A Functional Connection between the Circadian Clock and Hormonal Timing in Arabidopsis
Abstract
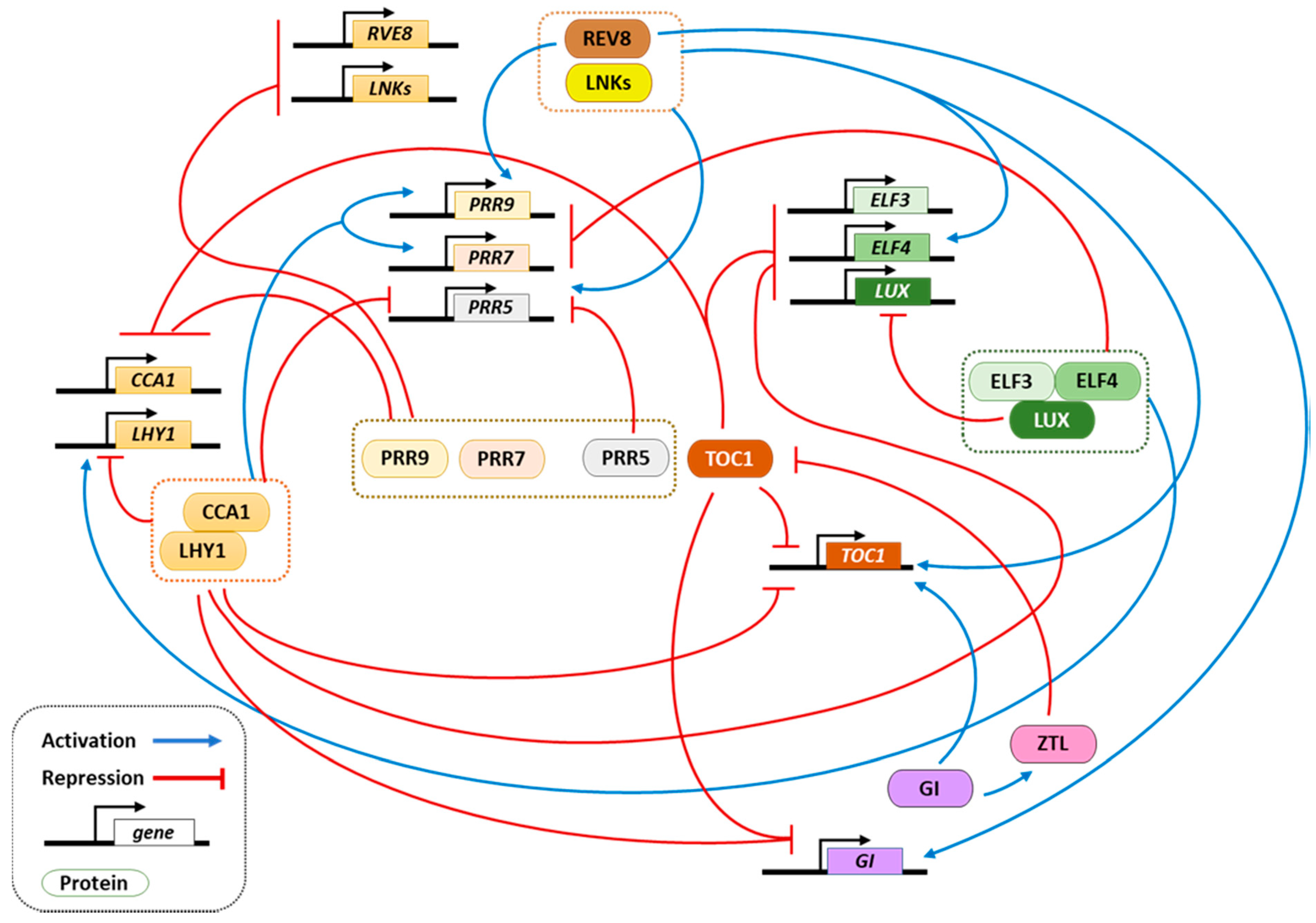
1. Circadian Clock Function in Arabidopsis thaliana
2. Interplay between Auxin and the Circadian Clock
3. Functional Connection of Cytokinins and the Circadian Clock
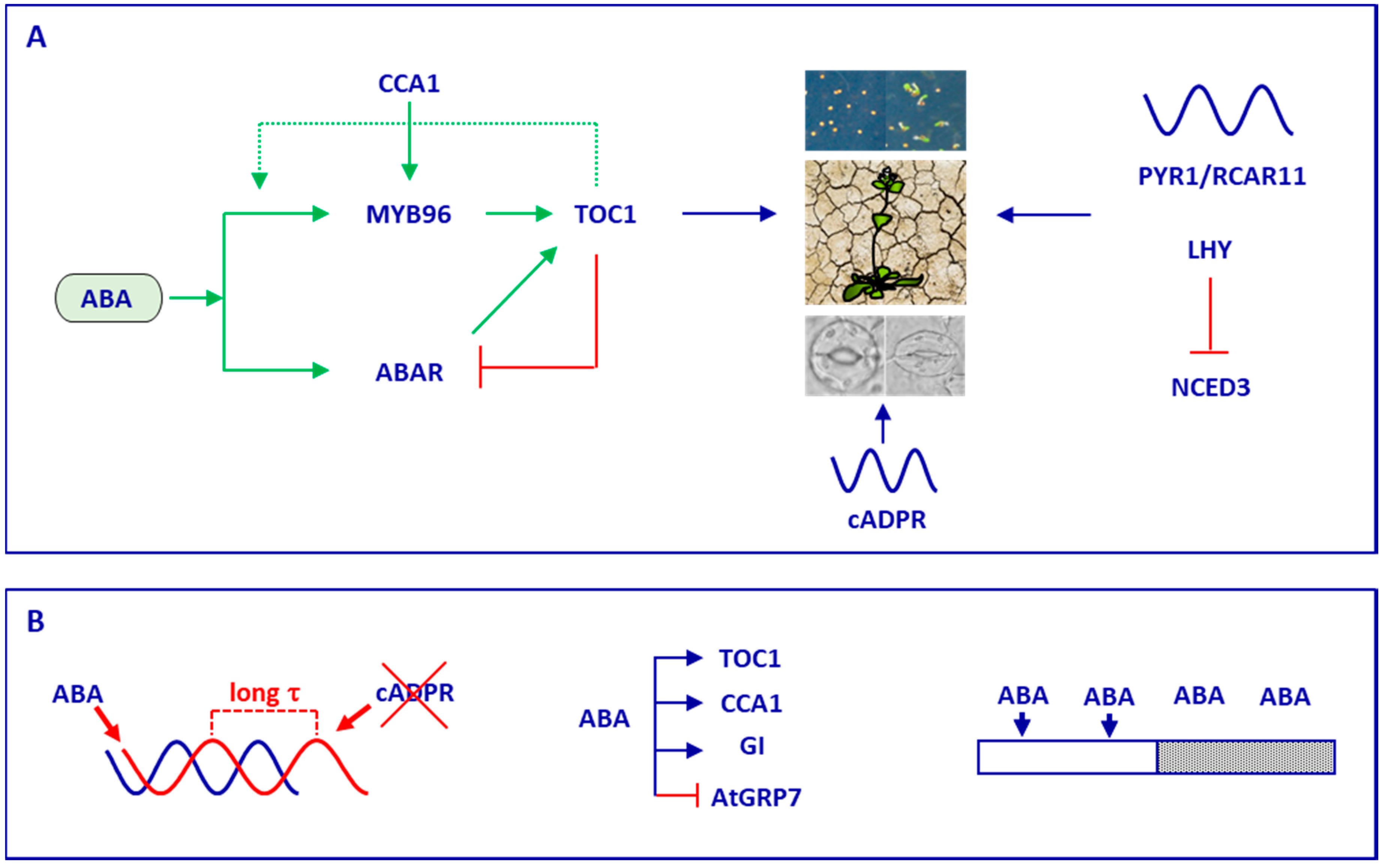
4. Timing by the Circadian Clock Controls Plant Responses to Abscisic Acid
5. Interconnection between Ethylene Signaling and the Circadian Clock
6. Circadian Gating of Gibberellin Signaling by the Clock
7. Perspectives
Funding
Acknowledgments
Conflicts of Interest
References
- Sanchez, S.E.; Kay, S.A. The plant circadian clock: From a simple timekeeper to a complex developmental manager. Cold Spring Harbor Perspect. Biol. 2016. [Google Scholar] [CrossRef] [PubMed]
- Greenham, K.; McClung, C.R. Integrating circadian dynamics with physiological processes in plants. Nat. Rev. Genet. 2015, 16, 598–610. [Google Scholar] [CrossRef] [PubMed]
- McClung, C.R. Beyond Arabidopsis: The circadian clock in non-model plant species. Semin. Cell Dev. Biol. 2013, 24, 430–436. [Google Scholar] [CrossRef] [PubMed]
- Anwer, M.U.; Davis, S.J. An overview of natural variation studies in the Arabidopsis thaliana circadian clock. Semin. Cell Dev. Biol. 2013, 24, 422–429. [Google Scholar] [CrossRef] [PubMed]
- Nagel, D.H.; Kay, S.A. Complexity in the wiring and regulation of plant circadian networks. Curr. Biol. 2012, 22, R648–R657. [Google Scholar] [CrossRef] [PubMed]
- Mas, P.; Yanovsky, M.J. Time for circadian rhythms: Plants get synchronized. Curr. Opin. Plant Biol. 2009, 12, 574–579. [Google Scholar] [CrossRef] [PubMed]
- Millar, A.J. The intracellular dynamics of circadian clocks reach for the light of ecology and evolution. Annu. Rev. Plant Biol. 2016, 67, 595–618. [Google Scholar] [CrossRef] [PubMed]
- Ouyang, Y.; Andersson, C.R.; Kondo, T.; Golden, S.S.; Johnson, C.H. Resonating circadian clocks enhance fitness in Cyanobacteria. Proc. Natl. Acad. Sci. USA 1998, 95, 8660–8664. [Google Scholar] [CrossRef] [PubMed]
- Green, R.M.; Tingay, S.; Wang, Z.-Y.; Tobin, E.M. Circadian rhythms confer a higher level of fitness to Arabidopsis plants. Plant Physiol. 2002, 129, 576–584. [Google Scholar] [CrossRef] [PubMed]
- Michael, T.P.; Salomé, P.A.; Hannah, J.Y.; Spencer, T.R.; Sharp, E.L.; McPeek, M.A.; Alonso, J.M.; Ecker, J.R.; McClung, C.R. Enhanced fitness conferred by naturally occurring variation in the circadian clock. Science 2003, 302, 1049–1053. [Google Scholar] [CrossRef] [PubMed]
- Dodd, A.N.; Salathia, N.; Hall, A.; Kévei, E.; Tóth, R.; Nagy, F.; Hibberd, J.M.; Millar, A.J.; Webb, A.A. Plant circadian clocks increase photosynthesis, growth, survival, and competitive advantage. Science 2005, 309, 630–633. [Google Scholar] [CrossRef] [PubMed]
- Mas, P. Circadian clock function in Arabidopsis thaliana: Time beyond transcription. Trends Cell Biol. 2008, 18, 273–281. [Google Scholar] [CrossRef] [PubMed]
- Kinmonth-Schultz, H.A.; Golembeski, G.S.; Imaizumi, T. Circadian clock-regulated physiological outputs: Dynamic responses in nature. Semin. Cell Dev. Biol. 2013, 24, 407–413. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.-Y.; Tobin, E.M. Constitutive expression of the CIRCADIAN CLOCK ASSOCIATED 1 (CCA1) gene disrupts circadian rhythms and suppresses its own expression. Cell 1998, 93, 1207–1217. [Google Scholar] [CrossRef]
- Schaffer, R.; Ramsay, N.; Samach, A.; Corden, S.; Putterill, J.; Carré, I.A.; Coupland, G. The late elongated hypocotyl mutation of Arabidopsis disrupts circadian rhythms and the photoperiodic control of flowering. Cell 1998, 93, 1219–1229. [Google Scholar] [CrossRef]
- Strayer, C.; Oyama, T.; Schultz, T.F.; Raman, R.; Somers, D.E.; Más, P.; Panda, S.; Kreps, J.A.; Kay, S.A. Cloning of the Arabidopsis clock gene TOC1, an autoregulatory response regulator homolog. Science 2000, 289, 768–771. [Google Scholar] [CrossRef] [PubMed]
