Segmental and Tandem Duplications Driving the Recent NBS-LRR Gene Expansion in the Asparagus Genome
Abstract
1. Introduction
2. Materials and Methods
2.1. Retrieval and Classification of Asparagus NBS-Encoding Genes
2.2. Conserved Domains, Sequence Alignment, and Phylogenetic Analyses
2.3. Distribution, Cluster Analysis, and Gene Duplications
2.4. Digital Expression Analysis
2.5. Identification and Analysis of the Promoter Regions
3. Results
3.1. Identification of NBS-Encoding Genes
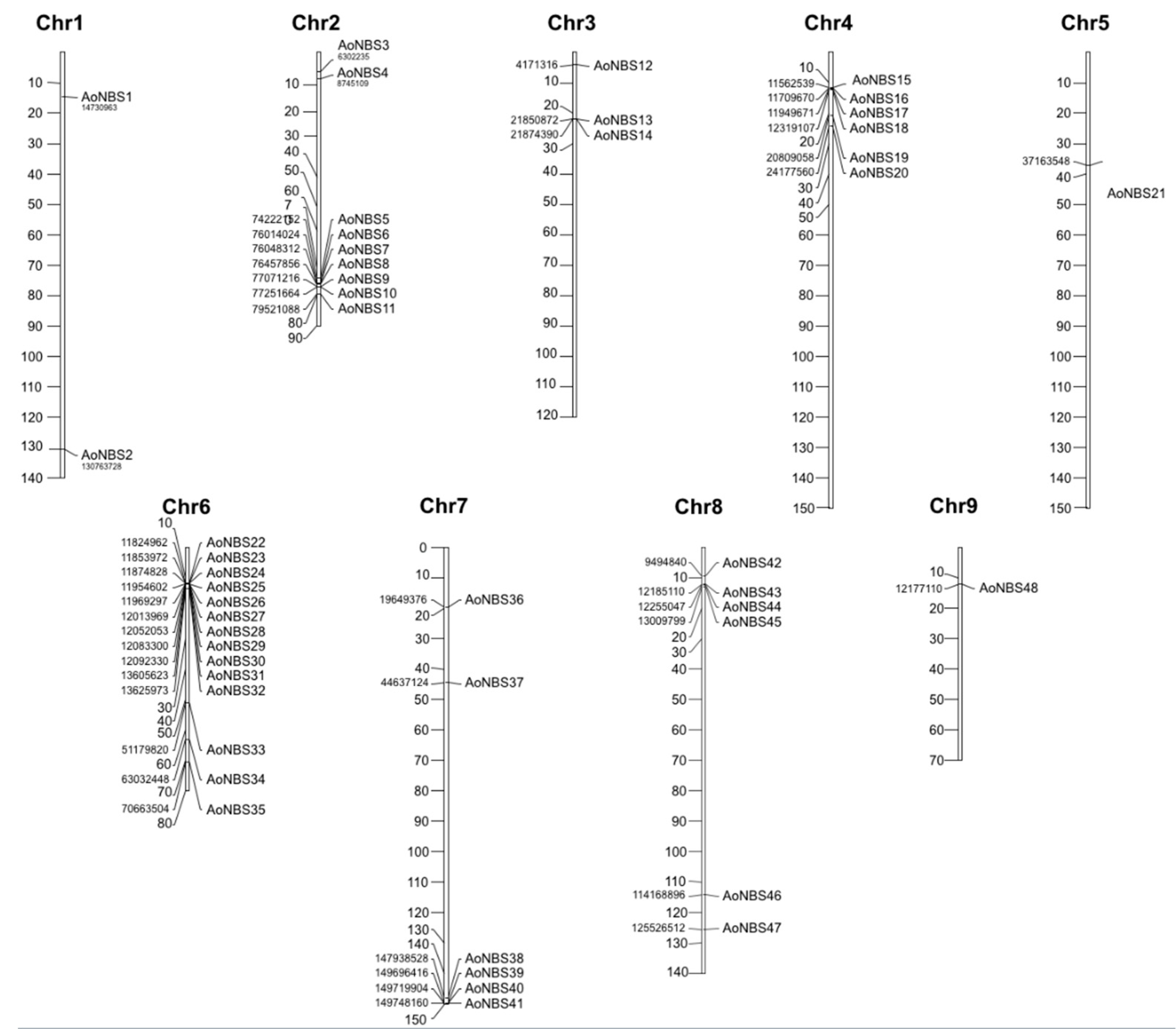
3.2. Distribution of NBS-Encoding Genes
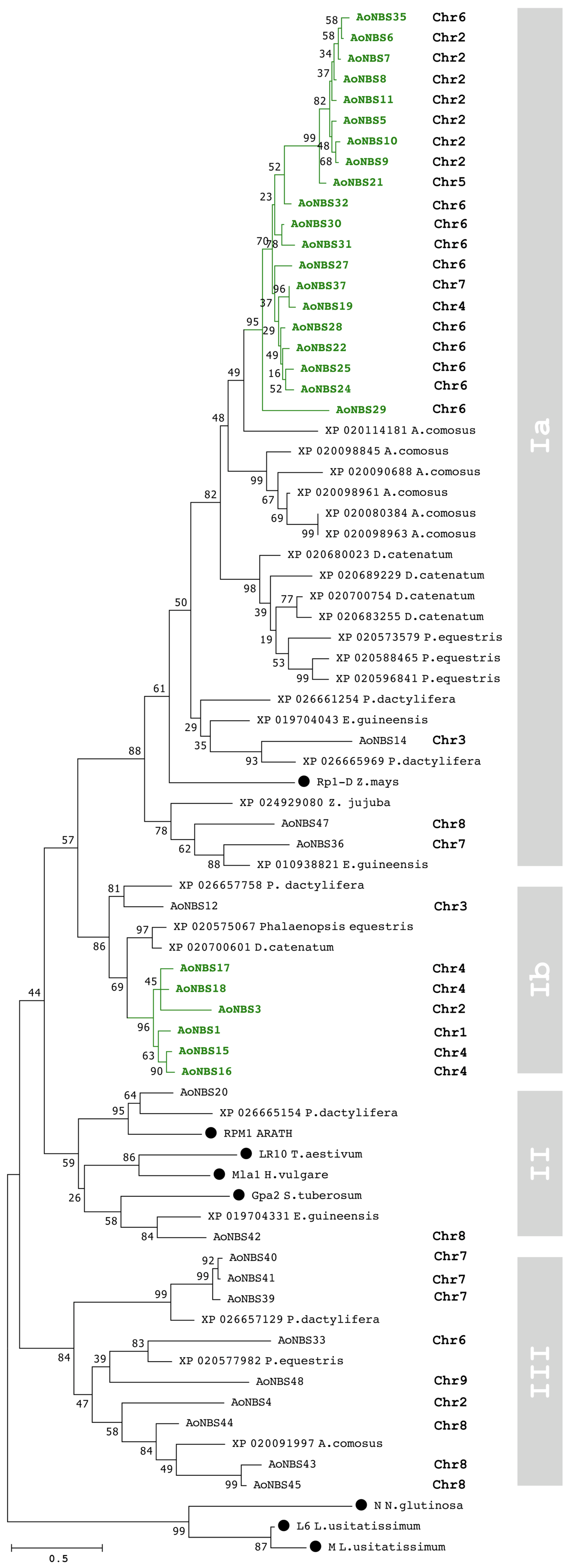
3.3. Sequence Alignment and Phylogenetic Analyses
