level of DNA methylation in the F2 population for all cytosine motifs, the DNA methylation level in region 1 varied considerably among individuals. In both regions, there might be a trans-chromosomal effect on the methylation level, possibly from the Col-allele chromosome to the Ler-allele’s. In region 1, methylation levels of Col and Ler alleles in F2 heterozygotes showed a positive correlation, suggesting that the elevation of methylation levels of Ler alleles could somewhat depend on the methylation levels of Col alleles. There could be a mechanism to balance the level of DNA methylation between alleles within a specific locus. In region 2, the methylation levels of Ler alleles were elevated in both F1 and F2, resulting in the same level of DNA methylation as in Col alleles.
A well-known trans-chromosomal epigenetic effect is paramutation, found in the maize red colour 1 (r1) locus involved in anthocyanin biosynthesis [40]. Several loci are reported as paramutable in maize and show 100% penetrance and heritability (e.g., red1, booster1, purple plant1). The paramutations and/or paramutation-like phenomena are also reported in other plants such as A. thaliana, tobacco, tomato, etc. [41]. The multiple epigenetic mechanism, including not only the RdDM pathway but also genes indirectly involved in DNA methylation such as DDM1, could be associated with the paramutation and paramutation-like phenomena [42]. Because there are several reads of small RNA that match the regions analyzed in this study, the RdDM mechanism might be involved in the methylation pattern of the two regions. Because a decrease in non–CpG DNA methylation in ago4 and dcl2/3/4 mutants was observed for region 1 (Table 3, Figure S5), the RdDM machinery may be involved in the change of DNA methylation observed at region 1 in F1 and F2. The involvement of RdDM machinery in region 1 could cause different trajectories between region 1 and 2. In addition, regions 1 and 2 both showed a substantial decrease in DNA methylation in the ddm1 and suvh4/kyp mutants (Table 3, Figure S5). This suggests that the genes related to the maintenance DNA methylation are involved in the DNA methylation in both regions. As suggested in a previous study [42], they could also be associated with the trans-chromosomal DNA methylation observed in regions 1 and 2. In contrast to region 1, region 2 showed little effect of RdDM machinery on the DNA methylation in the region. Although the details are unknown, this might suggest that the elevation of DNA methylation in region 2 is caused by another mechanism such as chromatin remodeling.
This study demonstrates that crossing individuals with different methylation levels can generate diversity of DNA methylation in the progenies, though the pattern is complex. In region 1, DNA methylation levels were decreased in F1 populations, especially for non-CpG motif cytosines. Then, however, the medians of methylation levels were increased with large variance in F2 populations. The trans-chromosomal methylation and demethylation in hybrids of A. thaliana were also shown in a previous study [17,18]. The difference between F1 and F2 might indicate the difference in trans-chromosomal effect associated with the expression of small RNAs in F1 and F2. The result implies the complexity of determining the DNA methylation level of the region after crossing different strains. The process of DNA methylation change was not completed in one generation; rather, it may take several or more generations to reach a new plateau of methylation. In the previous study of ddm1 mutant, ectopic accumulation of DNA methylation at many loci with the ddm1 mutant was observed after nine repeated self-pollinations of genome-wide hypomethylated ddm1 [43]. Instead of being a single state of plateau, the methylation level in this region could be diversified among progenies and retain multiple status of DNA methylation. Both Col and Ler alleles of homozygotes in F2 also showed diversified DNA methylation, indicating that the hybridization and succeeding selfing could cause allelic methylation differences. If the diversification observed in this study is rather common in plants, it could indicate the contribution of integrated organellar DNA-like sequences or other foreign DNA to generate the epigenetic diversity of the plant genome.