- Alabadí, D.; Oyama, T.; Yanovsky, M.J.; Harmon, F.G.; Más, P.; Kay, S.A. Reciprocal regulation between TOC1 and LHY/CCA1 within the Arabidopsis circadian clock. Science 2001, 293, 880–883. [Google Scholar] [CrossRef] [PubMed]
- Gendron, J.M.; Pruneda-Paz, J.L.; Doherty, C.J.; Gross, A.M.; Kang, S.E.; Kay, S.A. Arabidopsis circadian clock protein, TOC1, is a DNA-binding transcription factor. Proc. Natl. Acad. Sci. USA 2012, 109, 3167–3172. [Google Scholar] [CrossRef] [PubMed]
- Pokhilko, A.; Fernández, A.P.; Edwards, K.D.; Southern, M.M.; Halliday, K.J.; Millar, A.J. The clock gene circuit in Arabidopsis includes a repressilator with additional feedback loops. Mol. Syst. Biol. 2012, 8, 574. [Google Scholar] [CrossRef] [PubMed]
- Huang, W.; Pérez-García, P.; Pokhilko, A.; Millar, A.; Antoshechkin, I.; Riechmann, J.L.; Mas, P. Mapping the core of the Arabidopsis circadian clock defines the network structure of the oscillator. Science 2012, 336, 75–79. [Google Scholar] [CrossRef] [PubMed]
- Nakamichi, N.; Kiba, T.; Henriques, R.; Mizuno, T.; Chua, N.-H.; Sakakibara, H. PSEUDO-RESPONSE REGULATORS 9, 7, and 5 are transcriptional repressors in the Arabidopsis circadian clock. Plant Cell 2010, 22, 594–605. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Kim, J.; Somers, D.E. Transcriptional corepressor TOPLESS complexes with pseudoresponse regulator proteins and histone deacetylases to regulate circadian transcription. Proc. Natl. Acad. Sci. USA 2013, 110, 761–766. [Google Scholar] [CrossRef] [PubMed]
- Helfer, A.; Nusinow, D.A.; Chow, B.Y.; Gehrke, A.R.; Bulyk, M.L.; Kay, S.A. LUX ARRHYTHMO encodes a nighttime repressor of circadian gene expression in the Arabidopsis core clock. Curr. Biol. 2011, 21, 126–133. [Google Scholar] [CrossRef] [PubMed]
- Nusinow, D.A.; Helfer, A.; Hamilton, E.E.; King, J.J.; Imaizumi, T.; Schultz, T.F.; Farré, E.M.; Kay, S.A. The ELF4–ELF3–LUX complex links the circadian clock to diurnal control of hypocotyl growth. Nature 2011, 475, 398–402. [Google Scholar] [CrossRef] [PubMed]
- Locke, J.C.; Kozma-Bognár, L.; Gould, P.D.; Fehér, B.; Kevei, E.; Nagy, F.; Turner, M.S.; Hall, A.; Millar, A.J. Experimental validation of a predicted feedback loop in the multi-oscillator clock of Arabidopsis thaliana. Mol. Syst. Biol. 2006, 2, 59. [Google Scholar] [CrossRef] [PubMed]
- Más, P.; Kim, W.-Y.; Somers, D.E.; Kay, S.A. Targeted degradation of TOC1 by ZTL modulates circadian function in Arabidopsis thaliana. Nature 2003, 426, 567–570. [Google Scholar] [CrossRef] [PubMed]
- Kiba, T.; Henriques, R.; Sakakibara, H.; Chua, N.-H. Targeted degradation of PSEUDO-RESPONSE REGULATOR5 by an SCFZTL complex regulates clock function and photomorphogenesis in Arabidopsis thaliana. Plant Cell 2007, 19, 2516–2530. [Google Scholar] [CrossRef] [PubMed]
- Kim, W.-Y.; Fujiwara, S.; Suh, S.-S.; Kim, J.; Kim, Y.; Han, L.; David, K.; Putterill, J.; Nam, H.G.; Somers, D.E. ZEITLUPE is a circadian photoreceptor stabilized by GIGANTEA in blue light. Nature 2007, 449, 356–360. [Google Scholar] [CrossRef] [PubMed]
- Perales, M.; Mas, P. A functional link between rhythmic changes in chromatin structure and the Arabidopsis biological clock. Plant Cell. 2007, 19, 2111–2123. [Google Scholar] [CrossRef] [PubMed]
- Farinas, B.; Mas, P. Functional implication of the MYB transcription factor RVE8/LCL5 in the circadian control of histone acetylation. Plant J. 2011, 66, 318–329. [Google Scholar] [CrossRef] [PubMed]
- Rawat, R.; Takahashi, N.; Hsu, P.Y.; Jones, M.A.; Schwartz, J.; Salemi, M.R.; Phinney, B.S.; Harmer, S.L. REVEILLE8 and PSEUDO-REPONSE REGULATOR5 form a negative feedback loop within the Arabidopsis circadian clock. PLoS Genet. 2011, 7, e1001350. [Google Scholar] [CrossRef] [PubMed]
- Hsu, P.Y.; Devisetty, U.K.; Harmer, S.L. Accurate timekeeping is controlled by a cycling activator in Arabidopsis. Elife 2013, 2, e00473. [Google Scholar] [CrossRef] [PubMed]
- Xie, Q.; Wang, P.; Liu, X.; Yuan, L.; Wang, L.; Zhang, C.; Li, Y.; Xing, H.; Zhi, L.; Yue, Z. LNK1 and LNK2 are transcriptional coactivators in the Arabidopsis circadian oscillator. Plant Cell 2014, 2843–2857. [Google Scholar] [CrossRef] [PubMed]
