Transcriptomic Dynamics of Rice Varieties with Differential Cold Tolerance Under Low-Temperature Stress During Grain-Filling Stage
Abstract
1. Introduction
2. Materials and Methods
2.1. Plant Material
2.2. Temperature Treatments and Sampling
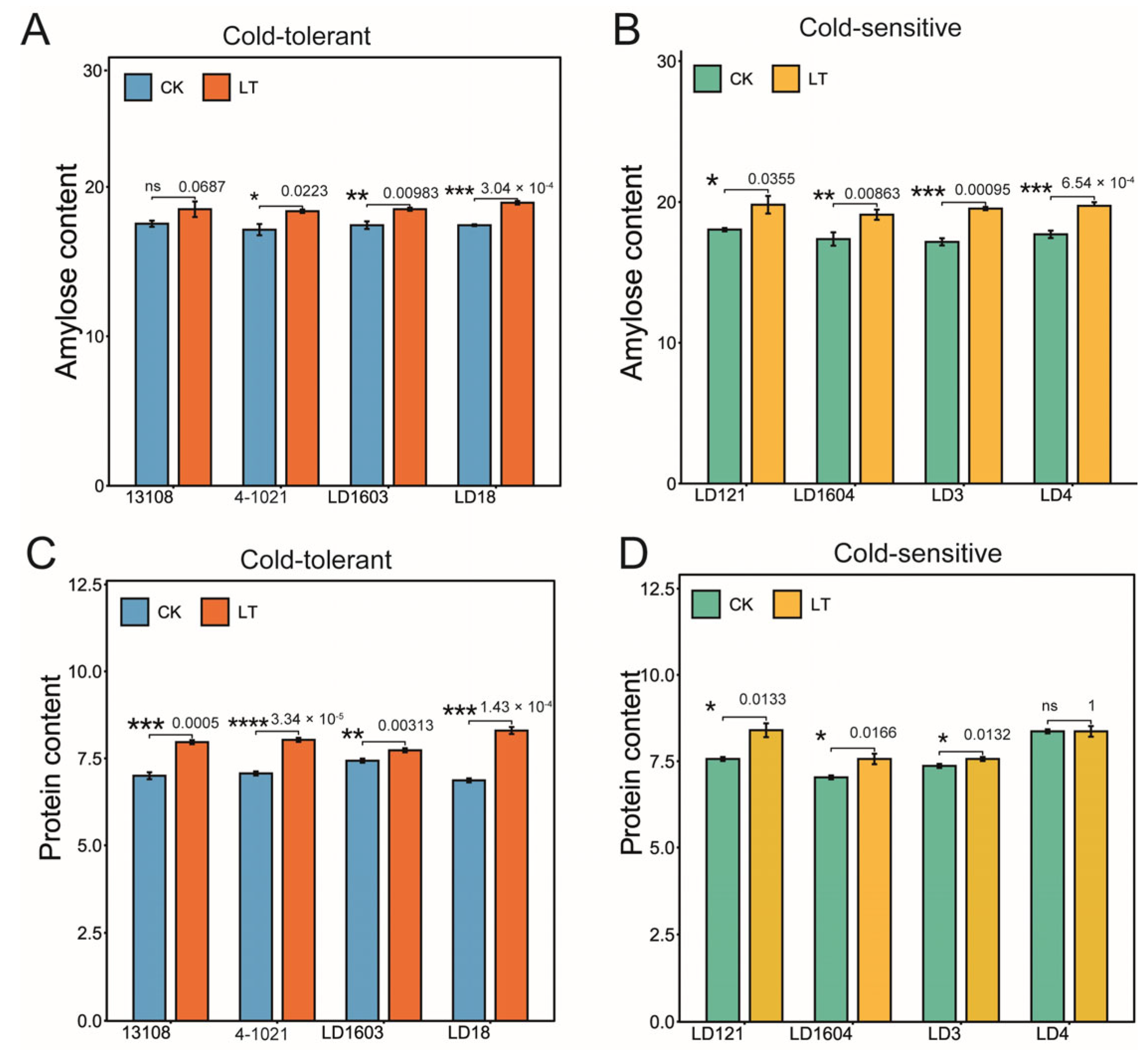
2.3. Determination of Amylose Content
2.4. Determination of Protein Content
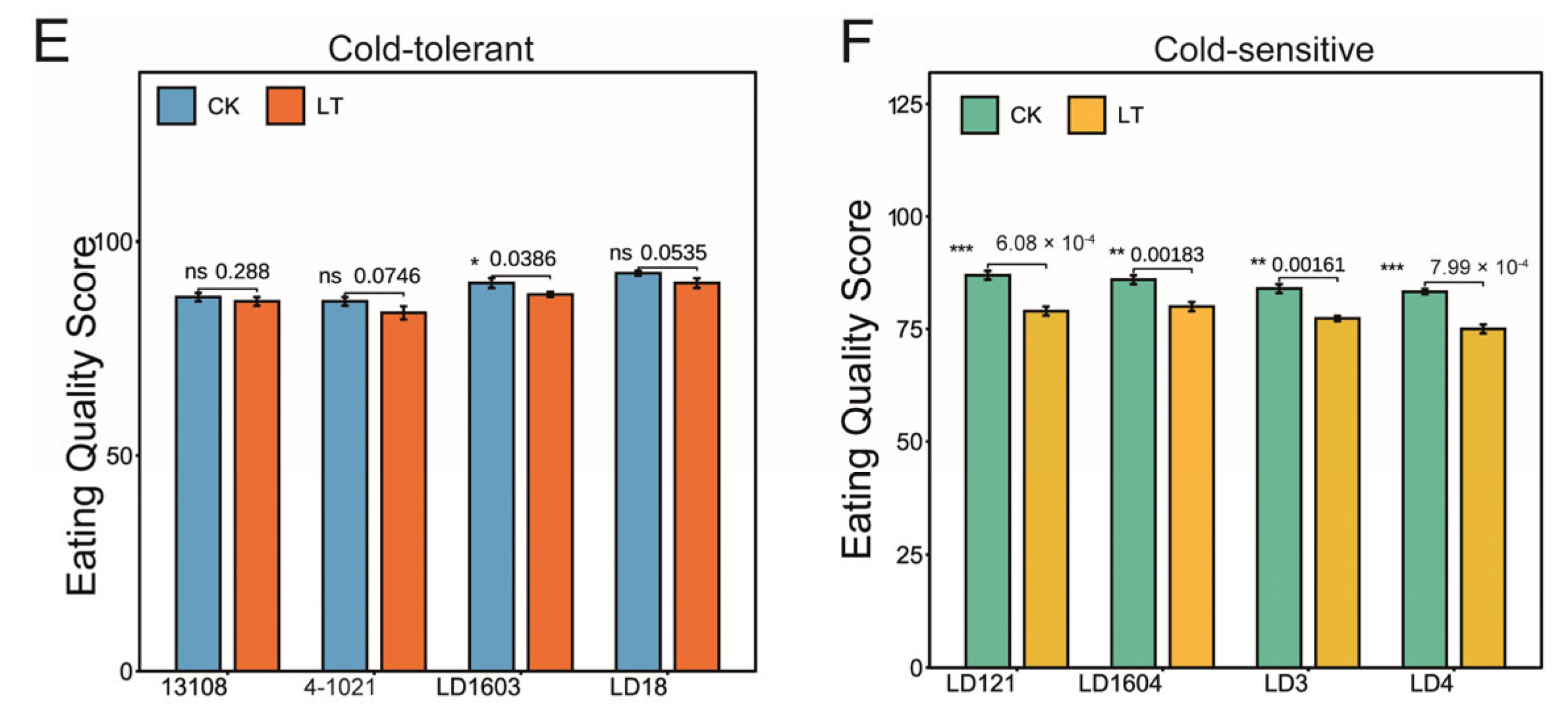
2.5. Taste Quality Evaluation Using Descriptive Analysis (DA)
2.6. RNA-Seq and Data Analysis
2.7. Correlation Analysis
2.8. Gene Co-Expression Network Analysis
3. Results
3.1. Low-Temperature Treatment Affected the Taste Quality of Rice in Both Cold-Tolerant and Cold-Sensitive Varieties
3.2. Quality Assessment and Summary Statistics of Transcriptome Data