TEs or repetitive sequences near genes can affect the gene expression. In A. thaliana and related species, DNA methylations on short interspersed nuclear elements (SINE)-related sequences at 5’ region control the expression of FWA [35,44,45]. In Arabidopsis-related species, there are intraspecific variations of DNA methylation at the 5’ regions of transcription start site that are associated with the expression of FWA, though there is no genetic variation in the region [35,45]. We might hypothesize that these DNA methylation variations within populations are incorporated or maintained by outcrossing between individuals, as previously proposed for population-level diversification via epigenetic phenomena [46]. For outcrossing species such as Arabidopsis lyrata, one of the close relatives of A. thaliana, the diversifying effect by outcrossing is relatively common, while a selfing species like A. thaliana might diversify the pattern of DNA methylation by occasional outcrossing. By using other natural strains containing intraspecific variations of integrated organellar DNA-like sequences near genes and tracking DNA methylation inheritance, the effect of crossing on methylation and expression could be clarified.
Supplementary Materials
The following are available online at http://www.mdpi.com/2073-4425/9/12/602/s1, Figure S1: DNA methylation at parental strains, Figure S2: Level of region 1 DNA methylation in parents, F1, and F2, Figure S3: Level of region 2 DNA methylation in parents, F1, and F2, Figure S4: Small RNA expression at parental strains, Figure S5: DNA methylation of region 1 in mutant lines, Figure S6: DNA methylation of region 2 in mutant lines, Table S1: Primers used in the amplicon-Seq, Table S2: Position and motif (CpG, CpHpG, and CpHpH) in analyzed sequence, Table S3: Mapped data of bisulfite amplicon-Seq, Table S4: Genotype of F2 populations, Table S5: Level of DNA methylation, Table S6: BLASTN search of region 1 & 2 in nuclear genome, Supplementary File 1: Sequences of analyzed regions, Supplementary File 2: Commands of data analysis.
Author Contributions
Conceptualization, T.Y., A.K., and T.K.; methodology, T.Y., A.K., Y.T., and T.K.; investigation, T.Y. and Y.T.; writing, T.Y. and A.K.
Funding
This work was supported by a NIG-JOINT (2015B-3) grant to A.K., a Research Fellow of Japan Society for the promotion of Science (201508708 to T.Y.), and a JSPS Grant-in-aid for Early-Career Scientists (18K14758 to T.Y.).
Conflicts of Interest
The authors declare no conflict of interest.
References
- Schmitz, R.J.; Schultz, M.D.; Urich, M.A.; Nery, J.R.; Pelizzola, M.; Libiger, O.; Alix, A.; McCosh, R.B.; Chen, H.; Schork, N.J.; et al. Patterns of population epigenomic diversity. Nature 2013, 495, 193–198. [Google Scholar] [CrossRef] [PubMed]
- Seymour, D.K.; Koenig, D.; Hagmann, J.; Becker, C.; Weigel, D. Evolution of DNA methylation patterns in the Brassicaceae is driven by differences in genome organization. PLoS Genet. 2014, 10, e1004785. [Google Scholar] [CrossRef] [PubMed]
- Niederhuth, C.E.; Bewick, A.J.; Ji, L.; Alabady, M.S.; Kim, K.; Li, Q.; Rohr, N.A.; Rambani, A.; Burke, J.M.; Udall, J.A.; et al. Widespread natural variation of DNA methylation within angiosperms. Genome Biol. 2016, 17, 194. [Google Scholar] [CrossRef] [PubMed]