- Liu, T.; Newton, L.; Liu, M.-J.; Shiu, S.-H.; Farré, E.M. A G-box-like motif is necessary for transcriptional regulation by circadian pseudo-response regulators in Arabidopsis. Plant Physiol. 2015, 170, 528–539. [Google Scholar] [CrossRef] [PubMed]
- Mizuno, T.; Kitayama, M.; Oka, H.; Tsubouchi, M.; Takayama, C.; Nomoto, Y.; Yamashino, T. The EC night-time repressor plays a crucial role in modulating circadian clock transcriptional circuitry by conservatively double-checking both warm-night and night-time-light signals in a synergistic manner in Arabidopsis thaliana. Plant Cell Physiol. 2014, 55, 2139–2151. [Google Scholar] [CrossRef] [PubMed]
- Rugnone, M.L.; Soverna, A.F.; Sanchez, S.E.; Schlaen, R.G.; Hernando, C.E.; Seymour, D.K.; Mancini, E.; Chernomoretz, A.; Weigel, D.; Más, P. LNK genes integrate light and clock signaling networks at the core of the Arabidopsis oscillator. Proc. Natl. Acad. Sci. USA 2013, 110, 12120–12125. [Google Scholar] [CrossRef] [PubMed]
- Mizuno, T.; Takeuchi, A.; Nomoto, Y.; Nakamichi, N.; Yamashino, T. The LNK1 night light-inducible and clock-regulated gene is induced also in response to warm-night through the circadian clock nighttime repressor in Arabidopsis thaliana. Plant Signal. Behav. 2014, 9, e28505. [Google Scholar] [CrossRef] [PubMed]
- Ma, Y.; Gil, S.; Grasser, K.D.; Mas, P. Targeted recruitment of the basal transcriptional machinery by LNK clock components controls the circadian rhythms of nascent RNAs in Arabidopsis. Plant Cell 2018, 30, 907–924. [Google Scholar] [CrossRef] [PubMed]
- Woodward, A.W.; Bartel, B. Auxin: Regulation, action, and interaction. Ann. Bot. 2005, 95, 707–735. [Google Scholar] [CrossRef] [PubMed]
- Davies, P.J. Plant Hormones: Physiology, Biochemistry and Molecular Biology; Springer Science & Business Media: Berlin, Germany, 2013. [Google Scholar]
- Paponov, I.A.; Paponov, M.; Teale, W.; Menges, M.; Chakrabortee, S.; Murray, J.A.H.; Palme, K. Comprehensive transcriptome analysis of auxin responses in Arabidopsis. Mol. Plant 2008, 1, 321–337. [Google Scholar] [CrossRef] [PubMed]
- Leyser, O. Auxin signaling. Plant Physiol. 2018, 176, 465–479. [Google Scholar] [CrossRef] [PubMed]
- Estelle, M.; Weijers, D.; Ljung, K.; Leyser, O. Auxin Signaling: From Synthesis to Systems Biology; Cold Spring Harbor Laboratory Press: New York, NY, USA, 2011; p. 245. [Google Scholar]
- Tiwari, S.B.; Hagen, G.; Guilfoyle, T.J. Aux/IAA proteins contain a potent transcriptional repression domain. Plant Cell 2004, 16, 533–543. [Google Scholar] [CrossRef] [PubMed]
- Szemenyei, H.; Hannon, M.; Long, J.A. TOPLESS mediates auxin-dependent transcriptional repression during Arabidopsis embryogenesis. Science 2008, 319, 1384–1386. [Google Scholar] [CrossRef] [PubMed]
- Shen-Miller, J. Rhythmicity in the basipetal transport of indoleacetic acid through coleoptiles. Plant Physiol. 1973, 51, 615–619. [Google Scholar] [CrossRef] [PubMed]
- Jouve, L.; Gaspar, T.; Kevers, C.; Greppin, H.; Degli Agosti, R. Involvement of indole-3-acetic acid in the circadian growth of the first internode of Arabidopsis. Planta 1999, 209, 136–142. [Google Scholar] [CrossRef] [PubMed]
- Covington, M.F.; Harmer, S.L. The circadian clock regulates auxin signaling and responses in Arabidopsis. PLoS Biol. 2007, 5, e222. [Google Scholar] [CrossRef] [PubMed]
- Rawat, R.; Schwartz, J.; Jones, M.A.; Sairanen, I.; Cheng, Y.; Andersson, C.R.; Zhao, Y.; Ljung, K.; Harmer, S.L. REVEILLE1, a Myb-like transcription factor, integrates the circadian clock and auxin pathways. Proc. Natl. Acad. Sci. USA 2009, 106, 16883–16888. [Google Scholar] [CrossRef] [PubMed]
- Voß, U.; Wilson, M.H.; Kenobi, K.; Gould, P.D.; Robertson, F.C.; Peer, W.A.; Lucas, M.; Swarup, K.; Casimiro, I.; Holman, T.J. The circadian clock rephases during lateral root organ initiation in Arabidopsis thaliana. Nat. Commun. 2015, 6, 7641. [Google Scholar] [CrossRef] [PubMed]