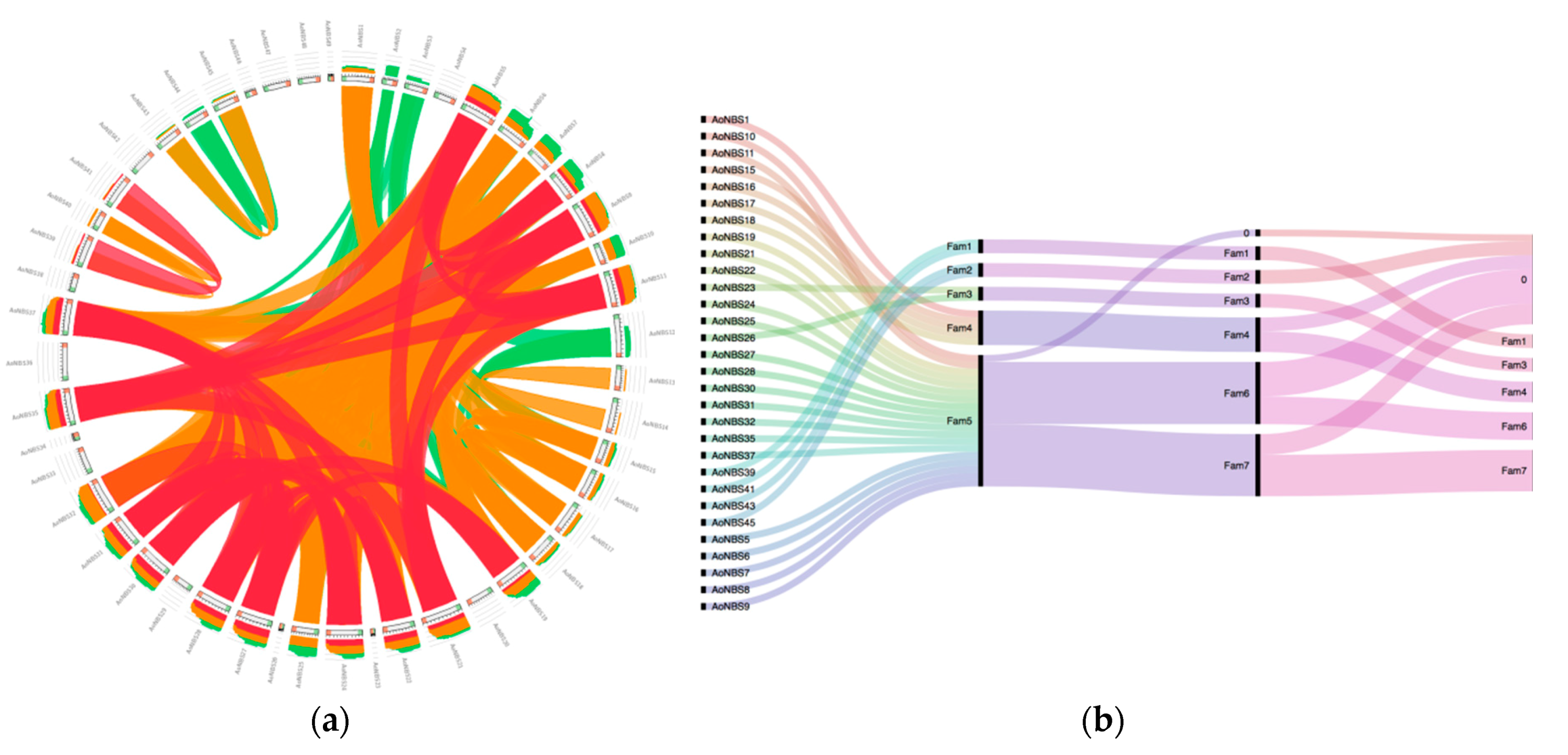
3.4. Gene Duplications
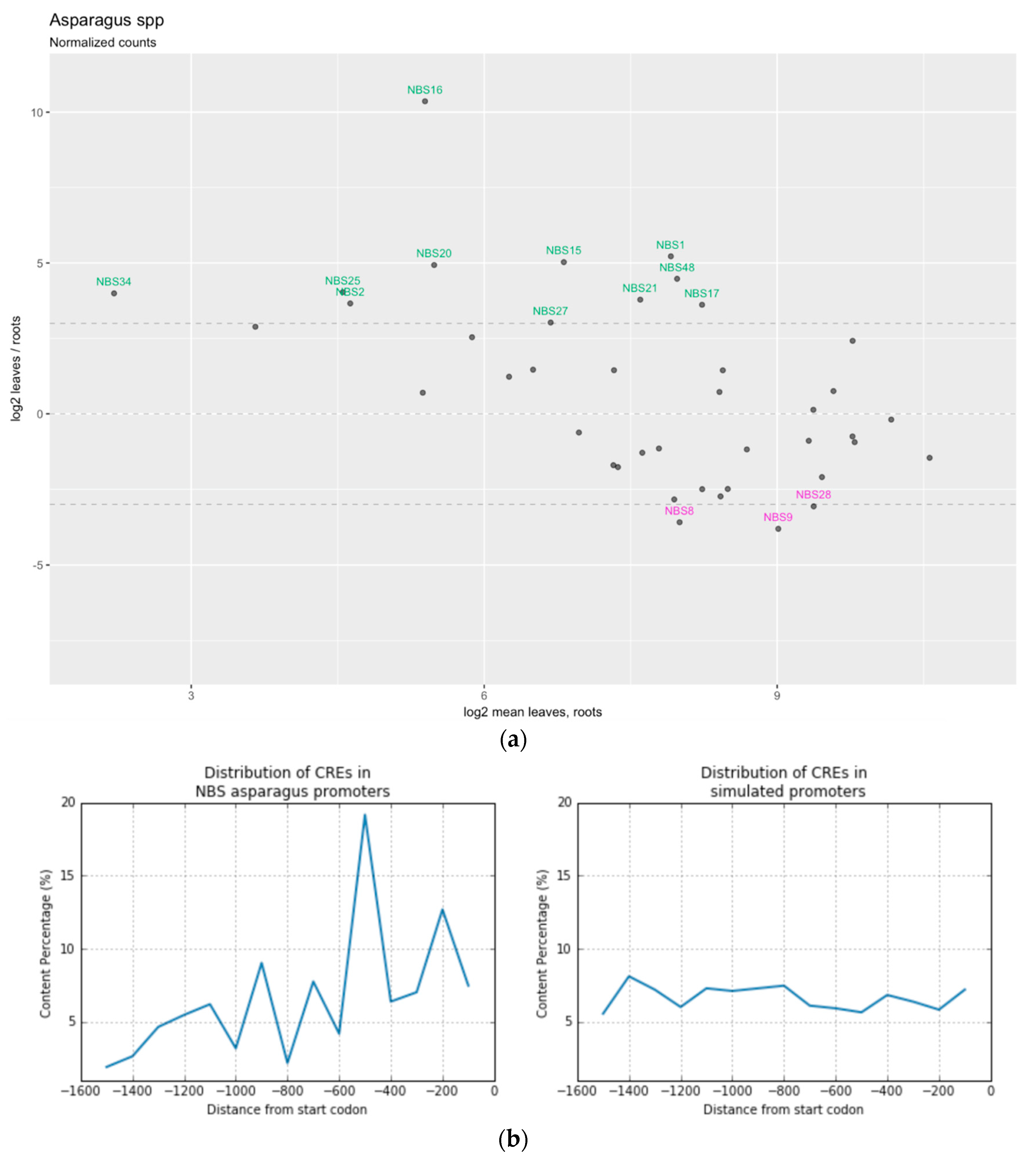
3.5. Digital Expression Profiling of the NBS-LRR Genes
3.6. Promoter Sequence Analysis
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
Data Availability
References
- FAOSTAT. Available online: http://faostat.fao.org (accessed on 2 September 2018).