- Kawakatsu, T.; Huang, S.-S.C.; Jupe, F.; Sasaki, E.; Schmitz, R.J.; Urich, M.A.; Castanon, R.; Nery, J.R.; Barragan, C.; He, Y.; et al. Epigenomic diversity in a global collection of Arabidopsis thaliana accessions. Cell 2016, 166, 492–505. [Google Scholar] [CrossRef] [PubMed]
- Law, J.A.; Jacobsen, S.E. Establishing, maintaining and modifying DNA methylation patterns in plants and animals. Nat. Rev. Genet. 2010, 11, 204–220. [Google Scholar] [CrossRef] [PubMed]
- Zemach, A.; Kim, M.; Hsieh, P.-H.H.; Coleman-Derr, D.; Eshed-Williams, L.; Thao, K.; Harmer, S.L.; Zilberman, D. The Arabidopsis nucleosome remodeler DDM1 allows DNA methyltransferases to access H1-containing heterochromatin. Cell 2013, 153, 193–205. [Google Scholar] [CrossRef]
- Matzke, M.A.; Mosher, R.A. RNA-directed DNA methylation: An epigenetic pathway of increasing complexity. Nat. Rev. Genet. 2014, 15, 394–408. [Google Scholar] [CrossRef] [PubMed]
- Du, J.; Johnson, L.M.; Jacobsen, S.E.; Patel, D.J. DNA methylation pathways and their crosstalk with histone methylation. Nat. Rev. Mol. Cell Biol. 2015, 16, 519–532. [Google Scholar] [CrossRef] [PubMed]
- Lister, R.; O’Malley, R.C.; Tonti-Filippini, J.; Gregory, B.D.; Berry, C.C.; Millar, A.; Ecker, J.R. Highly integrated single-base resolution maps of the epigenome in Arabidopsis. Cell 2008, 133, 523–536. [Google Scholar] [CrossRef] [PubMed]
- Zemach, A.; Niel, I.E.; Silva, P.; Zilberman, D. Genome-wide evolutionary analysis of eukaryotic DNA methylation. Science 2010, 328, 916–919. [Google Scholar] [CrossRef] [PubMed]
- Schulz, B.; Eckstein, R.; Durka, W. Scoring and analysis of methylation-sensitive amplification polymorphisms for epigenetic population studies. Mol. Ecol. Resour. 2013, 13, 642–653. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Yazaki, J.; Sundaresan, A.; Cokus, S.; Chan, S.; Chen, H.; Henderson, I.R.; Shinn, P.; Pellegrini, M.; Jacobsen, S.E.; et al. Genome-wide high-resolution mapping and functional analysis of DNA methylation in Arabidopsis. Cell 2006, 126, 1189–1201. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Clarenz, O.; Cokus, S.; Bernatavichute, Y.V.; Pellegrini, M.; Goodrich, J.; Jacobsen, S.E. Whole-genome analysis of histone H3 lysine 27 trimethylation in Arabidopsis. PLoS Biol. 2007, 5, e129. [Google Scholar] [CrossRef]
- Iwasaki, M.; Paszkowski, J. Identification of genes preventing transgenerational transmission of stress-induced epigenetic states. Proc. Natl. Acad. Sci. USA 2014, 111, 8547–8552. [Google Scholar] [CrossRef]
- Van der Graaf, A.; Wardenaar, R.; Neumann, D.A.; Taudt, A.; Shaw, R.G.; Jansen, R.C.; Schmitz, R.J.; Colomé-Tatché, M.; Johannes, F. Rate, spectrum, and evolutionary dynamics of spontaneous epimutations. Proc. Natl. Acad. Sci. USA 2015, 112, 6676–6681. [Google Scholar] [CrossRef] [PubMed]
- Quadrana, L.; Colot, V. Plant transgenerational epigenetics. Annu. Rev. Genet. 2015, 50, 467–491. [Google Scholar] [CrossRef]
- Greaves, I.K.; Groszmann, M.; Ying, H.; Taylor, J.M.; Peacock, W.; Dennis, E.S. Trans chromosomal methylation in Arabidopsis hybrids. Proc. Natl. Acad. Sci. USA 2012, 109, 3570–3575. [Google Scholar] [CrossRef]