- Hong, L.-W.; Yan, D.-W.; Liu, W.-C.; Chen, H.-G.; Lu, Y.-T. TIME FOR COFFEE controls root meristem size by changes in auxin accumulation in Arabidopsis. J. Exp. Bot. 2013, 65, 275–286. [Google Scholar] [CrossRef] [PubMed]
- Zha, P.; Jing, Y.; Xu, G.; Lin, R. PICKLE chromatin-remodeling factor controls thermosensory hypocotyl growth of Arabidopsis. Plant Cell Environ. 2017, 40, 2426–2436. [Google Scholar] [CrossRef] [PubMed]
- Meyer, K.; Köster, T.; Nolte, C.; Weinholdt, C.; Lewinski, M.; Grosse, I.; Staiger, D. Adaptation of iCLIP to plants determines the binding landscape of the clock-regulated RNA-binding protein AtGRP7. Genome Biol. 2017, 18, 204. [Google Scholar] [CrossRef] [PubMed]
- Hyunmo, C.; Eunkyoo, O. PIF4 integrates multiple environmental and hormonal signals for plant growth regulation in Arabidopsis. Mol. Cells 2016, 39, 587–593. [Google Scholar]
- Nozue, K.; Harmer, S.L.; Maloof, J.N. Genomic analysis of circadian clock-, light-, and growth-correlated genes reveals PHYTOCHROME-INTERACTING FACTOR5 as a modulator of auxin signaling in Arabidopsis. Plant Physiol. 2011, 156, 357–372. [Google Scholar] [CrossRef] [PubMed]
- Nomoto, Y.; Kubozono, S.; Yamashino, T.; Nakamichi, N.; Mizuno, T. Circadian clock-and PIF4-controlled plant growth: A coincidence mechanism directly integrates a hormone signaling network into the photoperiodic control of plant architectures in Arabidopsis thaliana. Plant Cell Physiol. 2012, 53, 1950–1964. [Google Scholar] [CrossRef] [PubMed]
- Kunihiro, A.; Yamashino, T.; Nakamichi, N.; Niwa, Y.; Nakanishi, H.; Mizuno, T. Phytochrome-interacting factor 4 and 5 (PIF4 and PIF5) activate the homeobox ATHB2 and auxin-inducible IAA29 genes in the coincidence mechanism underlying photoperiodic control of plant growth of Arabidopsis thaliana. Plant Cell Physiol. 2011, 52, 1315–1329. [Google Scholar] [CrossRef] [PubMed]
- Zhu, J.-Y.; Oh, E.; Wang, T.; Wang, Z.-Y. TOC1–PIF4 interaction mediates the circadian gating of thermoresponsive growth in Arabidopsis. Nat. Commun. 2016, 7, 13692. [Google Scholar] [CrossRef] [PubMed]
- Raschke, A.; Ibañez, C.; Ullrich, K.K.; Anwer, M.U.; Becker, S.; Glöckner, A.; Trenner, J.; Denk, K.; Saal, B.; Sun, X. Natural variants of ELF3 affect thermomorphogenesis by transcriptionally modulating PIF4-dependent auxin response genes. BMC Plant Biol. 2015, 15, 197. [Google Scholar] [CrossRef] [PubMed]
- Hanano, S.; Domagalska, M.A.; Nagy, F.; Davis, S.J. Multiple phytohormones influence distinct parameters of the plant circadian clock. Genes Cells 2006, 11, 1381–1392. [Google Scholar] [CrossRef] [PubMed]
- Hearn, T.J.; Marti, M.C.; Abdul-Awal, S.; Wimalasekera, R.; Stanton, C.R.; Haydon, M.J.; Theodoulou, F.L.; Hannah, M.A.; Webb, A.A. BIG regulates dynamic adjustment of circadian period in Arabidopsis thaliana. Plant Physiol. 2018, 178, 358–371. [Google Scholar] [CrossRef] [PubMed]
- Gil, P.; Dewey, E.; Friml, J.; Zhao, Y.; Snowden, K.C.; Putterill, J.; Palme, K.; Estelle, M.; Chory, J. BIG: A calossin-like protein required for polar auxin transport in Arabidopsis. Genes Dev. 2001, 15, 1985–1997. [Google Scholar] [CrossRef] [PubMed]
- Kieber, J.J.; Schaller, G.E. Cytokinins. In The Arabidopsis Book; American Society of Plant Biologists: Rockville, MD, USA, 2014; p. 12. [Google Scholar]
- Ha, S.; Vankova, R.; Yamaguchi-Shinozaki, K.; Shinozaki, K.; Tran, L.-S.P. Cytokinins: Metabolism and function in plant adaptation to environmental stresses. Trends Plant Sci. 2012, 17, 172–179. [Google Scholar] [CrossRef] [PubMed]
- Kakimoto, T. Biosynthesis of cytokinins. J. Plant Res. 2003, 116, 233–239. [Google Scholar] [CrossRef] [PubMed]
- Werner, T.; Köllmer, I.; Bartrina, I.; Holst, K.; Schmülling, T. New insights into the biology of cytokinin degradation. Plant Biol. 2006, 8, 371–381. [Google Scholar] [CrossRef] [PubMed]
- Inoue, T.; Higuchi, M.; Hashimoto, Y.; Seki, M.; Kobayashi, M.; Kato, T.; Tabata, S.; Shinozaki, K.; Kakimoto, T. Identification of CRE1 as a cytokinin receptor from Arabidopsis. Nature 2001, 409, 1060–1063. [Google Scholar] [CrossRef] [PubMed]