- Sneep, J. The significance of andromonoecism for the breeding of Asparagus officinalis L. II. Euphytica 1953, 2, 224–228. [Google Scholar] [CrossRef]
- Benson, B.L. Sex influence on foliar trait morphology in asparagus. HortScience 1982, 17, 625–627. [Google Scholar]
- Geoffriau, E.; Denoue, D.; Rameau, C. Assessment of genetic variation among asparagus (Asparagus officinalis L.) populations and cultivars: Agromorphological and isozymic data. Euphytica 1992, 61, 169–179. [Google Scholar] [CrossRef]
- Mercati, F.; Riccardi, P.; Harkess, A.; Sala, T.; Abenavoli, M.R.; Leebens-Mack, J.; Falavigna, A.; Sunseri, F. Single nucleotide polymorphism-based parentage analysis and population structure in garden asparagus, a worldwide genetic stock classification. Mol. Breed. 2015, 35, 59. [Google Scholar] [CrossRef]
- Moreno, R.; Espejo, J.A.; Cabrera, A.; Millán, T.; Gil, J. Ploidic and molecular analysis of ‘Morado de Huetor’ asparagus (Asparagus officinalis L.) population; a Spanish tetraploid landrace. Genet. Resour. Crop Evol. 2006, 53, 729–736. [Google Scholar] [CrossRef]
- Moreno, R.; Espejo, J.A.; Gil, J. Development of triploid hybrids in asparagus breeding employing a tetraploid landrace. Euphytica 2010, 173, 369–375. [Google Scholar] [CrossRef]
- Castro, P.; Gil, J.; Cabrera, A.; Moreno, R. Assessment of genetic diversity and phylogenetic relationships in Asparagus species related to Asparagus officinalis. Genet. Resour. Crop Evol. 2013, 60, 1275–1288. [Google Scholar] [CrossRef]
- Castro, P.; Rubio, J.; Gil, J.; Moreno, R. Introgression of new germplasm in current diploid cultivars of garden asparagus from a tetraploid Spanish landrace “Morado de Huétor”. Sci. Hortic. 2014, 168, 157–160. [Google Scholar] [CrossRef]
- Anido, F.L.; Cointry, E. Asparagus. In Vegetables II; Prohens, J., Nuez, F., Eds.; Springer: New York, NY, USA, 2008; pp. 87–119. [Google Scholar]
- Stephens, C.T.; de Vries, R.M.; Sink, K.C. Evaluation of Asparagus species for resistance to Fusarium oxysporum f.sp. asparagi and F. moniliforme. HortScience 1989, 24, 365–368. [Google Scholar]
- Broadhurst, P.G. Stemphylium disease tolerance in Asparagus officinalis L. Acta Hortic. 1996, 415, 387–391. [Google Scholar] [CrossRef]
- Iwato, M.; Kosaza, M.; Takeuchi, Y.; Matsumoto, M.; Inada, M.; Ozaki, Y.; Okubo, H. Stem blight resistance of Asparagus kiusianus and its hybrid with A. officinalis. Adv. Hortic. Sci. 2014, 28, 202–207. [Google Scholar]
- Abdelrahman, M.; Suzumura, N.; Mitoma, M.; Matsuo, S.; Ikeuchi, T.; Mori, M.; Murakami, K.; Ozaki, Y.; Matsumoto, M.; Uragami, A.; et al. Comparative de novo transcriptome profiles in Asparagus officinalis and A. kiusianus during the early stage of Phomopsis asparagi infection. Sci. Rep. 2017, 7, 2608. [Google Scholar] [CrossRef] [PubMed]
- Shao, Z.-Q.; Xue, J.-Y.; Wu, P.; Zhang, Y.-M.; Wu, Y.; Hang, Y.-Y.; Wang, B.; Chen, J.-Q. Large-scale analyses of angiosperm nucleotide-binding site-leucine-rich repeat genes reveal three anciently diverged classes with distinct evolutionary patterns. Plant Physiol. 2016, 170, 2095–2109. [Google Scholar] [CrossRef] [PubMed]
- Perazzolli, M.; Malacarne, G.; Baldo, A.; Righetti, L.; Bailey, A.; Fontana, P.; Velasco, R.; Malnoy, M. Characterization of resistance gene analogues (RGAs) in apple (Malus × domestica Borkh.) and their evolutionary history of the Rosaceae family. PLoS ONE 2014, 9, e83844. [Google Scholar] [CrossRef] [PubMed]
- Lozano, R.; Hamblin, M.T.; Prochnik, S.; Jannink, J.-L. Identification and distribution of the NBS-LRR gene family in the Cassava genome. BMC Genom. 2015, 16, 360. [Google Scholar] [CrossRef] [PubMed]
- Zhou, T.; Wang, Y.; Chen, J.Q.; Araki, H.; Jing, Z.; Jiang, K.; Shen, J.; Tian, D. Genome-wide identification of NBS genes in japonica rice reveals significant expansion of divergent non-TIR NBS-LRR genes. Mol. Genet. Genom. 2004, 271, 402–415. [Google Scholar]
