Endogenous Retroviruses in Host-Virus Coevolution: From Genomic Domestication to Functional Innovation
Abstract
1. Introduction
2. Genome Organization, Classification and Host Distribution of ERVs
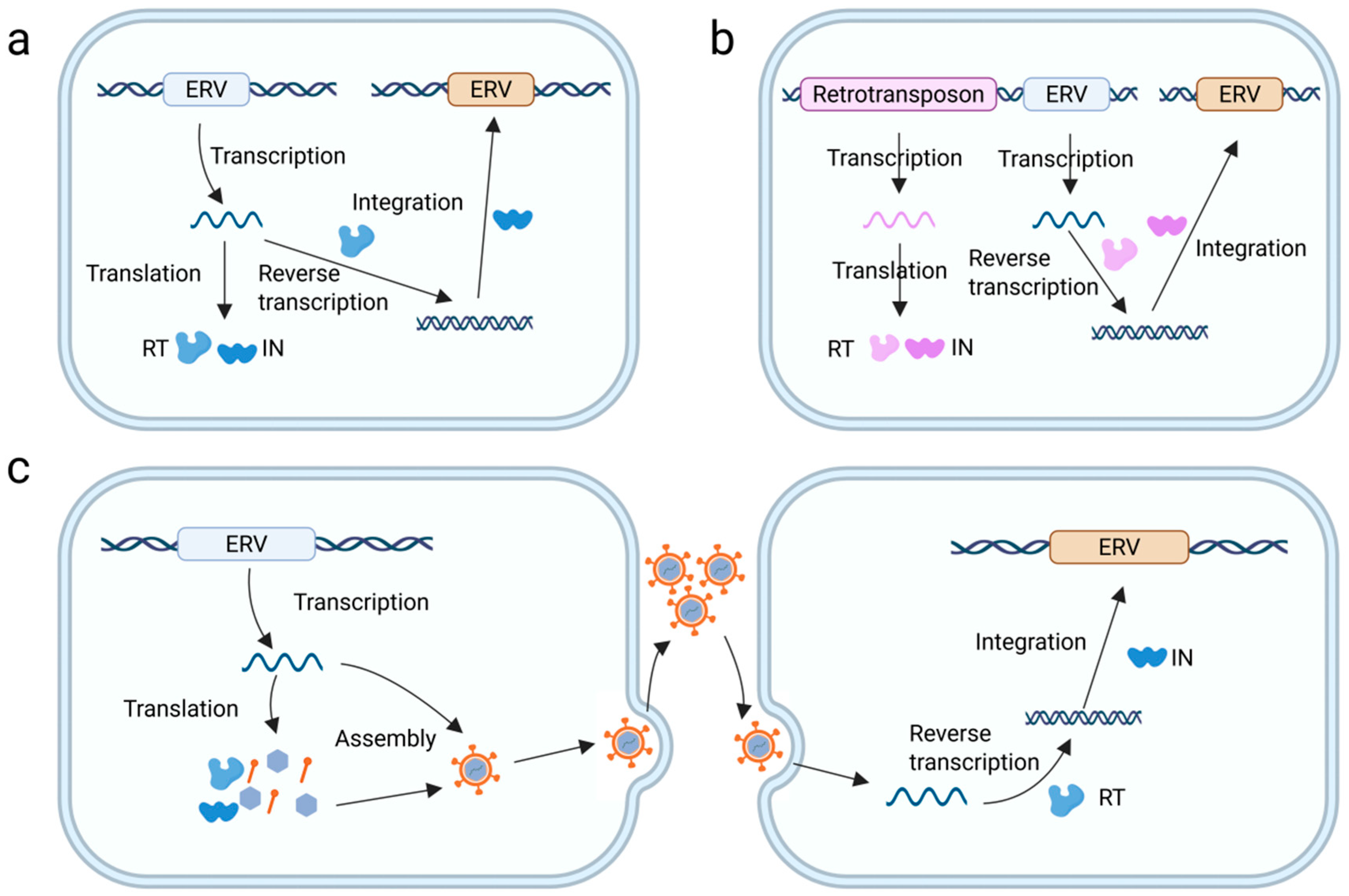
3. Genomic Integration, Amplification, and Epigenetic Silencing of ERVs
4. Interactions Between ERVs and XRVs
5. ERVs as Genomic Markers for Tracking Retroviral and Host Evolution
6. ERV Cross-Regional Spread and Host Population Dynamics
7. Pathways and Barriers in ERV Cross-Species Transmission
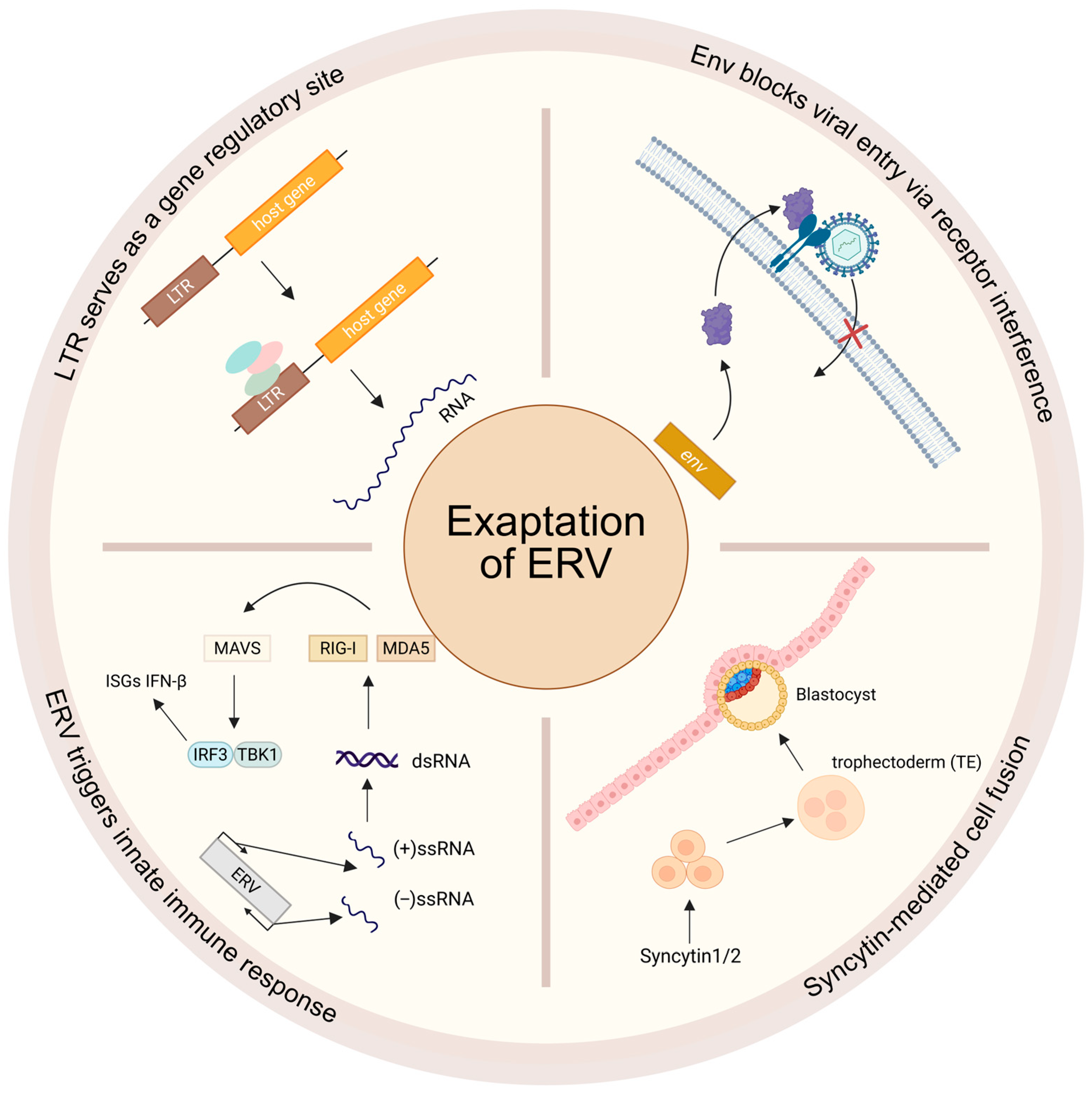
8. Exaptation of ERVs in Host Immunity and Development
9. Conclusions and Prospects
Author Contributions
Funding
Conflicts of Interest
References
- Weiss, R. Spontaneous Virus Production from Non-Virus Producing Rous Sarcoma Cells. Virology 1967, 32, 719. [Google Scholar] [CrossRef]
- Rowe, W.P.; Pincus, T. Quantitative Studies of Naturally Occurring Murine Leukemia-Virus Infection of Akr Mice. J. Exp. Med. 1972, 135, 429. [Google Scholar] [CrossRef]
- Weiss, R.A. The discovery of endogenous retroviruses. Retrovirology 2006, 3, 67. [Google Scholar] [CrossRef]
- Cohen, J.C.; Varmus, H.E. Endogenous Mammary Tumor Virus DNA Varies among Wild Mice and Segregates during Inbreeding. Nature 1979, 278, 418–423. [Google Scholar] [CrossRef]
- Bannert, N.; Kurth, R. Retroelements and the human genome: New perspectives on an old relation. Proc. Natl. Acad. Sci. USA 2004, 101, 14572–14579. [Google Scholar] [CrossRef] [PubMed]
- Hillier, L.W.; Miller, W.; Birney, E.; Warren, W.; Hardison, R.C.; Ponting, C.P.; Bork, P.; Burt, D.W.; Groenen, M.A.M.; Delany, M.E.; et al. Sequence and comparative analysis of the chicken genome provide unique perspectives on vertebrate evolution. Nature 2004, 432, 695–716. [Google Scholar] [CrossRef] [PubMed]
- Bai, J.; Yang, Z.Z.; Li, H.; Hong, Y.; Fan, D.D.; Lin, A.F.; Shao, J.-z. Genome-Wide Characterization of Zebrafish Endogenous Retroviruses Reveals Unexpected Diversity in Genetic Organizations and Functional Potentials. Microbiol. Spectr. 2021, 9, e02254-21. [Google Scholar] [CrossRef]
- Stoye, J.P. Endogenous retroviruses: Still active after all these years? Curr. Biol. 2001, 11, R914–R916. [Google Scholar] [CrossRef]
- Suntsova, M.; Garazha, A.; Ivanova, A.; Kaminsky, D.; Zhavoronkov, A.; Buzdin, A. Molecular functions of human endogenous retroviruses in health and disease. Cell. Mol. Life Sci. 2015, 72, 3653–3675. [Google Scholar] [CrossRef]
- Lefkowitz, E.J.; Dempsey, D.M.; Hendrickson, R.C.; Orton, R.J.; Siddell, S.G.; Smith, D.B. Virus taxonomy: The database of the International Committee on Taxonomy of Viruses (ICTV). Nucleic Acids Res. 2018, 46, D708–D717. [Google Scholar] [CrossRef] [PubMed]
- Katzourakis, A.; Rambaut, A.; Pybus, O.G. The evolutionary dynamics of endogenous retroviruses. Trends Microbiol. 2005, 13, 463–468. [Google Scholar] [CrossRef] [PubMed]
- Zheng, J.L.; Wei, Y.T.; Han, G.Z. The diversity and evolution of retroviruses: Perspectives from viral “fossils”. Virol. Sin. 2022, 37, 11–18. [Google Scholar] [CrossRef]
- Jern, P.; Greenwood, A.D. Wildlife endogenous retroviruses: Colonization, consequences, and cooption. Trends Genet. 2024, 40, 149–159. [Google Scholar] [CrossRef] [PubMed]
- Tsangaras, K.; Mayer, J.; Mirza, O.; Dayaram, A.; Higgins, D.P.; Bryant, B.; Campbell-Ward, M.; Sangster, C.; Casteriano, A.; Hoper, D. Evolutionarily Young African Rhinoceros Gammaretroviruses. J. Virol. 2023, 97, e01932-22. [Google Scholar] [CrossRef]
- Pramono, D.; Muto, Y.; Shimazu, Y.; Deshapriya, R.M.C.; Makundi, I.; Arnal, M.; de Luco, D.F.; Ngo, M.H.; Miyake, A.; Nishigaki, K. Endogenous retrovirus ERV-DC8 highly integrated in domestic cat populations is a replication-competent provirus. Biochem. Biophys. Res. Commun. 2024, 738, 150521. [Google Scholar] [CrossRef]
- Chu, L.; Su, F.; Han, G.-Z.; Wang, J. Jawless vertebrates do not escape retrovirus infection. Virology 2023, 583, 52–55. [Google Scholar] [CrossRef]
- Mason, A.S.; Lund, A.R.; Hocking, P.M.; Fulton, J.E.; Burt, D.W. Identification and characterisation of endogenous Avian Leukosis Virus subgroup E (ALVE) insertions in chicken whole genome sequencing data. Mob. DNA 2020, 11, 22. [Google Scholar] [CrossRef]
- Liu, M.Y.; Jia, L.; Li, H.P.; Liu, Y.J.; Han, J.W.; Wang, X.L.; Li, T.; Li, J.; Zhang, B.; Zhai, X. p53 Binding Sites in Long Terminal Repeat 5Hs (LTR5Hs) of Human Endogenous Retrovirus K Family (HML-2 Subgroup) Play Important Roles in the Regulation of LTR5Hs Transcriptional Activity. Microbiol. Spectr. 2022, 10, e00485-22. [Google Scholar] [CrossRef]
- Harding, E.F.; Mercer, L.K.; Yan, G.J.H.; Waters, P.D.; White, P.A. Invasion and Amplification of Endogenous Retroviruses in Dasyuridae Marsupial Genomes. Mol. Biol. Evol. 2024, 41, msae160. [Google Scholar] [CrossRef]
- Hron, T.; Farkasová, H.; Gifford, R.J.; Benda, P.; Hulva, P.; Görföl, T.; Paces, J.; Elleder, D. Remnants of an Ancient Deltaretrovirus in the Genomes of Horseshoe Bats (Rhinolophidae). Viruses 2018, 10, 185. [Google Scholar] [CrossRef] [PubMed]
- Farkasová, H.; Hron, T.; Paces, J.; Hulva, P.; Benda, P.; Gifford, R.J.; Elleder, D. Discovery of an endogenous Deltaretrovirus in the genome of long-fingered bats (Chiroptera: Miniopteridae). Proc. Natl. Acad. Sci. USA 2017, 114, 3145–3150. [Google Scholar] [CrossRef] [PubMed]