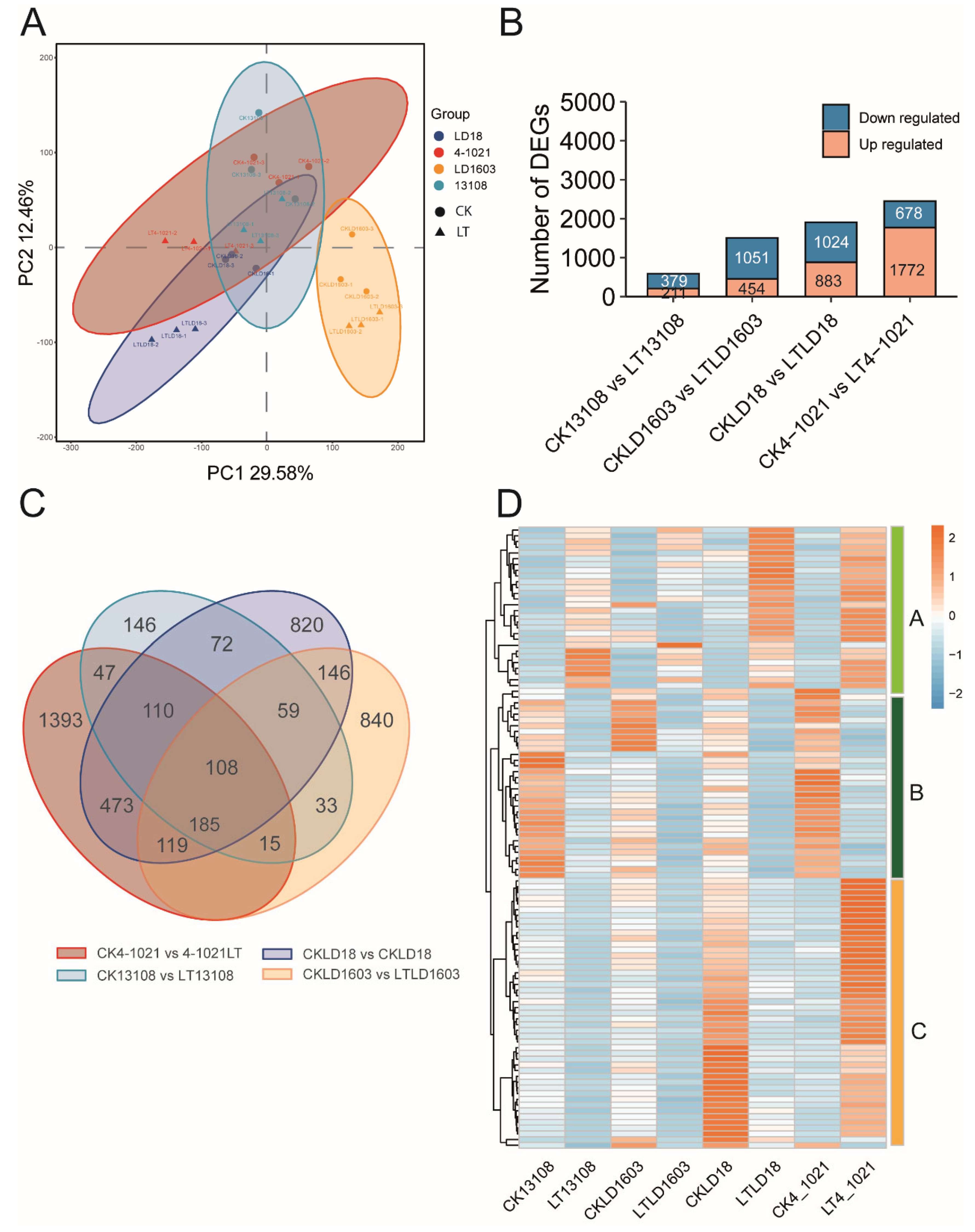
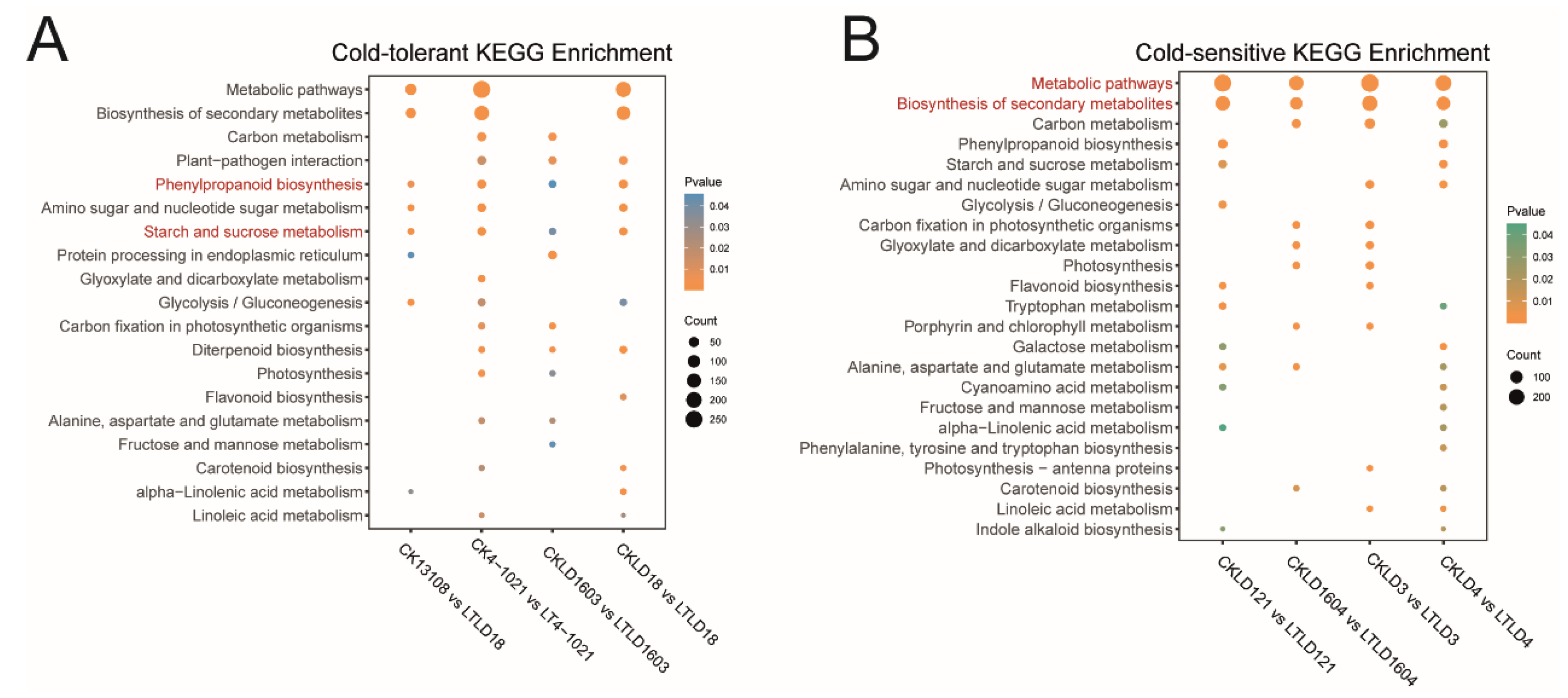
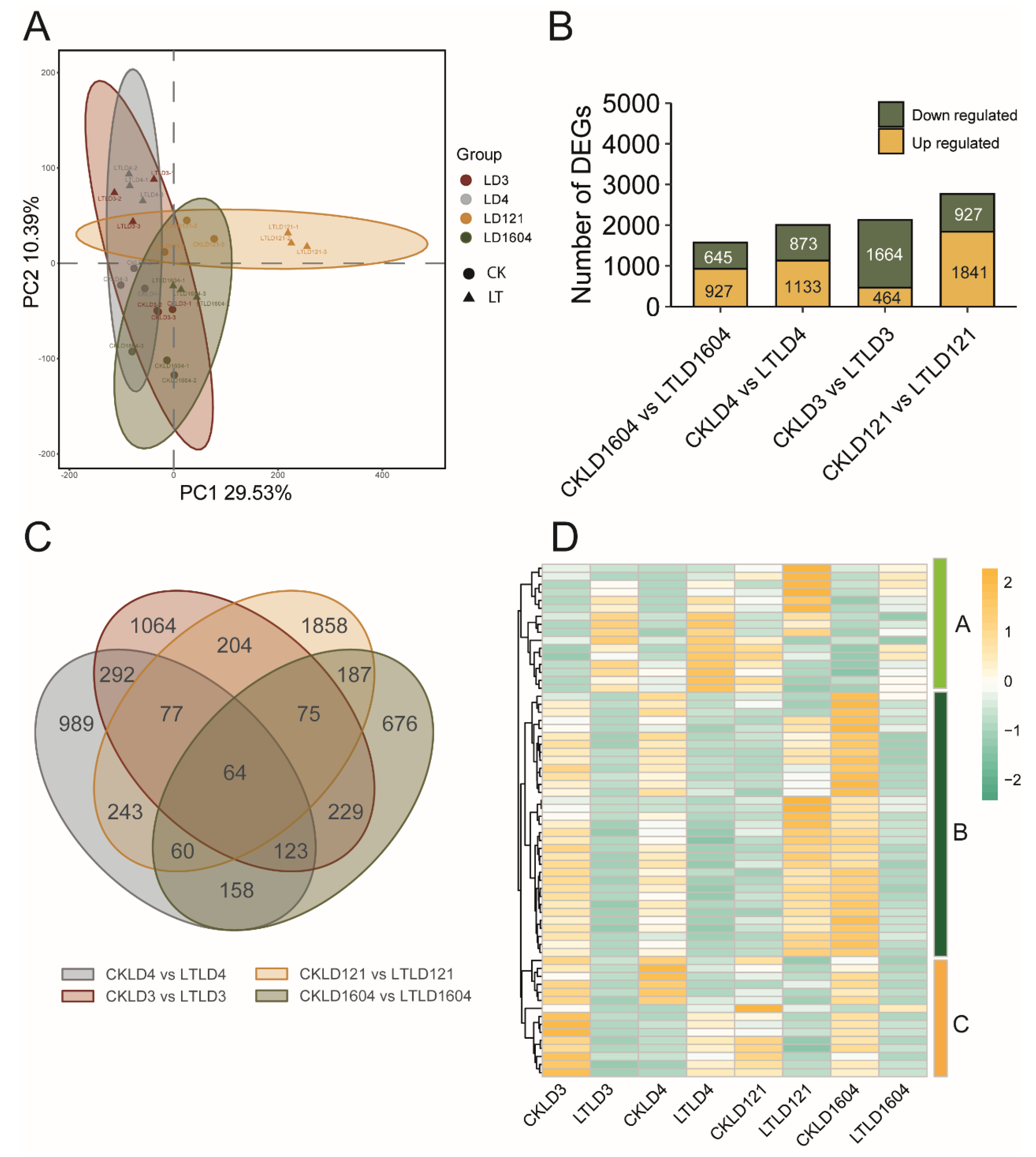
3.3. Comparative Transcriptomic Analysis of Cold-Tolerant Rice Varieties Between CK and LT Treatments
3.4. Comparative Transcriptomic Analysis of Cold-Sensitive Rice Varieties Between CK and LT Treatments
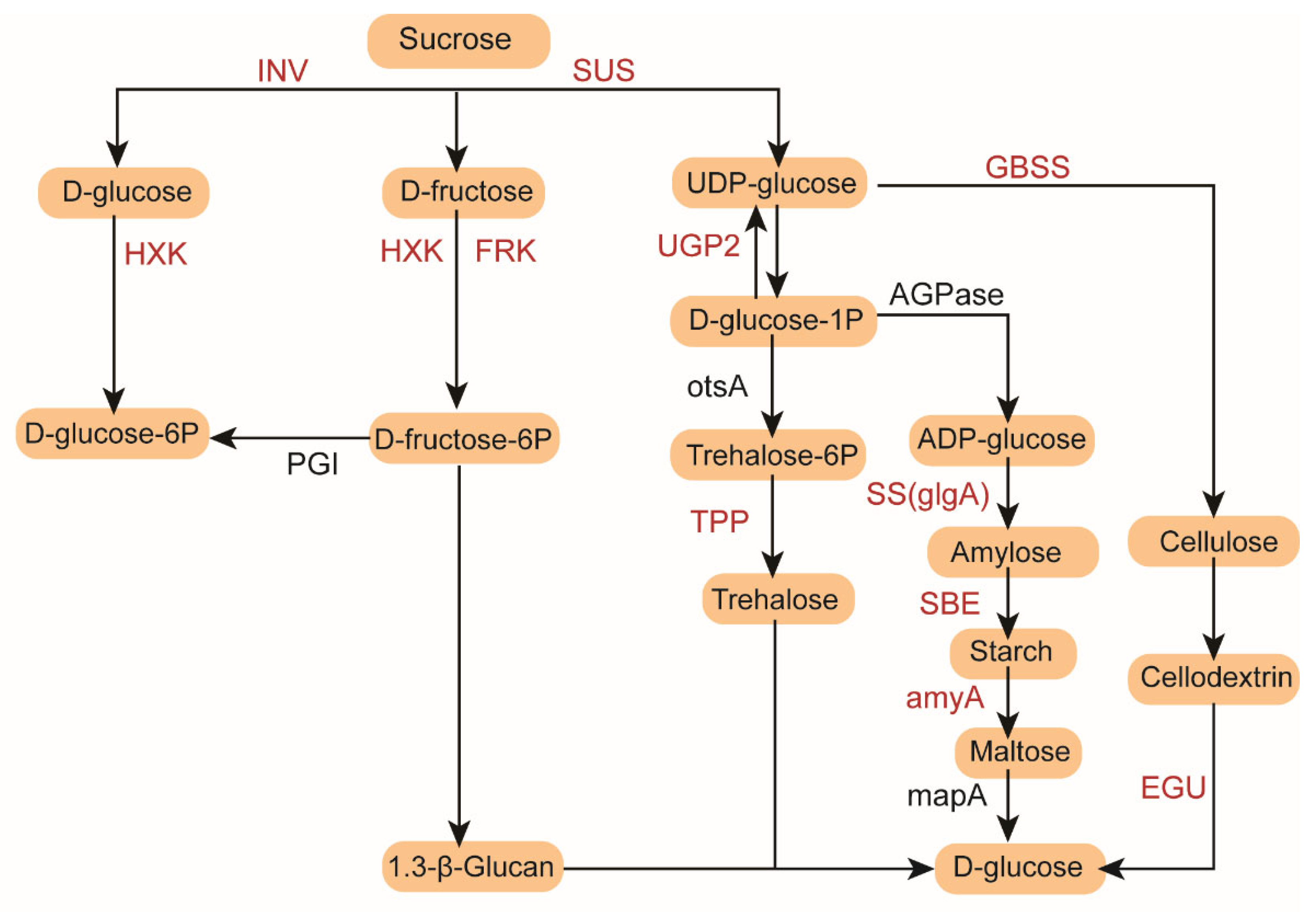
3.5. Transcriptomic Regulation of Starch and Sucrose Metabolism Pathways Under Low-Temperature Stress in Cold-Tolerant and Cold-Sensitive Rice Varieties
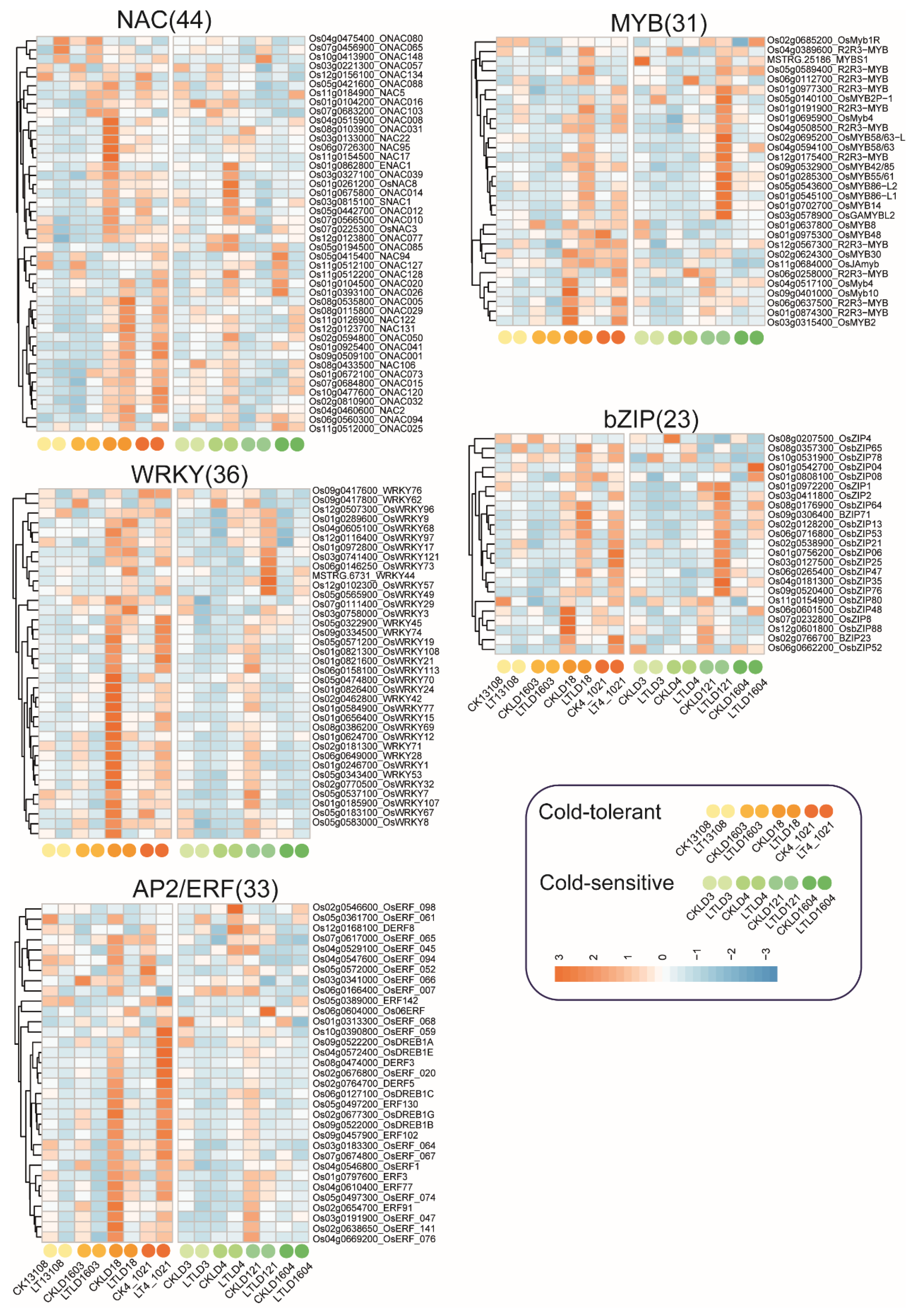
3.6. Differential Expression of Transcription Factor (TF) Families in Cold-Tolerant and Cold-Sensitive Rice Under Low-Temperature Stress
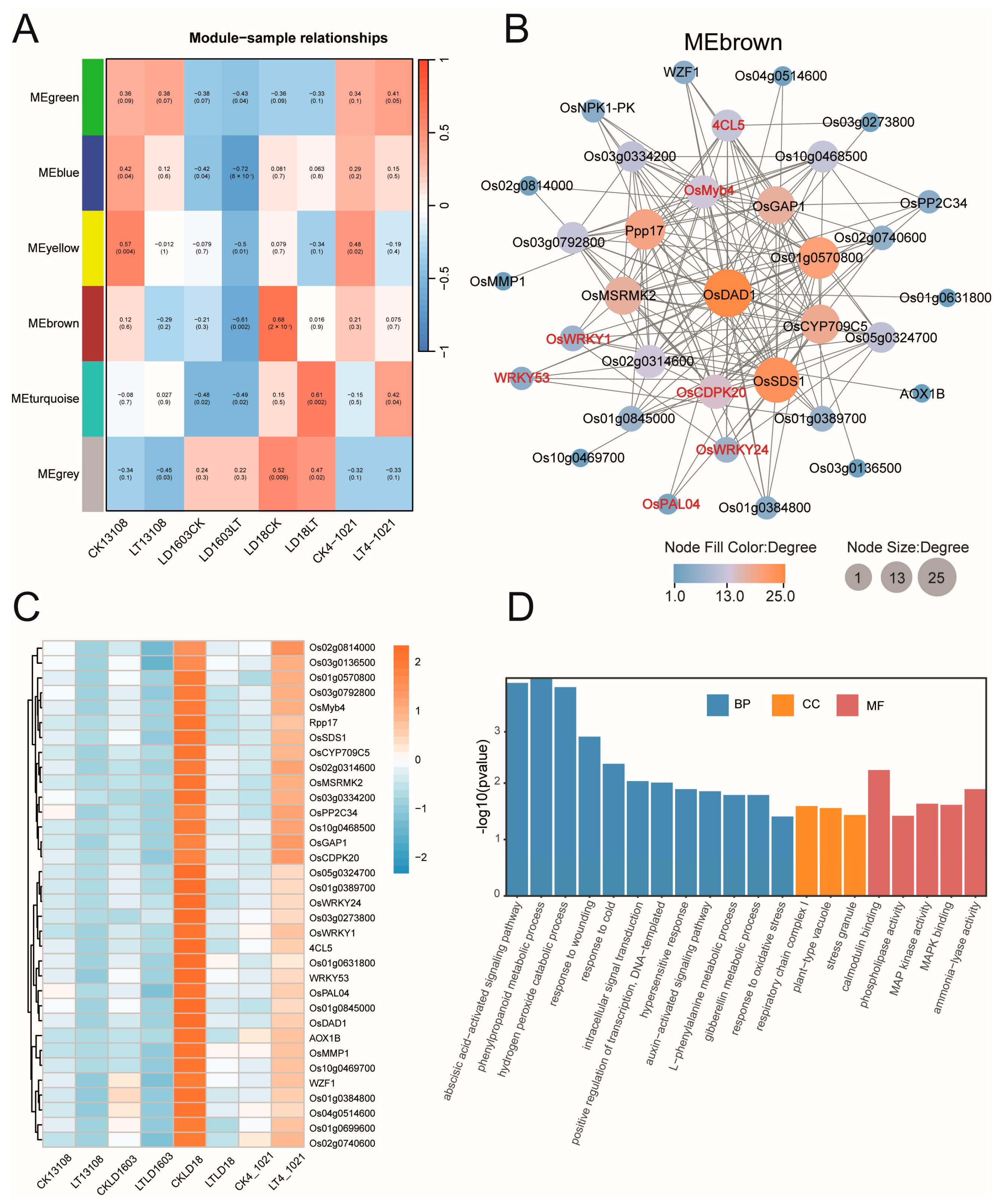