- Suzuki, T.; Miwa, K.; Ishikawa, K.; Yamada, H.; Aiba, H.; Mizuno, T. The Arabidopsis sensor His-kinase, AHK4, can respond to cytokinins. Plant Cell Physiol. 2001, 42, 107–113. [Google Scholar] [CrossRef] [PubMed]
- Heyl, A.; Riefler, M.; Romanov, G.A.; Schmülling, T. Properties, functions and evolution of cytokinin receptors. Eur. J. Cell Biol. 2012, 91, 246–256. [Google Scholar] [CrossRef] [PubMed]
- Heyl, A.; Schmülling, T. Cytokinin signal perception and transduction. Curr. Opin. Plant Biol. 2003, 6, 480–488. [Google Scholar] [CrossRef]
- Dortay, H.; Mehnert, N.; Bürkle, L.; Schmülling, T.; Heyl, A. Analysis of protein interactions within the cytokinin-signaling pathway of Arabidopsis thaliana. FEBS J. 2006, 273, 4631–4644. [Google Scholar] [CrossRef] [PubMed]
- Covington, M.F.; Maloof, J.N.; Straume, M.; Kay, S.A.; Harmer, S.L. Global transcriptome analysis reveals circadian regulation of key pathways in plant growth and development. Genome Biol. 2008, 9, R130. [Google Scholar] [CrossRef] [PubMed]
- Zheng, B.; Deng, Y.; Mu, J.; Ji, Z.; Xiang, T.; Niu, Q.W.; Chua, N.H.; Zuo, J. Cytokinin affects circadian-clock oscillation in a phytochrome B-and Arabidopsis response regulator 4-dependent manner. Physiol. Plant. 2006, 127, 277–292. [Google Scholar] [CrossRef]
- Nováková, M.; Motyka, V.; Dobrev, P.I.; Malbeck, J.; Gaudinová, A.; Vanková, R. Diurnal variation of cytokinin, auxin and abscisic acid levels in tobacco leaves. J. Exp. Bot. 2005, 56, 2877–2883. [Google Scholar] [CrossRef] [PubMed]
- Ezer, D.; Jung, J.-H.; Lan, H.; Biswas, S.; Gregoire, L.; Box, M.S.; Charoensawan, V.; Cortijo, S.; Lai, X.; Stöckle, D. The evening complex coordinates environmental and endogenous signals in Arabidopsis. Nat. Plants 2017, 3, 17087. [Google Scholar] [CrossRef] [PubMed]
- Ishida, K.; Yamashino, T.; Mizuno, T. Expression of the cytokinin-induced type-A response regulator gene ARR9 is regulated by the circadian clock in Arabidopsis thaliana. Biosci. Biotechnol. Biochem. 2008, 72, 3025–3029. [Google Scholar] [CrossRef] [PubMed]
- Putarjunan, A.; Rodermel, S. Gigantea suppresses immutans variegation by interactions with cytokinin and GA signaling pathways. Plant Physiol. 2014, 166, 2115–2132. [Google Scholar] [CrossRef] [PubMed]
- Salomé, P.A.; To, J.P.; Kieber, J.J.; McClung, C.R. Arabidopsis response regulators ARR3 and ARR4 play cytokinin-independent roles in the control of circadian period. Plant Cell 2006, 18, 55–69. [Google Scholar] [CrossRef] [PubMed]
- Nitschke, S.; Cortleven, A.; Iven, T.; Feussner, I.; Havaux, M.; Riefler, M.; Schmülling, T. Circadian stress regimes affect the circadian clock and cause jasmonic acid-dependent cell death in cytokinin-deficient Arabidopsis plants. Plant Cell 2016, 28, 1616–1639. [Google Scholar] [CrossRef] [PubMed]
- Salomé, P.A.; Michael, T.P.; Kearns, E.V.; Fett-Neto, A.G.; Sharrock, R.A.; McClung, C.R. The out of phase 1 mutant defines a role for PHYB in circadian phase control in Arabidopsis. Plant Physiol. 2002, 129, 1674–1685. [Google Scholar] [CrossRef] [PubMed]
- Leung, J.; Giraudat, J. Abscisic acid signal transduction. Annu. Rev. Plant Biol. 1998, 49, 199–222. [Google Scholar] [CrossRef] [PubMed]
- Finkelstein, R.R.; Gampala, S.S.; Rock, C.D. Abscisic acid signaling in seeds and seedlings. Plant Cell 2002, 14, S15–S45. [Google Scholar] [CrossRef] [PubMed]
- Thompson, A.J.; Mulholland, B.J.; Jackson, A.C.; McKee, J.M.; Hilton, H.W.; Symonds, R.C.; Sonneveld, T.; Burbidge, A.; Stevenson, P.; Taylor, I.B. Regulation and manipulation of ABA biosynthesis in roots. Plant Cell Environ. 2007, 30, 67–78. [Google Scholar] [CrossRef] [PubMed]
- Verslues, P.E.; Zhu, J.-K. New developments in abscisic acid perception and metabolism. Curr. Opin. Plant Biol. 2007, 10, 447–452. [Google Scholar] [CrossRef] [PubMed]
- Guo, J.; Yang, X.; Weston, D.J.; Chen, J.G. Abscisic acid receptors: Past, present and future F. J. Integr. Plant Biol. 2011, 53, 469–479. [Google Scholar] [CrossRef] [PubMed]
- Hirayama, T.; Shinozaki, K. Perception and transduction of abscisic acid signals: Keys to the function of the versatile plant hormone ABA. Trends Plant Sci. 2007, 12, 343–351. [Google Scholar] [CrossRef] [PubMed]