- Kambol, R.; Gatseva, A.; Gifford, R.J. An endogenous lentivirus in the germline of a rodent. Retrovirology 2022, 19, 30. [Google Scholar] [CrossRef]
- Katzourakis, A.; Tristem, M.; Pybus, O.G.; Gifford, R.J. Discovery and analysis of the first endogenous lentivirus. Proc. Natl. Acad. Sci. USA 2007, 104, 6261–6265. [Google Scholar] [CrossRef]
- Han, G.Z.; Worobey, M. An Endogenous Foamy Virus in the Aye-Aye (Daubentonia madagascariensis). J. Virol. 2012, 86, 7696–7698. [Google Scholar] [CrossRef]
- Han, G.Z.; Worobey, M. An Endogenous Foamy-like Viral Element in the Coelacanth Genome. PLoS Pathog. 2012, 8, e1002790. [Google Scholar] [CrossRef] [PubMed]
- Yedavalli, V.R.K.; Patil, A.; Parrish, J.; Kozak, C.A. A novel class III endogenous retrovirus with a class I envelope gene in African frogs with an intact genome and developmentally regulated transcripts in Xenopus tropicalis. Retrovirology 2021, 18, 20. [Google Scholar] [CrossRef]
- Xu, X.Y.; Zhao, H.Y.; Gong, Z.; Han, G.Z. Endogenous retroviruses of non-avian/mammalian vertebrates illuminate diversity and deep history of retroviruses. PLoS Pathog. 2018, 14, e1007072. [Google Scholar] [CrossRef]
- Hayward, A.; Cornwallis, C.; Jern, P. Pan-vertebrate comparative genomics unmasks retrovirus macroevolution. Proc. Natl. Acad. Sci. USA 2015, 112, 464–469. [Google Scholar] [CrossRef]
- Han, G.Z.; Worobey, M. A Primitive Endogenous Lentivirus in a Colugo: Insights into the Early Evolution of Lentiviruses. Mol. Biol. Evol. 2015, 32, 211–215. [Google Scholar] [CrossRef]
- Gifford, R.J.; Katzourakis, A.; Tristem, M.; Pybus, O.G.; Winters, M.; Shafer, R.W. A transitional endogenous lentivirus from the genome of a basal primate and implications for lentivirus evolution. Proc. Natl. Acad. Sci. USA 2008, 105, 20362–20367. [Google Scholar] [CrossRef] [PubMed]
- Cui, J.; Holmes, E.C. Endogenous Lentiviruses in the Ferret Genome. J. Virol. 2012, 86, 3383–3385. [Google Scholar] [CrossRef] [PubMed]
- Katzourakis, A.; Aiewsakun, P.; Jia, H.W.; Wolfe, N.D.; LeBreton, M.; Yoder, A.D.; Switzer, W.M. Discovery of prosimian and afrotherian foamy viruses and potential cross species transmissions amidst stable and ancient mammalian co-evolution. Retrovirology 2014, 11, 61. [Google Scholar] [CrossRef]
- Wang, X.J.; Chen, Y.C.J. Identification of Cartilaginous Fish Endogenous Foamy Virus Rooting to Vertebrate Counterparts. J. Virol. 2023, 97, e01816-22. [Google Scholar] [CrossRef]
- Chen, Y.C.; Zhang, Y.Y.; Wei, X.M.; Cui, J. Multiple Infiltration and Cross-Species Transmission of Foamy Viruses across the Paleozoic to the Cenozoic Era. J. Virol. 2021, 95, e0048421. [Google Scholar] [CrossRef] [PubMed]
- Wei, X.M.; Chen, Y.C.; Duan, G.Q.; Holmes, E.C.; Cui, J. A reptilian endogenous foamy virus sheds light on the early evolution of retroviruses. Virus Evol. 2019, 5, vez001. [Google Scholar] [CrossRef]
- Chen, Y.C.; Wei, X.M.; Zhang, G.J.; Holmes, E.C.; Cui, J. Identification and evolution of avian endogenous foamy viruses. Virus Evol. 2019, 5, vez049. [Google Scholar] [CrossRef] [PubMed]
- Prasad, V.M.; Leaman, D.P.; Lovendahl, K.N.; Croft, J.T.; Benhaim, M.A.; Hodge, E.A.; Zwick, M.B.; Lee, K.K. Cryo-ET of Env on intact HIV virions reveals structural variation and positioning on the Gag lattice. Cell 2022, 185, 641–653. [Google Scholar] [CrossRef]
- Das, K.; Martinez, S.E.; Bandwar, R.P.; Arnold, E. Structures of HIV-1 RT-RNA/DNA ternary complexes with dATP and nevirapine reveal conformational flexibility of RNA/DNA: Insights into requirements for RNase H cleavage. Nucleic Acids Res. 2014, 42, 8125–8137. [Google Scholar] [CrossRef]
- Cherepanov, P.; Maertens, G.; Proost, P.; Devreese, B.; Van Beeumen, J.; Engelborghs, Y.; De Clerq, E.; Debyser, Z. HIV-1 integrase forms stable tetramers and associates with LEDGF/p75 protein in human cells. J. Biol. Chem. 2003, 278, 372–381. [Google Scholar] [CrossRef]
- Chen, B. Molecular Mechanism of HIV-1 Entry. Trends Microbiol. 2019, 27, 878–891. [Google Scholar] [CrossRef]
- Sultana, T.; Zamborlini, A.; Cristofari, G.; Lesage, P. Integration site selection by retroviruses and transposable elements in eukaryotes. Nat. Rev. Genet. 2017, 18, 292–308. [Google Scholar] [CrossRef]
- Kvaratskhelia, M.; Sharma, A.; Larue, R.C.; Serrao, E.; Engelman, A. Molecular mechanisms of retroviral integration site selection. Nucleic Acids Res. 2014, 42, 10209–10225. [Google Scholar] [CrossRef]
- Asante-Appiah, E.; Skalka, A.M. Molecular mechanisms in retrovirus DNA integration. Antivir. Res. 1997, 36, 139–156. [Google Scholar] [CrossRef] [PubMed]
- Coffin, J.; Swanstrom, R. HIV Pathogenesis: Dynamics and Genetics of Viral Populations and Infected Cells. Cold Spring Harb. Perspect. Med. 2013, 3, a012526. [Google Scholar] [CrossRef] [PubMed]
- Nascimento, F.F.; Rodrigo, A.G. Computational Evaluation of the Strict Master and Random Template Models of Endogenous Retrovirus Evolution. PLoS ONE 2016, 11, e0162454. [Google Scholar] [CrossRef] [PubMed]
- Belshaw, R.; Pereira, V.; Katzourakis, A.; Talbot, G.; Paces, J.; Burt, A.; Tristem, M. Long-term reinfection of the human genome by endogenous retroviruses. Proc. Natl. Acad. Sci. USA 2004, 101, 4894–4899. [Google Scholar] [CrossRef]