3.7. WGCNA Revealed Key Genes in Cold-Tolerant Rice Under Cold Stress
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Ali, I.; Tang, L.; Dai, J.; Kang, M.; Zhu, Y. Responses of Grain Yield and Yield Related Parameters to Post-Heading Low-Temperature Stress in Japonica Rice. Plants 2021, 10, 1425. [Google Scholar] [CrossRef]
- Deng, F.; Li, Q.; Chen, H.; Zeng, Y.; Ren, W. Relationship between chalkiness and the structural and thermal properties of rice starch after shading during grain-filling stage. Carbohydr. Polym. 2021, 252, 117212. [Google Scholar] [CrossRef]
- Xu, C.; Yang, F.; Tang, X.; Lu, B.; Li, Z.; Liu, Z.; Ding, Y.; Ding, C.; Li, G. Super Rice With High Sink Activities Has Superior Adaptability to Low Filling Stage Temperature. Front. Plant Sci. 2021, 12, 729021. [Google Scholar] [CrossRef]
- Sharma, P.; Sharma, N.; Deswal, R. The molecular biology of the low-temperature response in plants. Bioessays 2010, 27, 1048–1059. [Google Scholar] [CrossRef]
- Oliver, S.N.; Dennis, E.S.; Dolferus, R. ABA Regulates Apoplastic Sugar Transport and is a Potential Signal for Cold-Induced Pollen Sterility in Rice. Plant Cell Physiol. 2007, 48, 1319–1330. [Google Scholar] [CrossRef] [PubMed]
- Aya, K.; Ueguchi-Tanaka, M.; Kondo, M.; Hamada, K.; Yano, K.; Nishimura, M.; Matsuoka, M. Gibberellin Modulates Anther Development in Rice via the Transcriptional Regulation of GAMYB. Plant Cell Online 2009, 21, 1453–1472. [Google Scholar] [CrossRef]