- Tan, S.; Wu, S. Genome wide analysis of nucleotide-binding site disease resistance genes in Brachypodium distachyon. Comput. Funct. Genom. 2012, 2012, 418208. [Google Scholar]
- Cheng, Y.; Li, X.; Jiang, H.; Ma, W.; Miao, W.; Yamada, T.; Zhang, M. Systematic analysis and comparison of nucleotide-binding site disease resistance genes in maize. FEBS J. 2012, 279, 2431–2443. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Liang, P.; Ming, R. Genome-wide identification and characterization of nucleotide-binding site (NBS) resistance genes in pineapple. Trop. Plant Biol. 2016, 9, 187–199. [Google Scholar] [CrossRef]
- Harkess, A.; Zhou, J.; Xu, C.; Bowers, J.E.; Van der Hulst, R.; Ayyampalayam, S.; Mercati, F.; Riccardi, P.; McKain, M.R.; Kakrana, A.; et al. The asparagus genome sheds light on the origin and evolution of a young Y chromosome. Nat. Commun. 2017, 8, 1279. [Google Scholar] [CrossRef] [PubMed]
- Rout, E.; Nanda, S.; Nayak, S.; Joshi, R.K. Molecular characterization of NBS encoding resistance genes and induction analysis of a putative candidate gene linked to Fusarium basal rot resistance in Allium sativum. Physiol. Mol. Plant Pathol. 2014, 85, 15–24. [Google Scholar] [CrossRef]
- Marchler-Bauer, A.; Bo, Y.; Han, L.; He, J.; Lanczycki, C.J.; Lu, S.; Chitsaz, F.; Derbyshire, M.K.; Geer, R.C.; Gonzales, N.R.; et al. CDD/SPARCLE: Functional classification of proteins via subfamily domain architectures. Nucleic Acids Res. 2017, 45, D200–D203. [Google Scholar] [CrossRef] [PubMed]
- Letunic, I.; Doerks, T.; Bork, P. SMART 7: Recent updates to the protein domain annotation resource. Nucleic Acids Res. 2012, 40, D302–D305. [Google Scholar] [CrossRef] [PubMed]
- Alva, V.; Nam, S.-Z.; Söding, J.; Lupas, A.N. The MPI bioinformatics Toolkit as an integrative platform for advanced protein sequence and structure analysis. Nucleic Acids Res. 2016, 44, W410–W415. [Google Scholar] [CrossRef] [PubMed]
- Li, W.; Cowley, A.; Uludag, M.; Gur, T.; McWilliam, H.; Squizzato, S.; Park, Y.M.; Buso, N.; Lopez, R. The EMBL-EBI bioinformatics web and programmatic tools framework. Nucleic Acids Res. 2015, 43, W580–W584. [Google Scholar] [CrossRef] [PubMed]
- Die, J.V. Refseqr: Common Computational Operations Working with GenBank. Zenodo. 2018. Available online: https://rdrr.io/github/jdieramon/refseqR/ (accessed on 21 November 2018).
- Tamura, K.; Stecher, G.; Peterson, D.; Filipski, A.; Kumar, S. MEGA6: Molecular Evolutionary Genetics Analysis version 6.0. Mol. Biol. Evol. 2013, 30, 2725–2729. [Google Scholar] [CrossRef] [PubMed]
- Jupe, F.; Pritchard, L.; Etherington, G.J.; Mackenzie, K.; Cock, P.J.A.; Wright, F.; Sharma, S.K.; Bolser, D.; Bryan, G.J.; Jones, J.D.G.; et al. Identification and localisation of the NB-LRR gene family within the potato genome. BMC Genom. 2012, 13, 75. [Google Scholar] [CrossRef] [PubMed]
- Jia, Y.; Yuan, Y.; Zhang, Y.; Yang, S.; Zhang, X. Extreme expansion of NBS-encoding genes in Rosaceae. BMC Genet. 2015, 16, 48. [Google Scholar] [CrossRef] [PubMed]
- Mace, E.; Tai, S.; Innes, D.; Godwin, I.; Hu, W.; Campbell, B.; Gilding, E.; Cruickshank, A.; Prentis, P.; Wang, J.; et al. The plasticity of NBS resistance genes in sorghum is driven by multiple evolutionary processes. BMC Plant Biol. 2014, 14, 253. [Google Scholar] [CrossRef] [PubMed]
- Darzentas, N. Circoletto: visualizing sequence similarity with Circos. Bioinformatics 2010, 26, 2620–2621. [Google Scholar] [CrossRef] [PubMed]
- Die, J.V. BSgenome. Aofficinalis. NCBI.V1: Asparagus Officinalis (Garden Asparagus) Full Genome (NCBI Version Aspof.V1). Bioconductor. 2018. Available online: http://bioconductor.org/packages/release/data/annotation/html/BSgenome.Aofficinalis.NCBI.V1.html (accessed on 21 November 2018).