- Greaves, I.K.; Groszmann, M.; Wang, A.; Peacock, J.W.; Dennis, E.S. Inheritance of trans chromosomal methylation patterns from Arabidopsis F1 hybrids. Proc. Natl. Acad. Sci. USA 2014, 111, 2017–2022. [Google Scholar] [CrossRef]
- Chodavarapu, R.K.; Feng, S.; Ding, B.; Simon, S.A.; Lopez, D.; Jia, Y.; Wang, G.-L.; Meyers, B.C.; Jacobsen, S.E.; Pellegrini, M. Transcriptome and methylome interactions in rice hybrids. Proc. Natl. Acad. Sci. USA 2012, 109, 12040–12045. [Google Scholar] [CrossRef]
- Zhu, W.; Hu, B.; Becker, C.; Doğan, E.; Berendzen, K.; Weigel, D.; Liu, C. Altered chromatin compaction and histone methylation drive non-additive gene expression in an interspecific Arabidopsis hybrid. Genome Biol. 2017, 18, 157. [Google Scholar] [CrossRef]
- Levin, H.L.; Moran, J.V. Dynamic interactions between transposable elements and their hosts. Nat. Rev. Genet. 2011, 12, 615–627. [Google Scholar] [CrossRef] [PubMed]
- Fedoroff, N.V. Transposable elements, epigenetics, and genome evolution. Science 2012, 338, 758–767. [Google Scholar] [CrossRef]
- Staginnus, C.; Richert-Pöggeler, K.R. Endogenous pararetroviruses: Two-faced travelers in the plant genome. Trends Plant Sci. 2006, 11, 485–491. [Google Scholar] [CrossRef]
- Timmis, J.N.; Ayliffe, M.A.; Huang, C.Y.; Martin, W. Endosymbiotic gene transfer: Organelle genomes forge eukaryotic chromosomes. Nat. Rev. Genet. 2004, 5, 123–135. [Google Scholar] [CrossRef]
- Noutsos, C.; Richly, E.; Leister, D. Generation and evolutionary fate of insertions of organelle DNA in the nuclear genomes of flowering plants. Genome Res. 2005, 15, 616–628. [Google Scholar] [CrossRef]
- Huang, C.Y.; Grünheit, N.; Ahmadinejad, N.; Timmis, J.N.; Martin, W. Mutational decay and age of chloroplast and mitochondrial genomes transferred recently to angiosperm nuclear chromosomes. Plant Physiol. 2005, 138, 1723–1733. [Google Scholar] [CrossRef] [PubMed]
- Lough, A.N.; Faries, K.M.; Koo, D.-H.; Hussain, A.; Roark, L.M.; Langewisch, T.L.; Backes, T.; Kremling, K.A.; Jiang, J.; Birchler, J.A.; et al. Cytogenetic and sequence analyses of mitochondrial DNA insertions in nuclear chromosomes of maize. G3 Genes Genomes Genet. 2015, 5, 2229–2239. [Google Scholar] [CrossRef]
- Yoshida, T.; Furihata, H.Y.; Kawabe, A. Patterns of genomic integration of nuclear chloroplast DNA fragments in plant species. DNA Res. 2014, 21, 127–140. [Google Scholar] [CrossRef] [PubMed]
- Yoshida, T.; Furihata, H.; Kawabe, A. Analysis of nuclear mitochondrial DNAs and factors affecting patterns of integration in plant species. Genes Genet. Syst. 2017, 92, 27–33. [Google Scholar] [CrossRef]
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef]
- Krueger, F.; Andrews, S.R. Bismark: A flexible aligner and methylation caller for Bisulfite-Seq applications. Bioinformatics 2011, 27, 1571–1572. [Google Scholar] [CrossRef]
- Schultz, M.D.; Schmitz, R.J.; Ecker, J.R. “Leveling” the playing field for analyses of single-base resolution DNA methylomes. Trends Genet. 2012, 28, 583–585. [Google Scholar] [CrossRef]