- Oda, S.; Kaneko, F.; Yano, K.; Fujioka, T.; Masuko, H.; Park, J.I.; Kikuchi, S.; Hamada, K.; Endo, M.; Nagano, K. Morphological and gene expression analysis under cool temperature conditions in rice anther development. Genes. Genet. Syst. 2010, 85, 107. [Google Scholar] [CrossRef]
- Sakata, T.; Oda, S.; Tsunaga, Y.; Shomura, H.; Kawagishi-Kobayashi, M.; Aya, K.; Saeki, K.; Endo, T.; Nagano, K.; Kojima, M. Reduction of Gibberellin by Low Temperature Disrupts Pollen Development in Rice. Plant Physiol. 2014, 164, 2011–2019. [Google Scholar] [CrossRef] [PubMed]
- Oliver, S.N.; Dongen, J.T.V.; Alfred, S.C.; Mamun, E.A.; Dolferus, R. Cold-induced repression of the rice anther-specific cell wall invertase gene OSINV4 is correlated with sucrose accumulation and pollen sterility. Plant Cell Environ. 2010, 28, 1534–1551. [Google Scholar] [CrossRef]
- Liu, C.-T.; Wang, W.; Mao, B.-G.; Chu, C. Cold stress tolerance in rice: Physiological changes, molecular mechanism, and future prospects. Hereditas 2018, 40, 171–185. [Google Scholar] [PubMed]
- Huang, J.; Sun, S.; Xu, D.; Lan, H.; Sun, H.; Wang, Z.; Bao, Y.; Wang, J.; Tang, H.; Zhang, H. A TFIIIA-type zinc finger protein confers multiple abiotic stress tolerances in transgenic rice (Oryza sativa L.). Plant Mol. Biol. 2012, 80, 337–350. [Google Scholar] [CrossRef]
- Bonnecarrère, V.; Borsani, O.; Díaz, P.; Capdevielle, F.; Blanco, P.; Monza, J. Response to photoxidative stress induced by cold in japonica rice is genotype dependent. Plant Sci. 2011, 180, 726–732. [Google Scholar] [CrossRef]