- Wei, Y.; Chang, Y.; Zeng, H.; Liu, G.; He, C.; Shi, H. RAV transcription factors are essential for disease resistance against cassava bacterial blight via activation of melatonin biosynthesis genes. J. Pineal Res. 2018, 64. [Google Scholar] [CrossRef] [PubMed]
- Conforte, A.J.; Guimarães-Dias, F.; Neves-Borges, A.C.; Bencke-Malato, M.; Felix-Whipps, D.; Alves-Ferreira, M. Isolation and characterization of a promoter responsive to salt, osmotic and dehydration stresses in soybean. Genet. Mol. Biol. 2017, 40, 226–237. [Google Scholar] [CrossRef] [PubMed]
- Liu, H.; Zhong, Y.; Guo, C.; Wang, X.-L.; Xiong, J.; Cheng, Q.; Cheng, Z.-M. Genome-wide analysis and evolution of the bZIP transcription factor gene family in six Fragaria species. Plant Syst. Evol. 2017, 303, 1–13. [Google Scholar] [CrossRef]
- Hernandez-Garcia, C.M.; Finer, J.J. Identification and validation of promoters and cis-acting regulatory elements. Plant Sci. 2014, 217–218, 109–119. [Google Scholar] [CrossRef] [PubMed]
- Porter, B.W.; Paidi, M.; Ming, R.; Alam, M.; Nishijima, W.T.; Zhu, Y.J. Genome-wide analysis of Carica papaya reveals a small NBS resistance gene family. Mol. Genet. Genom. 2009, 281, 609–626. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Xia, R.; Kuang, H.; Meyers, B.C. The diversification of plant NBS-LRR defense genes directs the evolution of microRNAs that target them. Mol. Biol. Evol. 2016, 33, 2692–2705. [Google Scholar] [CrossRef] [PubMed]
- Lai, Y.; Eulgem, T. Transcript-level expression control of plant NLR genes. Mol. Plant Pathol. 2018, 19, 1267–1281. [Google Scholar] [CrossRef] [PubMed]
- Wan, H.; Yuan, W.; Bo, K.; Shen, J.; Pang, X.; Chen, J. Genome-wide analysis of NBS-encoding disease resistance genes in Cucumis sativus and phylogenetic study of NBS-encoding genes in Cucurbitaceae crops. BMC Genom. 2013, 14, 109. [Google Scholar] [CrossRef] [PubMed]
- Ming, R.; Hou, S.; Feng, Y.; Yu, Q.; Dionne-Laporte, A.; Saw, J.H.; Senin, P.; Wang, W.; Ly, B.V.; Lewis, K.L.T.; et al. The draft genome of the transgenic tropical fruit tree papaya (Carica papaya Linnaeus). Nature 2008, 452, 991–996. [Google Scholar] [CrossRef] [PubMed]
- Sagi, M.S.; Deokar, A.A.; Tar’an, B. Genetic analysis of NBS-LRR gene family in chickpea and their expression profiles in response to ascochyta blight infection. Front. Plant Sci. 2017, 8, 838. [Google Scholar] [CrossRef] [PubMed]
- Zhong, Y.; Yin, H.; Sargent, D.J.; Malnoy, M.; Cheng, Z.-M. M. Species-specific duplications driving the recent expansion of NBS-LRR genes in five Rosaceae species. BMC Genomics 2015, 16, 77. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Kapos, P.; Zhang, Y. NLRs in plants. Curr. Opin. Immunol. 2015, 32, 114–121. [Google Scholar] [CrossRef] [PubMed]
- Mondragón-Palomino, M.; Meyers, B.C.; Michelmore, R.W.; Gaut, B.S. Patterns of positive selection in the complete NBS-LRR gene family of Arabidopsis thaliana. Genome Res. 2002, 12, 1305–1315. [Google Scholar] [CrossRef] [PubMed]
- Huang, S.; Ding, J.; Deng, D.; Tang, W.; Sun, H.; Liu, D.; Zhang, L.; Niu, X.; Zhang, X.; Meng, M.; et al. Draft genome of the kiwifruit Actinidia chinensis. Nat. Commun. 2013, 4, 2640. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Zhong, Y.A.N.; Huang, K.; Cheng, Z.-M. Genomewide analysis of NBS-encoding genes in kiwi fruit (Actinidia chinensis). J. Genet. 2016, 95, 997–1001. [Google Scholar] [CrossRef] [PubMed]