- Stroud, H.; Greenberg, M.V.; Feng, S.; Bernatavichute, Y.V.; Jacobsen, S.E. Comprehensive analysis of silencing mutants reveals complex regulation of the Arabidopsis methylome. Cell 2013, 152, 352–364. [Google Scholar] [CrossRef]
- Lira-Medeiros, C.; Parisod, C.; Fernandes, R.; Mata, C.; Cardoso, M.; Ferreira, P. Epigenetic variation in mangrove plants occurring in contrasting natural environment. PLoS ONE 2010, 5, e10326. [Google Scholar] [CrossRef]
- Fujimoto, R.; Sasaki, T.; Kudoh, H.; Taylor, J.M.; Kakutani, T.; Dennis, E.S. Epigenetic variation in the FWA gene within the genus Arabidopsis. Plant J. 2011, 66, 831–843. [Google Scholar] [CrossRef]
- Sáez-Laguna, E.; Guevara, M.-Á.; Díaz, L.-M.; Sánchez-Gómez, D.; Collada, C.; Aranda, I.; Cervera, M.-T.T. Epigenetic variability in the genetically uniform forest tree species Pinus pinea L. PLoS ONE 2014, 9, e103145. [Google Scholar] [CrossRef]
- Shen, X.; Jonge, J.; Forsberg, S.K.; Pettersson, M.E.; Sheng, Z.; Hennig, L.; Carlborg, Ö. Natural CMT2 variation is associated with genome-wide methylation changes and temperature seasonality. PLoS Genet. 2014, 10, e1004842. [Google Scholar] [CrossRef]
- Dubin, M.J.; Zhang, P.; Meng, D.; Remigereau, M.-S.; Osborne, E.J.; Casale, F.; Drewe, P.; Kahles, A.; Jean, G.; Vilhjálmsson, B.; et al. DNA methylation in Arabidopsis has a genetic basis and shows evidence of local adaptation. eLife 2015, 4, e05255. [Google Scholar] [CrossRef]
- Zoldoš, V.; Biruš, I.; Muratović, E.; Šatović, Z.; Vojta, A.; Robin, O.; Pustahija, F.; Bogunić, F.; Bočkor, V.V.; Siljak-Yakovlev, S. Epigenetic differentiation of natural populations of Lilium bosniacum associated with contrasting habitat conditions. Genome Biol. Evol. 2018, 10, 291–303. [Google Scholar] [CrossRef]
- Brink, A.R. A genetic change associated with the R locus in maize which is directed and potentially reversible. Genetics 1956, 41, 872. [Google Scholar]
- Pilu, R. Paramutation phenomena in plants. Semin. Cell Dev. Biol. 2015, 44, 2–10. [Google Scholar] [CrossRef]
- Zheng, Z.; Yu, H.; Miki, D.; Jin, D.; Zhang, Q.; Ren, Z.; Gong, Z.; Zhang, H.; Zhu, J.-K. Involvement of multiple gene-silencing pathways in a paramutation-like phenomenon in Arabidopsis. Cell Rep. 2015, 11, 1160–1167. [Google Scholar] [CrossRef]
- Ito, T.; Tarutani, Y.; To, T.K.; Kassam, M.; Duvernois-Berthet, E.; Cortijo, S.; Takashima, K.; Saze, H.; Toyoda, A.; Fujiyama, A.; et al. Genome-wide negative feedback drives transgenerational DNA methylation dynamics in Arabidopsis. PLoS Genet. 2015, 11, e1005154. [Google Scholar] [CrossRef]
- Kinoshita, Y.; Saze, H.; Kinoshita, T.; Miura, A.; Soppe, W.J.; Koornneef, M.; Kakutani, T. Control of FWA gene silencing in Arabidopsis thaliana by SINE-related direct repeats. Plant J. 2007, 49, 38–45. [Google Scholar] [CrossRef]
- Fujimoto, R.; Kinoshita, Y.; Kawabe, A.; Kinoshita, T.; Takashima, K.; Nordborg, M.; Nasrallah, M.E.; Shimizu, K.K.; Kudoh, H.; Kakutani, T. Evolution and control of imprinted FWA genes in the genus Arabidopsis. PLoS Genet. 2008, 4, e1000048. [Google Scholar] [CrossRef]
- Rapp, R.A.; Wendel, J.F. Epigenetics and plant evolution. New Phytol. 2005, 168, 81–91. [Google Scholar] [CrossRef]
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).