- Bannert, N.; Kurth, R. The evolutionary dynamics of human endogenous retroviral families. Annu. Rev. Genom. Hum. Genet. 2006, 7, 149–173. [Google Scholar] [CrossRef]
- Magiorkinis, G.; Gifford, R.J.; De Katzourakis, A.; Ranter, J.; Belshaw, R. Env-less endogenous retroviruses are genomic superspreaders. Proc. Natl. Acad. Sci. USA 2012, 109, 7385–7390. [Google Scholar] [CrossRef]
- Pavlícek, A.; Paces, J.; Elleder, D.; Hejnar, J. Processed Pseudogenes of human endogenous retroviruses generated by LINEs: Their integration, stability, and distribution. Genome Res. 2002, 12, 391–399. [Google Scholar] [CrossRef]
- Ernst, C.; Odom, D.T.; Kutter, C. The emergence of piRNAs against transposon invasion to preserve mammalian genome integrity. Nat. Commun. 2017, 8, 1411. [Google Scholar] [CrossRef]
- Aravin, A.A.; Sachidanandam, R.; Bourc’his, D.; Schaefer, C.; Pezic, D.; Toth, K.F.; Bestor, T.; Hannon, G.J. A piRNA pathway primed by individual transposons is linked to de novo DNA methylation in mice. Mol. Cell 2008, 31, 785–799. [Google Scholar] [CrossRef] [PubMed]
- Maksakova, I.A.; Goyal, P.; Bullwinkel, J.; Brown, J.P.; Bilenky, M.; Mager, D.L.; Singh, P.B.; Lorincz, M.C. H3K9me3-binding proteins are dispensable for SETDB1/H3K9me3-dependent retroviral silencing. Epigenet. Chromatin 2011, 4, 12. [Google Scholar] [CrossRef] [PubMed]
- Chelmicki, T.; Roger, E.; Teissandier, A.; Dura, M.; Bonneville, L.; Rucli, S.; Dossin, F.; Fouassier, C.; Lameiras, S.; Bourc′his, D. m6A RNA methylation regulates the fate of endogenous retroviruses. Nature 2021, 591, 312–316. [Google Scholar] [CrossRef] [PubMed]
- Guallar, D.; Bi, X.J.; Pardavila, J.A.; Huang, X.; Saenz, C.; Shi, X.L.; Zhou, H.; Faiola, F.; Ding, J.; Haruehanroengra, P. RNA-dependent chromatin targeting of TET2 for endogenous retrovirus control in pluripotent stem cells. Nat. Genet. 2018, 50, 443–451. [Google Scholar] [CrossRef]
- Stolz, P.; Mantero, A.S.; Tvardovskiy, A.; Ugur, E.; Wange, L.E.; Mulholland, C.B.; Cheng, Y.; Wierer, M.; Enard, W.; Schneider, R.; et al. TET1 regulates gene expression and repression of endogenous retroviruses independent of DNA demethylation. Nucleic Acids Res. 2022, 50, 8491–8511. [Google Scholar] [CrossRef]
- Spencer, T.E.; Palmarini, M. Endogenous Retroviruses of Sheep: A Model System for Understanding Physiological Adaptation to an Evolving Ruminant Genome. J. Reprod. Dev. 2012, 58, 33–37. [Google Scholar] [CrossRef]
- Wang, X.J.; Liu, S.Y. Endogenous Jaagsiekte sheep retrovirus envelope protein promotes sheep trophoblast cell fusion by activating PKA/MEK/ERK1/2 signaling. Theriogenology 2022, 193, 58–67. [Google Scholar] [CrossRef]
- Cumer, T.; Pompanon, F.; Boyer, F. Old origin of a protective endogenous retrovirus (enJSRV) in the Ovis genus. Heredity 2019, 122, 187–194. [Google Scholar] [CrossRef]
- Arnaud, F.; Caporale, M.; Varela, M.; Biek, R.; Chessa, B.; Alberti, A.; Golder, M.; Mura, M.; Zhang, Y.-p.; Yu, L. A paradigm for virus-host coevolution: Sequential counter-adaptations between endogenous and exogenous retroviruses. PLoS Pathog. 2007, 3, 1716–1729. [Google Scholar] [CrossRef]
- Mura, M.; Murcia, P.; Caporale, M.; Spencer, T.E.; Nagashima, K.; Rein, A.; Palmarini, M. Late viral interference induced by transdominant Gag of an endogenous retrovirus. Proc. Natl. Acad. Sci. USA 2004, 101, 11117–11122. [Google Scholar] [CrossRef]
- Murcia, P.R.; Arnaud, F.; Palmarini, M. The transdominant endogenous retrovirus enJS56A1 associates with and blocks intracellular trafficking of Jaagsiekte sheep retrovirus Gag. J. Virol. 2007, 81, 1762–1772. [Google Scholar] [CrossRef]
- Chiu, E.S.; VandeWoude, S. Endogenous Retroviruses Drive Resistance and Promotion of Exogenous Retroviral Homologs. Annu. Rev. Anim. Biosci. 2021, 9, 225–248. [Google Scholar] [CrossRef] [PubMed]
- Ngo, M.H.; AbuEed, L.; Kawasaki, J.; Oishi, N.; Pramono, D.; Kimura, T.; Sakurai, M.; Watanabe, K.; Mizukami, Y.; Ochi, H. Multiple recombination events between endogenous retroviral elements and feline leukemia virus. J. Virol. 2024, 98, e01400-23. [Google Scholar] [CrossRef]
- Shoji, H.; Tsukasa, Y.; Kitao, K.; Miyazawa, T. Characterization of ferret Pit1 as a receptor of feline leukemia virus subgroup B. J. Vet. Med. Sci. 2023, 85, 326–328. [Google Scholar] [CrossRef] [PubMed]
- Erbeck, K.; Gagne, R.B.; Kraberger, S.; Chiu, E.S.; Roelke-Parker, M.; VandeWoude, S. Feline Leukemia Virus (FeLV) Endogenous and Exogenous Recombination Events Result in Multiple FeLV-B Subtypes during Natural Infection. J. Virol. 2021, 95, e0035321. [Google Scholar] [CrossRef]
- Tandon, R.; Cattori, V.; Pepin, A.C.; Riond, B.; Meli, M.L.; McDonald, M.; Doherr, M.G.; Lutz, H.; Hoffman-Lehmann, R. Association between endogenous feline leukemia virus loads and exogenous feline leukemia virus infection in domestic cats. Virus Res. 2008, 135, 136–143. [Google Scholar] [CrossRef]
- Young, G.R.; Terry, S.N.; Manganaro, L.; Cuesta-Dominguez, A.; Deikus, G.; Bernal-Rubio, D.; Campisi, L.; Fernandez-Sesma, A.; Sebra, R.; Simon, V.; et al. HIV-1 Infection of Primary CD4+T Cells Regulates the Expression of Specific Human Endogenous Retrovirus HERV-K (HML-2) Elements. J. Virol. 2018, 92, e01507-17. [Google Scholar] [CrossRef] [PubMed]