- Park, S.-Y.; Fung, P.; Nishimura, N.; Jensen, D.R.; Fujii, H.; Zhao, Y.; Lumba, S.; Santiago, J.; Rodrigues, A.; Tsz-fung, F.C. Abscisic acid inhibits type 2C protein phosphatases via the PYR/PYL family of START proteins. Science 2009, 324, 1068–1071. [Google Scholar] [CrossRef] [PubMed]
- Ma, Y.; Szostkiewicz, I.; Korte, A.; Moes, D.; Yang, Y.; Christmann, A.; Grill, E. Regulators of PP2C phosphatase activity function as abscisic acid sensors. Science 2009, 324, 1064–1068. [Google Scholar] [CrossRef] [PubMed]
- Kobayashi, Y.; Murata, M.; Minami, H.; Yamamoto, S.; Kagaya, Y.; Hobo, T.; Yamamoto, A.; Hattori, T. Abscisic acid-activated SNRK2 protein kinases function in the gene-regulation pathway of ABA signal transduction by phosphorylating ABA response element-binding factors. Plant J. 2005, 44, 939–949. [Google Scholar] [CrossRef] [PubMed]
- Hubbard, K.E.; Nishimura, N.; Hitomi, K.; Getzoff, E.D.; Schroeder, J.I. Early abscisic acid signal transduction mechanisms: Newly discovered components and newly emerging questions. Genes Dev. 2010, 24, 1695–1708. [Google Scholar] [CrossRef] [PubMed]
- Mizuno, T.; Yamashino, T. Comparative transcriptome of diurnally oscillating genes and hormone-responsive genes in Arabidopsis thaliana: Insight into circadian clock-controlled daily responses to common ambient stresses in plants. Plant Cell Physiol. 2008, 49, 481–487. [Google Scholar] [CrossRef] [PubMed]
- Adams, S.; Grundy, J.; Veflingstad, S.R.; Dyer, N.P.; Hannah, M.A.; Ott, S.; Carré, I.A. Circadian control of abscisic acid biosynthesis and signalling pathways revealed by genome-wide analysis of LHY binding targets. New Phytol. 2018, 220, 893–907. [Google Scholar] [CrossRef] [PubMed]
- Choudhary, M.K.; Nomura, Y.; Wang, L.; Nakagami, H.; Somers, D.E. Quantitative circadian phosphoproteomic analysis of Arabidopsis reveals extensive clock control of key components in physiological, metabolic and signaling pathways. Mol. Cell. Proteom. 2015, 14, 2243–2260. [Google Scholar] [CrossRef] [PubMed]
- Greenham, K.; Guadagno, C.R.; Gehan, M.A.; Mockler, T.C.; Weinig, C.; Ewers, B.E.; McClung, C.R. Temporal network analysis identifies early physiological and transcriptomic indicators of mild drought in Brassica rapa. eLife 2017, 6, e29655. [Google Scholar] [CrossRef] [PubMed]
- Legnaioli, T.; Cuevas, J.; Mas, P. TOC1 functions as a molecular switch connecting the circadian clock with plant responses to drought. EMBO J. 2009, 28, 3745–3757. [Google Scholar] [CrossRef] [PubMed]
- Robertson, F.C.; Skeffington, A.W.; Gardner, M.J.; Webb, A.A. Interactions between circadian and hormonal signalling in plants. Plant Mol. Biol. 2009, 69, 419. [Google Scholar] [CrossRef] [PubMed]
- Kurup, S.; Jones, H.D.; Holdsworth, M.J. Interactions of the developmental regulator ABI3 with proteins identified from developing Arabidopsis seeds. Plant J. 2000, 21, 143–155. [Google Scholar] [CrossRef] [PubMed]
- Lee, H.G.; Mas, P.; Seo, P.J. MYB96 shapes the circadian gating of ABA signaling in Arabidopsis. Sci. Rep. 2016, 6, 17754. [Google Scholar] [CrossRef] [PubMed]
- Dodd, A.N.; Gardner, M.J.; Hotta, C.T.; Hubbard, K.E.; Dalchau, N.; Love, J.; Assie, J.-M.; Robertson, F.C.; Jakobsen, M.K.; Gonçalves, J. The Arabidopsis circadian clock incorporates a cADPR-based feedback loop. Science 2007, 318, 1789–1792. [Google Scholar] [CrossRef] [PubMed]
- Sánchez, J.P.; Duque, P.; Chua, N.H. ABA activates ADPR cyclase and cADPR induces a subset of ABA-responsive genes in Arabidopsis. Plant J. 2004, 38, 381–395. [Google Scholar] [CrossRef] [PubMed]
- Fukushima, A.; Kusano, M.; Nakamichi, N.; Kobayashi, M.; Hayashi, N.; Sakakibara, H.; Mizuno, T.; Saito, K. Impact of clock-associated Arabidopsis pseudo-response regulators in metabolic coordination. Proc. Natl. Acad. Sci. USA 2009, 106, 7251–7256. [Google Scholar] [CrossRef] [PubMed]