- Dangl, J.L.; Jones, J.D. Plant pathogens and integrated defence responses to infection. Nature 2001, 411, 826–833. [Google Scholar] [CrossRef] [PubMed]
- Shao, Z.-Q.; Zhang, Y.-M.; Hang, Y.-Y.; Xue, J.-Y.; Zhou, G.-C.; Wu, P.; Wu, X.-Y.; Wu, X.-Z.; Wang, Q.; Wang, B.; et al. Long-term evolution of nucleotide-binding site-leucine-rich repeat genes: Understanding gained from and beyond the legume family. Plant Physiol. 2014, 166, 217–234. [Google Scholar] [CrossRef] [PubMed]
- Huang, S.; Li, R.; Zhang, Z.; Li, L.; Gu, X.; Fan, W.; Lucas, W.J.; Wang, X.; Xie, B.; Ni, P.; et al. The genome of the cucumber, Cucumis sativus L. Nat. Genet. 2009, 41, 1275–1281. [Google Scholar] [CrossRef] [PubMed]
- Guo, S.; Zhang, J.; Sun, H.; Salse, J.; Lucas, W.J.; Zhang, H.; Zheng, Y.; Mao, L.; Ren, Y.; Wang, Z.; et al. The draft genome of watermelon (Citrullus lanatus) and resequencing of 20 diverse accessions. Nat. Genet. 2013, 45, 51–58. [Google Scholar] [CrossRef] [PubMed]
- Cheng, X.; Jiang, H.; Zhao, Y.; Qian, Y.; Zhu, S.; Cheng, B. A genomic analysis of disease-resistance genes encoding nucleotide binding sites in Sorghum bicolor. Genet. Mol. Biol. 2010, 33, 292–297. [Google Scholar] [CrossRef] [PubMed]
- Dinesh-Kumar, S.P.; Baker, B.J. Alternatively spliced N resistance gene transcripts: Their possible role in tobacco mosaic virus resistance. Proc. Natl. Acad. Sci. USA 2000, 97, 1908–1913. [Google Scholar] [CrossRef] [PubMed]
- Mandadi, K.K.; Scholthof, K.-B.G. Genome-wide analysis of alternative splicing landscapes modulated during plant-virus interactions in Brachypodium distachyon. Plant Cell 2015, 27, 71–85. [Google Scholar] [CrossRef] [PubMed]
- Guo, C.; Sun, X.; Chen, X.; Yang, S.; Li, J.; Wang, L.; Zhang, X. Cloning of novel rice blast resistance genes from two rapidly evolving NBS-LRR gene families in rice. Plant Mol. Biol. 2016, 90, 95–105. [Google Scholar] [CrossRef] [PubMed]
- Marone, D.; Russo, M.A.; Laidò, G.; De Leonardis, A.M.; Mastrangelo, A.M. Plant nucleotide binding site-leucine-rich repeat (NBS-LRR) genes: Active guardians in host defense responses. Int. J. Mol. Sci. 2013, 14, 7302–7326. [Google Scholar] [CrossRef] [PubMed]
- Boratyn, G.M.; Thierry-Mieg, J.; Thierry-Mieg, D.; Busby, B.; Madden, T.L. Magic-BLAST, an accurate DNA and RNA-seq aligner for long and short reads. BioRxiv 2018. [Google Scholar] [CrossRef]
- Wang, L.; Guo, Z.; Zhang, Y.; Wang, Y.; Yang, G.; Yang, L.; Wang, L.; Wang, R.; Xie, Z. Overexpression of LhSorNPR1, a NPR1-like gene from the oriental hybrid lily ’Sorbonne’, conferred enhanced resistance to Pseudomonas syringae pv. tomato DC3000 in Arabidopsis. Physiol. Mol. Biol. Plants 2017, 23, 793–808. [Google Scholar] [CrossRef] [PubMed]
- Zhong, Y.; Li, Y.; Huang, K.; Cheng, Z.-M. Species-specific duplications of NBS-encoding genes in Chinese chestnut (Castanea mollissima). Sci. Rep. 2015, 5, 16638. [Google Scholar] [CrossRef] [PubMed]
- Die, J.V.; Román, B.; Qi, X.; Rowland, L.J. Genome-scale examination of NBS-encoding genes in blueberry. Sci. Rep. 2018, 8, 3429. [Google Scholar] [CrossRef] [PubMed]
- Asano, T.; Hayashi, N.; Kobayashi, M.; Aoki, N.; Miyao, A.; Mitsuhara, I.; Ichikawa, H.; Komatsu, S.; Hirochika, H.; Kikuchi, S.; et al. A rice calcium-dependent protein kinase OsCPK12 oppositely modulates salt-stress tolerance and blast disease resistance. Plant J. 2012, 69, 26–36. [Google Scholar] [CrossRef] [PubMed]