- Fasching, L.; Kapopoulou, A.; Sachdeva, R.; Petri, R.; Jönsson, M.E.; Männe, C.; Turelli, P.; Jern, P.; Cammas, F.; Trono, D.; et al. TRIM28 Represses Transcription of Endogenous Retroviruses in Neural Progenitor Cells. Cell Rep. 2015, 10, 20–28. [Google Scholar] [CrossRef]
- Gonzalez-Hernandez, M.J.; Swanson, M.D.; Contreras-Galindo, R.; Cookinham, S.; King, S.; Noel, R.J.; Kaplan, M.H.; Markovitz, D.M. Expression of Human Endogenous Retrovirus Type K (HML-2) Is Activated by the Tat Protein of HIV-1. J. Virol. 2012, 86, 7790–7805. [Google Scholar] [CrossRef]
- Gonzalez-Hernandez, M.J.; Cavalcoli, J.; Sartor, M.A.; Contreras-Galindo, R.; Meng, F.; Dai, M.H.; Dube, D.; Saha, A.S.; Gitlin, S.D.; Omenn, G.S.; et al. Regulation of the Human Endogenous Retrovirus K (HML-2) Transcriptome by the HIV-1 Tat Protein. J. Virol. 2014, 88, 8924–8935. [Google Scholar] [CrossRef]
- Kiessling, M.; Cole, J.J.; Kübel, S.; Klein, P.; Korn, K.; Henry, A.R.; Labourne, F.; Fourati, S.; Harrer, E.; Harrer, T.; et al. Chronic inflammation degrades CD4 T cell immunity to prior vaccines in treated HIV infection. Nat. Commun. 2024, 15, 10200. [Google Scholar] [CrossRef]
- Russ, E.; Mikhalkevich, N.; Iordanskiy, S. Expression of Human Endogenous Retrovirus Group K (HERV-K) HML-2 Correlates with Immune Activation of Macrophages and Type I Interferon Response. Microbiol. Spectr. 2023, 11, e04438-22. [Google Scholar] [CrossRef]
- Curty, G.; Iniguez, L.P.; Soares, M.A.; Nixon, D.F.; Rougvie, M.D. Off-Target Effect of Activation of NF-κB by HIV Latency Reversal Agents on Transposable Elements Expression. Viruses 2022, 14, 1571. [Google Scholar] [CrossRef] [PubMed]
- Monde, K.; Terasawa, H.; Nakano, Y.; Soheilian, F.; Nagashima, K.; Maeda, Y.; Ono, A. Molecular mechanisms by which HERV-K Gag interferes with HIV-1 Gag assembly and particle infectivity. Retrovirology 2017, 14, 27. [Google Scholar] [CrossRef] [PubMed]
- Jones, R.B.; Garrison, K.E.; Mujib, S.; Mihajlovic, V.; Aidarus, N.; Hunter, D.V.; Martin, E.; John, V.M.; Zhan, W.; Faruk, N.F.; et al. HERV-K-specific T cells eliminate diverse HIV-1/2 and SIV primary isolates. J. Clin. Investig. 2012, 122, 4473–4489. [Google Scholar] [CrossRef]
- Johnson, W.E. Origins and evolutionary consequences of ancient endogenous retroviruses. Nat. Rev. Microbiol. 2019, 17, 355–370. [Google Scholar] [CrossRef]
- Wang, J.H.; Han, G.Z. A Sister Lineage of Sampled Retroviruses Corroborates the Complex Evolution of Retroviruses. Mol. Biol. Evol. 2021, 38, 1031–1039. [Google Scholar] [CrossRef]
- Martins, H.; Villesen, P. Improved Integration Time Estimation of Endogenous Retroviruses with Phylogenetic Data. PLoS ONE 2011, 6, e14745. [Google Scholar] [CrossRef]
- Blanco-Melo, D.; Gifford, R.J.; Bieniasz, P.D. Reconstruction of a replication-competent ancestral murine endogenous retrovirus-L. Retrovirology 2018, 15, 34. [Google Scholar] [CrossRef]
- Lipson, M.; Loh, P.R.; Sankararaman, S.; Patterson, N.; Berger, B.; Reich, D. Calibrating the Human Mutation Rate via Ancestral Recombination Density in Diploid Genomes. PLoS Genet. 2015, 11, e1005550. [Google Scholar] [CrossRef]
- Jarosz, A.S.; Halo, J.V. Transcription of Endogenous Retroviruses: Broad and Precise Mechanisms of Control. Viruses 2024, 16, 1312. [Google Scholar] [CrossRef]
- van der Kuyl, A.C. Mutation Rate Variation and Other Challenges in 2-LTR Dating of Primate Endogenous Retrovirus Integrations. J. Mol. Evol. 2024, 93, 62–82. [Google Scholar] [CrossRef]
- Holloway, J.R.; Williams, Z.H.; Freeman, M.M.; Bulow, U.; Coffin, J.M. Gorillas have been infected with the HERV-K (HML-2) endogenous retrovirus much more recently than humans and chimpanzees. Proc. Natl. Acad. Sci. USA 2019, 116, 1337–1346. [Google Scholar] [CrossRef] [PubMed]
- Halo, J.; Pendleton, A.L.; Jarosz, A.S.; Gifford, R.J.; Day, M.L.; Kidd, J.M. Origin and recent expansion of an endogenous gammaretroviral lineage in domestic and wild canids. Retrovirology 2019, 16, 6. [Google Scholar] [CrossRef] [PubMed]
- Stocking, C.; Kozak, C.A. Murine endogenous retroviruses. Cell. Mol. Life Sci. 2008, 65, 3383–3398. [Google Scholar] [CrossRef]
- Hanke, K.; Hohn, O.; Bannert, N. HERV-K(HML-2), a seemingly silent subtenant-but still waters run deep. Apmis 2016, 124, 67–87. [Google Scholar] [CrossRef]
- Wildschutte, J.H.; Williams, Z.H.; Montesion, M.; Subramanian, R.P.; Kidd, J.M.; Coffin, J.M. Discovery of unfixed endogenous retrovirus insertions in diverse human populations. Proc. Natl. Acad. Sci. USA 2016, 113, E2326–E2334. [Google Scholar] [CrossRef]
- Subramanian, R.P.; Wildschutte, J.H.; Russo, C.; Coffin, J.M. Identification, characterization, and comparative genomic distribution of the HERV-K (HML-2) group of human endogenous retroviruses. Retrovirology 2011, 8, 90. [Google Scholar] [CrossRef]
- Belshaw, R.; Dawson, A.L.A.; Woolven-Allen, J.; Redding, J.; Burt, A.; Tristem, M. Genomewide screening reveals high levels of insertional polymorphism in the human endogenous retrovirus family HERV-K(HML2): Implications for present-day activity. J. Virol. 2005, 79, 12507–12514. [Google Scholar] [CrossRef] [PubMed]
- Chen, M.Y.; Yang, C.Q.; Zhai, X.L.; Wang, C.L.; Liu, M.Y.; Zhang, B.H.; Guo, X.; Wang, Y.L.; Li, H.P.; Liu, Y.J.; et al. Comprehensive Identification and Characterization of HML-9 Group in Chimpanzee Genome. Viruses 2024, 16, 892. [Google Scholar] [CrossRef]