- Sanchez-Villarreal, A.; Shin, J.; Bujdoso, N.; Obata, T.; Neumann, U.; Du, S.X.; Ding, Z.; Davis, A.M.; Shindo, T.; Schmelzer, E. TIME FOR COFFEE is an essential component in the maintenance of metabolic homeostasis in Arabidopsis thaliana. Plant J. 2013, 76, 188–200. [Google Scholar] [PubMed]
- Mir, R.; Hernández, M.L.; Abou-Mansour, E.; Martínez-Rivas, J.M.; Mauch, F.; Métraux, J.-P.; León, J. Pathogen and Circadian Controlled 1 (PCC1) regulates polar lipid content, ABA-related responses, and pathogen defence in Arabidopsis thaliana. J. Exp. Bot. 2013, 64, 3385–3395. [Google Scholar] [CrossRef] [PubMed]
- Footitt, S.; Ölçer-Footitt, H.; Hambidge, A.J.; Finch-Savage, W.E. A laboratory simulation of Arabidopsis seed dormancy cycling provides new insight into its regulation by clock genes and the dormancy-related genes DOG1, MFT, CIPK23 and PHYA. Plant Cell Environ. 2017, 40, 1474–1486. [Google Scholar] [CrossRef] [PubMed]
- Penfield, S.; Hall, A. A role for multiple circadian clock genes in the response to signals that break seed dormancy in Arabidopsis. Plant Cell 2009, 21, 1722–1732. [Google Scholar] [CrossRef] [PubMed]
- Cao, S.; Jiang, L.; Song, S.; Jing, R.; Xu, G. AtGRP7 is involved in the regulation of abscisic acid and stress responses in Arabidopsis. Cell. Mol. Biol. Lett. 2006, 11, 526. [Google Scholar] [CrossRef] [PubMed]
- Riboni, M.; Robustelli Test, A.; Galbiati, M.; Tonelli, C.; Conti, L. ABA-dependent control of GIGANTEA signalling enables drought escape via up-regulation of FLOWERING LOCUS T in Arabidopsis thaliana. J. Exp. Bot. 2016, 67, 6309–6322. [Google Scholar] [CrossRef] [PubMed]
- Broekgaarden, C.; Caarls, L.; Vos, I.A.; Pieterse, C.M.; Van Wees, S.C. Ethylene: Traffic controller on hormonal crossroads to defense. Plant Physiol. 2015, 169, 2371–2379. [Google Scholar] [CrossRef] [PubMed]
- Broekaert, W.F.; Delauré, S.L.; De Bolle, M.F.; Cammue, B.P. The role of ethylene in host-pathogen interactions. Annu. Rev. Phytopathol. 2006, 44, 393–416. [Google Scholar] [CrossRef] [PubMed]
- Dubois, M.; Van den Broeck, L.; Inzé, D. The pivotal role of ethylene in plant growth. Trends Plant Sci. 2018, 23, 311–323. [Google Scholar] [CrossRef] [PubMed]
- Barry, C.S.; Giovannoni, J.J. Ethylene and fruit ripening. J. Plant Growth Regul. 2007, 26, 143. [Google Scholar] [CrossRef]
- Zhong, S.; Shi, H.; Xue, C.; Wang, L.; Xi, Y.; Li, J.; Quail, P.H.; Deng, X.W.; Guo, H. A molecular framework of light-controlled phytohormone action in Arabidopsis. Curr. Biol. 2012, 22, 1530–1535. [Google Scholar] [CrossRef] [PubMed]
- García, M.J.; Romera, F.J.; Lucena, C.; Alcántara, E.; Pérez-Vicente, R. Ethylene and the regulation of physiological and morphological responses to nutrient deficiencies. Plant Physiol. 2015, 169, 51–60. [Google Scholar] [CrossRef] [PubMed]
- Clark, K.L.; Larsen, P.B.; Wang, X.; Chang, C. Association of the Arabidopsis CTR1 Raf-like kinase with the ETR1 and ERS ethylene receptors. Proc. Natl. Acad. Sci. USA 1998, 95, 5401–5406. [Google Scholar] [CrossRef] [PubMed]
- Shakeel, S.; Gao, Z.; Amir, M.; Chen, Y.-F.; Rai, M.I.; Haq, N.U.; Schaller, G.E. Ethylene regulates levels of ethylene-receptor/CTR1 signaling complexes in Arabidopsis thaliana. J. Biol. Chem. 2015, 290, 12415–12424. [Google Scholar] [CrossRef] [PubMed]
- Potuschak, T.; Lechner, E.; Parmentier, Y.; Yanagisawa, S.; Grava, S.; Koncz, C.; Genschik, P. EIN3-dependent regulation of plant ethylene hormone signaling by two Arabidopsis F box proteins: EBF1 and EBF2. Cell 2003, 115, 679–689. [Google Scholar] [CrossRef]
- Guo, H.; Ecker, J.R. Plant responses to ethylene gas are mediated by SCFEBF1/EBF2 dependent proteolysis of EIN3 transcription factor. Cell 2003, 115, 667–677. [Google Scholar] [CrossRef]
- Thain, S.C.; Vandenbussche, F.; Laarhoven, L.J.; Dowson-Day, M.J.; Wang, Z.-Y.; Tobin, E.M.; Harren, F.J.; Millar, A.J.; Van Der Straeten, D. Circadian rhythms of ethylene emission in Arabidopsis. Plant Physiol. 2004, 136, 3751–3761. [Google Scholar] [CrossRef] [PubMed]
- Ellison, C.T.; Vandenbussche, F.; Van Der Straeten, D.; Harmer, S.L. XAP5 CIRCADIAN TIMEKEEPER regulates ethylene responses in aerial tissues of Arabidopsis. Plant Physiol. 2010, 155, 988–999. [Google Scholar] [CrossRef] [PubMed]