- Wu, J.; Zhang, Y.; Yin, L.; Qu, J.; Lu, J. Linkage of cold acclimation and disease resistance through plant-pathogen interaction pathway in Vitis amurensis grapevine. Funct. Integr. Genom. 2014, 14, 741–755. [Google Scholar] [CrossRef] [PubMed]
- Wu, J.; Folta, K.M.; Xie, Y.; Jiang, W.; Lu, J.; Zhang, Y. Overexpression of Muscadinia rotundifolia CBF2 gene enhances biotic and abiotic stress tolerance in Arabidopsis. Protoplasma 2017, 254, 239–251. [Google Scholar] [CrossRef] [PubMed]
| Predicted Protein Domain | Letter Code | No. of Genes |
|---|---|---|
| NBS-LRR-type genes | ||
| TIR-NBS-LRR subclass | TNL | 0 |
| non-TIR-NBS-LRR subclass | ||
| CC-NBS-LRR | CNL | 29 |
| NBS-LRR | NL | 6 |
| RPW8-NBS-LRR | RNL | 4 |
| XCC-NBS-LRR | XNL | 3 |
| NBS-type genes | ||
| NBS | N | 4 |
| CC-NBS | CN | 2 |
| XCC-NBS | XN | 1 |
| Total genes | 49 |
| Cluster | Cluster Size (kb) | No. of NBS Genes | No. of Genes Neighboring | Chr. | Gene ID | NBS Class |
|---|---|---|---|---|---|---|
| 1 | 31 | 2 | 0 | 2 | AoNBS6, AoNBS7 | CNL |
| 2 | 180.45 | 2 | 7 | 2 | AoNBS9 AoNBS10 | CNL, CN |
| 3 | 3.43 | 2 | 0 | 3 | AoNBS13, AoNBS14 | CNL |
| 4 | 144.67 | 2 | 3 | 4 | AoNBS15, AoNBS16 | RNL |
| 5 | 126.09 | 5 | 6 | 6 | AoNBS22, AoNBS23, AoNBS24, AoNBS25, AoNBS26 | CNL, N |
| 6 | 66.09 | 4 | 2 | 6 | AoNBS27, AoNBS28, AoNBS29, AoNBS30 | CNL, NL |
| 7 | 16.61 | 2 | 2 | 6 | AoNBS31, AoNBS32 | CNL |
| 8 | 36.92 | 3 | 2 | 7 | AoNBS39, AoNBS40, AoNBS41 | CNL, NL |
| 9 | 60.08 | 2 | 1 | 8 | AoNBS43, AoNBS44 | NL, CNL |
| CRE | Motif | Promoters Observed | Occurr. Observed | Avg. Occurrs. per Promoter | Occurrs. Expected | Enrichment Factor | p-Value |
|---|---|---|---|---|---|---|---|
| CBF1/CRT | TGGCCGAC | 3 | 3 | 1 | 0 | ∞ | 0.00150 |
| AtBH5 | CAATNATTG | 12 | 15 | 1.3 | 3 | 5 | 0.00444 |
| SURE1STPAT21 | AATAGAAAA | 7 | 7 | 1 | 2 | 3.5 | 0.00505 |
| ABR | ACGTGTC | 6 | 6 | 1 | 2 | 3 | 0.00774 |
| RAV1-B | CACCTG | 9 | 12 | 1.3 | 6 | 2 | 0.01440 |
| MYCATERD22 | CACATG | 15 | 23 | 1.5 | 12 | 1.9 | 0.01559 |
| WBBOXPCWRKY1 | TTTGACY | 16 | 19 | 1.2 | 11 | 1.7 | 0.01408 |
| GT1GM4SCAM4 | GAAAAA | 30 | 70 | 2.3 | 45 | 1.6 | 0.02178 |
| MYCATERD1 | CATGTG | 12 | 17 | 1.4 | 12 | 1.4 | 0.02038 |
| RAV1AAT | CAACA | 36 | 94 | 2.4 | 66 | 1.4 | 0.02179 |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Die, J.V.; Castro, P.; Millán, T.; Gil, J. Segmental and Tandem Duplications Driving the Recent NBS-LRR Gene Expansion in the Asparagus Genome. Genes 2018, 9, 568. https://doi.org/10.3390/genes9120568
Die JV, Castro P, Millán T, Gil J. Segmental and Tandem Duplications Driving the Recent NBS-LRR Gene Expansion in the Asparagus Genome. Genes. 2018; 9(12):568. https://doi.org/10.3390/genes9120568
Chicago/Turabian StyleDie, Jose V., Patricia Castro, Teresa Millán, and Juan Gil. 2018. "Segmental and Tandem Duplications Driving the Recent NBS-LRR Gene Expansion in the Asparagus Genome" Genes 9, no. 12: 568. https://doi.org/10.3390/genes9120568
APA StyleDie, J. V., Castro, P., Millán, T., & Gil, J. (2018). Segmental and Tandem Duplications Driving the Recent NBS-LRR Gene Expansion in the Asparagus Genome. Genes, 9(12), 568. https://doi.org/10.3390/genes9120568