- Zhu, H.N.; Gifford, R.J.; Murcia, P.R. Distribution, Diversity, and Evolution of Endogenous Retroviruses in Perissodactyl Genomes. J. Virol. 2018, 92, e00927-18. [Google Scholar] [CrossRef]
- Olival, K.J.; Hosseini, P.R.; Zambrana-Torrelio, C.; Ross, N.; Bogich, T.L.; Daszak, P. Host and viral traits predict zoonotic spillover from mammals. Nature 2017, 548, 612. [Google Scholar] [CrossRef]
- Boso, G.; Kozak, C.A. Retroviral Restriction Factors and Their Viral Targets: Restriction Strategies and Evolutionary Adaptations. Microorganisms 2020, 8, 1965. [Google Scholar] [CrossRef]
- Lloyd-Smith, J.O.; Schreiber, S.J.; Kopp, P.E.; Getz, W.M. Superspreading and the effect of individual variation on disease emergence. Nature 2005, 438, 355–359. [Google Scholar] [CrossRef]
- Wang, J.H.; Han, G.Z. Genome mining shows that retroviruses are pervasively invading vertebrate genomes. Nat. Commun. 2023, 14, 4968. [Google Scholar] [CrossRef]
- Löber, U.; Hobbs, M.; Dayaram, A.; Tsangaras, K.; Jones, K.; Alquezar-Planas, D.E.; Ishida, Y.; Meers, J.; Mayer, J.; Quedenau, C.; et al. Degradation and remobilization of endogenous retroviruses by recombination during the earliest stages of a germ-line invasion. Proc. Natl. Acad. Sci. USA 2018, 115, 8609–8914. [Google Scholar] [CrossRef]
- Blyton, M.D.J.; Young, P.R.; Moore, B.; Chappell, K.J. Geographic patterns of koala retrovirus genetic diversity, endogenization, and subtype distributions. Proc. Natl. Acad. Sci. USA 2022, 120, e2220541120. [Google Scholar] [CrossRef]
- Stephenson, T.; Speight, N.; Low, W.Y.; Woolford, L.; Tearle, R.; Hemmatzadeh, F. Molecular Diagnosis of Koala Retrovirus (KoRV) in South Australian Koalas (Phascolarctos cinereus). Animals 2021, 11, 1477. [Google Scholar] [CrossRef] [PubMed]
- Akter, L.; Hashem, M.; Kayesh, M.E.H.; Hossain, M.A.; Maetani, F.; Akhter, R.; Hossain, K.A.; Rashid, M.H.O.; Sakurai, H.; Asai, T.; et al. A preliminary study of gene expression changes in Koalas Infected with Koala Retrovirus (KoRV) and identification of potential biomarkers for KoRV pathogenesis. BMC Vet. Res. 2024, 20, 496. [Google Scholar] [CrossRef]
- Xu, W.Q.; Stadler, C.; Gorman, K.; Jensen, N.; Kim, D.; Zheng, H.Q.; Tang, S.H.; Switzer, W.M.; Pye, G.W.; Eiden, M.V. An exogenous retrovirus isolated from koalas with malignant neoplasias in a US zoo. Proc. Natl. Acad. Sci. USA 2013, 110, 11547–11552. [Google Scholar] [CrossRef] [PubMed]
- Chappell, K.J.; Brealey, J.C.; Amarilla, A.A.; Watterson, D.; Hulse, L.; Palmieri, C.; Johnston, S.D.; Holmes, E.C.; Meers, J.; Young, P.R. Phylogenetic Diversity of Koala Retrovirus within a Wild Koala Population. J. Virol. 2017, 91, e01820-16. [Google Scholar] [CrossRef]
- Joyce, B.A.; Blyton, M.D.J.; Johnston, S.D.; Young, P.R.; Chappell, K.J. Koala retrovirus genetic diversity and transmission dynamics within captive koala populations. Proc. Natl. Acad. Sci. USA 2021, 118, e2024021118. [Google Scholar] [CrossRef]
- Xu, W.Q.; Gorman, K.; Santiago, J.C.; Kluska, K.; Eiden, M.V. Genetic Diversity of Koala Retroviral Envelopes. Viruses 2015, 7, 1258–1270. [Google Scholar] [CrossRef] [PubMed]
- Legione, A.R.; Patterson, J.L.S.; Whiteley, P.; Firestone, S.M.; Curnick, M.; Bodley, K.; Lynch, M.; Gilkerson, J.R.; Sansom, F.M.; Devlin, J.M. Koala retrovirus genotyping analyses reveal a low prevalence of KoRV-A in Victorian koalas and an association with clinical disease. J. Med. Microbiol. 2017, 66, 236–244. [Google Scholar] [CrossRef]
- Simmons, G.S.; Young, P.R.; Hanger, J.J.; Jones, K.; Clarke, D.; McKee, J.J.; Meers, J. Prevalence of koala retrovirus in geographically diverse populations in Australia. Aust. Vet. J. 2012, 90, 404–409. [Google Scholar] [CrossRef]
- Tarlinton, R.E.; Legione, A.R.; Sarker, N.; Fabijan, J.; Meers, J.; McMichael, L.; Simmons, G.; Owen, H.; Seddon, J.M.; Dick, G.; et al. Differential and defective transcription of koala retrovirus indicates the complexity of host and virus evolution. J. Gen. Virol. 2022, 103, 001749. [Google Scholar] [CrossRef]
- Sarker, N.; Fabijan, J.; Owen, H.; Seddon, J.; Simmons, G.; Speight, N.; Kaler, J.; Woolford, L.; Emes, R.D.; Hemmatzadeh, F.; et al. Koala retrovirus viral load and disease burden in distinct northern and southern koala populations. Sci. Rep. 2020, 10, 263. [Google Scholar] [CrossRef] [PubMed]
- Sarker, N.; Fabijan, J.; Seddon, J.; Tarlinton, R.; Owen, H.; Simmons, G.; Thia, J.; Blanchard, A.M.; Speight, N.; Kaler, J.; et al. Genetic diversity of Koala retrovirus env gene subtypes: Insights into northern and southern koala populations. J. Gen. Virol. 2019, 100, 1328–1339. [Google Scholar] [CrossRef] [PubMed]
- Menkhorst, P. Hunted, Marooned, Re-Introduced, Contracepted: A History of Koala Management in Victoria; Too Close for Comfort: Contentious Issues in Human-Wildlife Encounters; Royal Zoological Society of New South Wales: Sydney, Australia, 2008; Volume 34, pp. 73–92. [Google Scholar]
- Hobbs, M.; King, A.; Salinas, R.; Chen, Z.L.; Tsangaras, K.; Greenwood, A.D.; Johnson, R.N.; Belov, K.; Wilkins, M.R.; Timms, P. Long-read genome sequence assembly provides insight into ongoing retroviral invasion of the koala germline. Sci. Rep. 2017, 7, 15838. [Google Scholar] [CrossRef]