- Xu, Y.-J.; Lei, Y.; Li, R.; Zhang, L.-L.; Zhao, Z.-X.; Zhao, J.-H.; Fan, J.; Li, Y.; Yang, H.; Shang, J. XAP5 CIRCADIAN TIMEKEEPER positively regulates RESISTANCE TO POWDERY MILDEW8. 1–mediated immunity in Arabidopsis. Front. Plant Sci. 2017, 8, 2044. [Google Scholar] [CrossRef] [PubMed]
- Song, Q.; Ando, A.; Xu, D.; Fang, L.; Zhang, T.; Huq, E.; Qiao, H.; Deng, X.W.; Chen, Z.J. Diurnal down-regulation of ethylene biosynthesis mediates biomass heterosis. Proc. Natl. Acad. Sci. USA 2018, 115, 5606–5611. [Google Scholar] [CrossRef] [PubMed]
- Haydon, M.J.; Mielczarek, O.; Frank, A.; Román, Á.; Webb, A.A. Sucrose and ethylene signaling interact to modulate the circadian clock. Plant Physiol. 2017, 175, 947–958. [Google Scholar] [CrossRef] [PubMed]
- Davière, J.-M.; Achard, P. Gibberellin signaling in plants. Development 2013, 140, 1147–1151. [Google Scholar] [CrossRef] [PubMed]
- Bolle, C. The role of GRAS proteins in plant signal transduction and development. Planta 2004, 218, 683–692. [Google Scholar] [CrossRef] [PubMed]
- Davière, J.-M.; Achard, P. A pivotal role of DELLAs in regulating multiple hormone signals. Mol. Plant 2016, 9, 10–20. [Google Scholar] [CrossRef] [PubMed]
- Feng, S.; Martinez, C.; Gusmaroli, G.; Wang, Y.; Zhou, J.; Wang, F.; Chen, L.; Yu, L.; Iglesias-Pedraz, J.M.; Kircher, S. Coordinated regulation of Arabidopsis thaliana development by light and gibberellins. Nature 2008, 451, 475–479. [Google Scholar] [CrossRef] [PubMed]
- Sun, T.-P. The molecular mechanism and evolution of the GA–GID1–DELLA signaling module in plants. Curr. Biol. 2011, 21, R338–R345. [Google Scholar] [CrossRef] [PubMed]
- Ueguchi-Tanaka, M.; Ashikari, M.; Nakajima, M.; Itoh, H.; Katoh, E.; Kobayashi, M.; Chow, T.-Y.; Yue-ie, C.H.; Kitano, H.; Yamaguchi, I. GIBBERELLIN INSENSITIVE DWARF1 encodes a soluble receptor for gibberellin. Nature 2005, 437, 693–698. [Google Scholar] [CrossRef] [PubMed]
- Ariizumi, T.; Murase, K.; Sun, T.-P.; Steber, C.M. Proteolysis-independent downregulation of DELLA repression in Arabidopsis by the gibberellin receptor GIBBERELLIN INSENSITIVE DWARF1. Plant Cell 2008, 20, 2447–2459. [Google Scholar] [CrossRef] [PubMed]
- Arana, M.V.; Marín-de la Rosa, N.; Maloof, J.N.; Blázquez, M.A.; Alabadí, D. Circadian oscillation of gibberellin signaling in Arabidopsis. Proc. Natl. Acad. Sci. USA 2011, 108, 9292–9297. [Google Scholar] [CrossRef] [PubMed]
- Filo, J.; Wu, A.; Eliason, E.; Richardson, T.; Thines, B.C.; Harmon, F.G. Gibberellin driven growth in elf3 mutants requires PIF4 and PIF5. Plant Signal. Behav. 2015, 10, e992707. [Google Scholar] [CrossRef] [PubMed]
- Zheng, H.; Zhang, F.; Wang, S.; Su, Y.; Jiang, P.; Cheng, R.; Ji, X.; Hou, S.; Ding, Y. MLK1 and MLK2 coordinate RGA and CCA1 activity to regulate hypocotyl elongation in Arabidopsis thaliana. Plant Cell 2017, 30, 67–82. [Google Scholar] [CrossRef] [PubMed]
- Tseng, T.-S.; Salomé, P.A.; McClung, C.R.; Olszewski, N.E. SPINDLY and GIGANTEA interact and act in Arabidopsis thaliana pathways involved in light responses, flowering, and rhythms in cotyledon movements. Plant Cell 2004, 16, 1550–1563. [Google Scholar] [CrossRef] [PubMed]
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Singh, M.; Mas, P. A Functional Connection between the Circadian Clock and Hormonal Timing in Arabidopsis. Genes 2018, 9, 567. https://doi.org/10.3390/genes9120567
Singh M, Mas P. A Functional Connection between the Circadian Clock and Hormonal Timing in Arabidopsis. Genes. 2018; 9(12):567. https://doi.org/10.3390/genes9120567
Chicago/Turabian StyleSingh, Manjul, and Paloma Mas. 2018. "A Functional Connection between the Circadian Clock and Hormonal Timing in Arabidopsis" Genes 9, no. 12: 567. https://doi.org/10.3390/genes9120567
APA StyleSingh, M., & Mas, P. (2018). A Functional Connection between the Circadian Clock and Hormonal Timing in Arabidopsis. Genes, 9(12), 567. https://doi.org/10.3390/genes9120567