- Reperant, L.A.; Kuiken, T.; Osterhaus, A.D.M.E. Adaptive pathways of zoonotic influenza viruses: From exposure to establishment in humans. Vaccine 2012, 30, 4419–4434. [Google Scholar] [CrossRef]
- Dunemann, S.M.; Wasmuth, J.D. Horizontal transfer of a retrotransposon between parasitic nematodes and the common shrew. Mob. DNA 2019, 10, 24. [Google Scholar] [CrossRef]
- Brook, C.E.; Boots, M.; Chandran, K.; Dobson, A.P.; Drosten, C.; Graham, A.L.; Grenfell, B.T.; Müller, M.A.; Ng, M.; Wang, L.F.; et al. Accelerated viral dynamics in bat cell lines, with implications for zoonotic emergence. Elife 2020, 9, e48401. [Google Scholar] [CrossRef]
- French, R.K.; Anderson, S.; Cain, K.; Digby, A.; Greene, T.C.; Miskelly, C.; Muller, C.G.; Taylor, M.W.; Team, K.R.; Geoghegan, J.L.; et al. Diversity and cross-species transmission of viruses in a remote island ecosystem: Implications for wildlife conservation. Virus Evol. 2025, 11, veae113. [Google Scholar] [CrossRef] [PubMed]
- Hayward, A.; Grabherr, M.; Jern, P. Broad-scale phylogenomics provides insights into retrovirus-host evolution. Proc. Natl. Acad. Sci. USA 2013, 110, 20146–20151. [Google Scholar] [CrossRef] [PubMed]
- Cui, J.; Tachedjian, G.; Wang, L.F. Bats and Rodents Shape Mammalian Retroviral Phylogeny. Sci. Rep. 2015, 5, 16561. [Google Scholar] [CrossRef]
- Holmes, E.C. The Evolutionary Genetics of Emerging Viruses. Annu. Rev. Ecol. Evol. Syst. 2009, 40, 353–372. [Google Scholar] [CrossRef]
- Verneret, M.; Leroux, C.; Faraut, T.; Navratil, V.; Lerat, E.; Turpin, J. A genome-wide study of ruminants uncovers two endogenous retrovirus families recently active in goats. Mob. DNA 2025, 16, 4. [Google Scholar] [CrossRef]
- Zhuo, X.Y.; Feschotte, C. Cross-Species Transmission and Differential Fate of an Endogenous Retrovirus in Three Mammal Lineages. PLoS Pathog. 2015, 11, e1005279. [Google Scholar] [CrossRef]
- Aiewsakun, P.; Simmonds, P.; Katzourakis, A. The First Co-Opted Endogenous Foamy Viruses and the Evolutionary History of Reptilian Foamy Viruses. Viruses 2019, 11, 641. [Google Scholar] [CrossRef]
- Wang, Y.M.; Liska, F.; Gosele, C.; Sedová, L.; Kren, V.; Krenová, D.; Ivics, Z.; Hubner, N.; Izsvák, Z. A novel active endogenous retrovirus family contributes to genome variability in rat inbred strains. Genome Res. 2010, 20, 19–27. [Google Scholar] [CrossRef] [PubMed]
- Shojima, T.; Yoshikawa, R.; Hoshino, S.; Shimode, S.; Nakagawa, S.; Ohata, T.; Nakaoka, R.; Miyazawaa, T. Identification of a Novel Subgroup of Koala Retrovirus from Koalas in Japanese Zoos. J. Virol. 2013, 87, 9943–9948. [Google Scholar] [CrossRef] [PubMed]
- Simmons, G.; Clarke, D.; McKee, J.; Young, P.; Meers, J. Discovery of a Novel Retrovirus Sequence in an Australian Native Rodent (Melomys burtoni): A Putative Link between Gibbon Ape Leukemia Virus and Koala Retrovirus. PLoS ONE 2014, 9, e106954. [Google Scholar] [CrossRef]
- Alfano, N.; Michaux, J.; Morand, S.; Aplin, K.; Tsangaras, K.; Löber, U.; Fabre, P.H.; Fitriana, Y.; Semiadi, G.; Ishida, Y.; et al. Endogenous Gibbon Ape Leukemia Virus Identified in a Rodent (Melomys burtoni subsp.) from Wallacea (Indonesia). J. Virol. 2016, 90, 8169–8180. [Google Scholar] [CrossRef]
- Fabre, P.H.; Fitriana, Y.S.; Semiadi, G.; Pagès, M.; Aplin, K.; Supriatna, N.; Helgen, K.M. New record of Melomys burtoni (Mammalia; Rodentia; Murinae) from Halmahera (North Moluccas; Indonesia): A review of Moluccan. Mammalia 2018, 82, 218–247. [Google Scholar] [CrossRef]
- Hayward, J.A.; Tachedjian, M.; Kohl, C.; Johnson, A.; Dearnley, M.; Jesaveluk, B.; Langer, C.; Solymosi, P.D.; Hille, G.; Nitsche, A.; et al. Infectious KoRV-related retroviruses circulating in Australian bats. Proc. Natl. Acad. Sci. USA 2020, 117, 9529–9536. [Google Scholar] [CrossRef] [PubMed]
- McMichael, L.; Smith, C.; Gordon, A.; Agnihotri, K.; Meers, J.; Oakey, J. A novel Australian flying-fox retrovirus shares an evolutionary ancestor with Koala; Gibbon and Melomys gamma-retroviruses. Virus Genes 2019, 55, 421–424. [Google Scholar] [CrossRef]
- Denner, J. Monitoring for PERV Following Xenotransplantation. Transpl. Int. 2024, 37, 13491. [Google Scholar] [CrossRef]
- Ericsson, T.A.; Takeuchi, Y.; Templin, C.; Quinn, G.; Farhadian, S.F.; Wood, J.C.; Oldmixon, B.A.; Suling, K.M.; Ishii, J.K.; Kitagawa, Y.; et al. Identification of receptors for pig endogenous retrovirus. Proc. Natl. Acad. Sci. USA 2003, 100, 6759–6764. [Google Scholar] [CrossRef]
- Denner, J. Recombinant porcine endogenous retroviruses (PERV-A/C): A new risk for xenotransplantation? Arch. Virol. 2008, 153, 1421–1426. [Google Scholar] [CrossRef]
- Specke, V.; Rubant, S.; Denner, J. Productive infection of human primary cells and cell lines with porcine endogenous retroviruses. Virology 2001, 285, 177–180. [Google Scholar] [CrossRef]
- Irgang, M.; Sauer, I.M.; Karlas, A.; Zeilinger, K.; Gerlach, J.C.; Kurth, R.; Neuhaus, P.; Denner, J. Porcine endogenous retroviruses: No infection in patients treated with a bioreactor based on porcine liver cells. J. Clin. Virol. 2003, 28, 141–154. [Google Scholar] [CrossRef]
- Kassiotis, G.; Stoye, J.P. Immune responses to endogenous retroelements: Taking the bad with the good. Nat. Rev. Immunol. 2016, 16, 207–219. [Google Scholar] [CrossRef]
- Nair, V.P.; Liu, H.; Ciceri, G.; Jungverdorben, J.; Frishman, G.; Tchieu, J.; Cederquist, G.Y.; Rothenaigner, I.; Schorpp, K.; Klepper, L.; et al. Activation of HERV-K(HML-2) disrupts cortical patterning and neuronal differentiation by increasing NTRK3. Cell Stem Cell 2021, 28, 1671–1673. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.H.; Han, G.Z. Frequent Retroviral Gene Co-option during the Evolution of Vertebrates. Mol. Biol. Evol. 2020, 37, 3232–3242. [Google Scholar] [CrossRef] [PubMed]
- Rebollo, R.; Romanish, M.T.; Mager, D.L. Transposable Elements: An Abundant and Natural Source of Regulatory Sequences for Host Genes. Annu. Rev. Genet. 2012, 46, 21–42. [Google Scholar] [CrossRef]
- Mi, S.; Lee, X.; Li, X.P.; Veldman, G.M.; Finnerty, H.; Racie, L.; LaVallie, E.; Tang, X.Y.; Edouard, P.; Howes, S.; et al. Syncytin is a captive retroviral envelope protein involved in human placental morphogenesis. Nature 2000, 403, 785–789. [Google Scholar] [CrossRef] [PubMed]
- Yu, J.D.; Qiu, P.S.; Ai, J.W.; Liu, B.; Han, G.Z.; Zhu, F.; Zhang, W.H.; Cui, J. Endogenous retrovirus activation: Potential for immunology and clinical applications. Natl. Sci. Rev. 2024, 11, nwae034. [Google Scholar] [CrossRef]
- Di Giorgio, E.; Xodo, L.E. Endogenous Retroviruses (ERVs): Does RLR (RIG-I-Like Receptors)-MAVS Pathway Directly Control Senescence and Aging as a Consequence of ERV De-Repression? Front. Immunol. 2022, 13, 917998. [Google Scholar] [CrossRef]
- Zhao, Y.H.; Li, W.J.; Xu, J.W.; Bao, L.J.; Wu, K.Y.; Shan, R.P.; Hu, X.Y.; Fu, Y.H.; Zhao, C.J. Endogenous retroviruses modulate the susceptibility of mice to- Staphylococcus aureus induced mastitis by activating cGAS-STING signaling. Int. Immunopharmacol. 2024, 142, 113171. [Google Scholar] [CrossRef]
- Liu, J.; Min, S.J.; Kim, D.; Park, J.; Park, E.; Koh, Y.; Shin, D.Y.; Kim, T.K.; Byun, J.M.; Yoon, S.S.; et al. Epigenetic priming improves salvage chemotherapy in diffuse large B-cell lymphoma via endogenous retrovirus-induced cGAS-STING activation. Clin. Epigenet. 2023, 15, 75. [Google Scholar] [CrossRef]
- Chen, S.H.; Hu, X.M.; Cui, I.H.; Wu, S.G.; Dou, C.F.; Liu, Y.Y.; Sun, Z.; Xue, S.L.; Geng, T.Y.; Liu, Z.P.; et al. An endogenous retroviral element exerts an antiviral innate immune function via the derived lncRNA lnc-ALVE1-AS1. Antivir. Res. 2019, 170, 104571. [Google Scholar] [CrossRef]
- Zhou, B.; Qi, F.; Wu, F.Y.; Nie, H.B.; Song, Y.F.; Shao, L.; Han, J.X.; Wu, Z.; Saiyin, H.X.G.; Wei, G.; et al. Endogenous Retrovirus-Derived Long Noncoding RNA Enhances Innate Immune Responses via Derepressing RELA Expression. mBio 2019, 10, e00937-19. [Google Scholar] [CrossRef]
- Chuong, E.B.; Elde, N.C.; Feschotte, C. Regulatory evolution of innate immunity through co-option of endogenous retroviruses. Science 2016, 351, 1083–1087. [Google Scholar] [CrossRef]
- Jin, X.Y.; Li, X.Y.; Guan, F.; Zhang, J.H. Human Endogenous Retroviruses and Toll-Like Receptors. Viral Immunol. 2023, 36, 73–82. [Google Scholar] [CrossRef]
- Dembny, P.; Newman, A.G.; Singh, M.; Hinz, M.; Szczepek, M.; Krüger, C.; Adalbert, R.; Dzaye, O.; Trimbuch, T.; Wallach, T.; et al. Human endogenous retrovirus HERV-K(HML-2) RNA causes neurodegeneration through Toll-like receptors. JCI Insight 2020, 5, e131093. [Google Scholar] [CrossRef] [PubMed]
- Bergallo, M.; Loiacono, E.; Gambarino, S.; Montanari, P.; Galliano, I.; Coppo, R. Toll-like receptors are essential for the control of endogenous retrovirus expression in Idiopathic Nephrotic Syndrome. Minerva Urol. Nefrol. 2017, 69, 201–208. [Google Scholar] [CrossRef]
- Benit, L.; DeParseval, N.; Casella, J.F.; Callebaut, I.; Cordonnier, A.; Heidmann, T. Cloning of a new murine endogenous retrovirus; MuERV-L, with strong similarity to the human HERV-L element and with a gag coding sequence closely related to the Fv1 restriction gene. J. Virol. 1997, 71, 5652–5657. [Google Scholar] [CrossRef]
- Goldstone, D.C.; Walker, P.A.; Calder, L.J.; Coombs, P.J.; Kirkpatrick, J.; Ball, N.J.; Hilditch, L.; Yap, M.W.; Rosenthal, P.B.; Stoye, J.P.; et al. Structural studies of postentry restriction factors reveal antiparallel dimers that enable avid binding to the HIV-1 capsid lattice. Proc. Natl. Acad. Sci. USA 2014, 111, 9609–9614. [Google Scholar] [CrossRef] [PubMed]
- Yap, M.W.; Young, G.R.; Varnaite, R.; Morand, S.; Stoye, J.P. Duplication and divergence of the retrovirus restriction gene in Mus caroli allows protection from multiple retroviruses. PLoS Genet. 2020, 16, e1008471. [Google Scholar] [CrossRef] [PubMed]
- Jolicoeur, P.; Baltimore, D. Effect of Fv-1 Gene Product on Proviral DNA Formation and Integration in Cells Infected with Murine Leukemia Viruses. Proc. Natl. Acad. Sci. USA 1976, 73, 2236–2240. [Google Scholar] [CrossRef]
- Boso, G.; Lam, O.; Bamunusinghe, D.; Oler, A.J.; Wollenberg, K.; Liu, Q.P.; Shaffer, E.; Kozak, C.A. Patterns of Coevolutionary Adaptations across Time and Space in Mouse Gammaretroviruses and Three Restrictive Host Factors. Viruses 2021, 13, 1864. [Google Scholar] [CrossRef]
- Bassin, R.H.; Ruscetti, S.; Ali, I.; Haapala, D.K.; Rein, A. Normal DBA/2 mouse cells synthesize a glycoprotein which interferes with MCF virus infection. Virology 1982, 123, 139–151. [Google Scholar] [CrossRef]
- Wu, T.Y.; Yan, Y.H.; Kozak, C.A. Rmcf2, a xenotropic provirus in the Asian mouse species Mus castaneus, blocks infection by polytropic mouse gammaretroviruses. J. Virol. 2005, 79, 9677–9684. [Google Scholar] [CrossRef] [PubMed]
- Blond, J.L.; Lavillette, D.; Cheynet, V.; Bouton, O.; Oriol, G.; Chapel-Fernandes, S.; Mandrand, B.; Mallet, F.; Cosset, F.L. An envelope glycoprotein of the human endogenous retrovirus HERV-W is expressed in the human placenta and fuses cells expressing the type D mammalian retrovirus receptor. J. Virol. 2000, 74, 3321–3329. [Google Scholar] [CrossRef] [PubMed]
- Lavillette, D.; Marin, M.; Ruggieri, A.; Mallet, F.; Cosset, F.L.; Kabat, D. The envelope glycoprotein of human endogenous retrovirus type W uses a divergent family of amino acid transporters/cell surface receptors. J. Virol. 2002, 76, 6442–6452. [Google Scholar] [CrossRef]
- Heidmann, O.; Béguin, A.; Paternina, J.; Berthier, R.; Deloger, M.; Bawa, O.; Heidmann, T. HEMO, an ancestral endogenous retroviral envelope protein shed in the blood of pregnant women and expressed in pluripotent stem cells and tumors. Proc. Natl. Acad. Sci. USA 2017, 114, E6642–E6651. [Google Scholar] [CrossRef]
- Bischof, P.; Irminger-Finger, I. The human cytotrophoblastic cell, a mononuclear chameleon. Int. J. Biochem. Cell Biol. 2005, 37, 1–16. [Google Scholar] [CrossRef]
- Dupressoir, A.; Lavialle, C.; Heidmann, T. From ancestral infectious retroviruses to bona fide cellular genes: Role of the captured syncytins in placentation. Placenta 2012, 33, 663–671. [Google Scholar] [CrossRef] [PubMed]
- Barzoki, M.G.; Malekshahi, S.S.; Heydarifard, Z.; Mahmodi, M.J.; Soltanghoraee, H. The important biological roles of Syncytin-1 of human endogenous retrovirus W (HERV-W) and Syncytin-2 of HERV-FRD in the human placenta development. Mol. Biol. Rep. 2023, 50, 7901–7907. [Google Scholar] [CrossRef] [PubMed]
- Soe, K.; Andersen, T.L.; Hobolt-Pedersen, A.S.; Bjerregaard, B.; Larsson, L.; Delaissé, J.M. Involvement of human endogenous retroviral syncytin-1 in human osteoclast fusion. Bone 2011, 48, 837–846. [Google Scholar] [CrossRef]
- Coudert, A.E.; Redelsperger, F.; Chabbi-Achengli, Y.; Vernochet, C.; Marty, C.; Decrouy, X.; Heidmann, T.; de Vernejoul, M.C.; Dupressoir, A. Role of the captured retroviral envelope syncytin-B gene in the fusion of osteoclast and giant cell precursors and in bone resorption, analyzed ex vivo and in vivo in syncytin-B knockout mice. Bone Rep. 2019, 11, 100214. [Google Scholar] [CrossRef] [PubMed]
- Kodama, J.; Kaito, T. Osteoclast Multinucleation: Review of Current Literature. Int. J. Mol. Sci. 2020, 21, 5685. [Google Scholar] [CrossRef] [PubMed]
| ERV Type | Retrovirus Type | Species | Virus |
|---|---|---|---|
| Class I ERVs | γ-retroviruses | Ceratotherium simum simum | SimumERV [14] |
| Felis silvestris catus | ERV-DC8/FeLV [15] | ||
| ε-retroviruses | Eptatretus burgeri | EbuERV [16] | |
| Class II ERVs | α-retroviruses | Gallus gallus | ALVE [17] |
| β-retroviruses | Homo sapiens | HML-2/HERVK [18] | |
| Tasmanian devil | MEBrv1/2/4 [19] | ||
| δ-retroviruses | rhinolophid bats | ChirDelta2 [20] | |
| Miniopterus natalensis | MINERVa [21] | ||
| lentiviruses | Pedetes capensis | SpELV [22] | |
| Oryctolagus cuniculus | RELIK [23] | ||
| Class III ERVs | spumaviruses | Daubentonia madagascariensis | PSFVaye [24] |
| Latimeria chalumnae | CoeEFV [25] | ||
| Xenopus tropicalis | XtERV-S [26] |
| Names of ERVs in Original Host Species | Original Host Species | Names of ERVs in New Host Species | New Host Species | Transmission Pathways | Nucleotide Similarity |
|---|---|---|---|---|---|
| RfRV | Tupaia belangeri | RfRV | Pangolin | Bats or rodents | Not reported [116] |
| enJSRV | Ovis aries | enJSRV | Capra hircus | Horizontal transmission | 90% [118] |
| MLERV1 | Myotis lucifugus | FcERV_γ6 | Felis catus | Direct contact or predation | 85% [119] |
| Panthera tigris | |||||
| MLERV1 | Myotis lucifugus | MPERV1 | Manis pentadactyla | Direct contact | 85% [119] |
| ERV-Spuma-Spu | Sphenodon punctatus | ERV-Spuma-Ppi ERV-Spuma-Gja | Paroedura picta | Unknown non- reptile host | Not reported [120] |
| Gekko japonicus | |||||
| RnERV-K8e | Rattus norvegicus | MmERV-K10c | Mus musculus | Not reported | Partial similarity [121] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Jiang, R.; Zhou, J.; Liu, Y.; Zhou, G.; Fan, D.; Xiang, L.; Chen, Y.; Shao, J. Endogenous Retroviruses in Host-Virus Coevolution: From Genomic Domestication to Functional Innovation. Genes 2025, 16, 964. https://doi.org/10.3390/genes16080964
Jiang R, Zhou J, Liu Y, Zhou G, Fan D, Xiang L, Chen Y, Shao J. Endogenous Retroviruses in Host-Virus Coevolution: From Genomic Domestication to Functional Innovation. Genes. 2025; 16(8):964. https://doi.org/10.3390/genes16080964
Chicago/Turabian StyleJiang, Ruqi, Jingjun Zhou, Yue Liu, Guanjin Zhou, Dongdong Fan, Lixin Xiang, Ye Chen, and Jianzhong Shao. 2025. "Endogenous Retroviruses in Host-Virus Coevolution: From Genomic Domestication to Functional Innovation" Genes 16, no. 8: 964. https://doi.org/10.3390/genes16080964
APA StyleJiang, R., Zhou, J., Liu, Y., Zhou, G., Fan, D., Xiang, L., Chen, Y., & Shao, J. (2025). Endogenous Retroviruses in Host-Virus Coevolution: From Genomic Domestication to Functional Innovation. Genes, 16(8), 964. https://doi.org/10.3390/genes16080964