- Xu, Y.; Wang, R.; Wang, Y.; Zhang, L.; Yao, S. A point mutation in LTT1 enhances cold tolerance at the booting stage in rice. Plant Cell Environ. 2020, 43, 992–1007. [Google Scholar] [CrossRef]
- Guo, Z.; Cai, L.; Liu, C.; Huang, C.; Guo, T. Global analysis of differentially expressed genes between two Japonica rice varieties induced by low temperature during the booting stage by RNA-Seq. R. Soc. Open Sci. 2020, 7, 192243. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Wu, X.; Chen, Y.; Wu, C.; Long, W.; Zhu, S. Transcriptomic profiling of the cold stress and recovery responsiveness of two contrasting Guizhou HE rice genotypes. Genes. Genom. 2023, 45, 401–412. [Google Scholar] [CrossRef]
- Lei, Z. Transcriptome Analysis Revealed the Dynamic and Rapid Transcriptional Reprogramming Involved in Cold Stress and Related Core Genes in the Rice Seedling Stage. Int. J. Mol. Sci. 2023, 24, 1914. [Google Scholar] [CrossRef]
- Hong, W.J.; Jiang, X.; Ahn, H.R.; Choi, J.; Jung, K.H. Systematic Analysis of Cold Stress Response and Diurnal Rhythm Using Transcriptome Data in Rice Reveals the Molecular Networks Related to Various Biological Processes. Int. J. Mol. Sci. 2020, 21, 6872. [Google Scholar] [CrossRef] [PubMed]
- Zeng, Z.; Zhang, S.; Li, W.; Chen, B.; Li, W. Gene-coexpression network analysis identifies specific modules and hub genes related to cold stress in rice. BMC Genom. 2022, 23, 251. [Google Scholar] [CrossRef]
- Zhang, G.Q. Prospects of utilization of inter-subspecific heterosis between indica and japonica rice. J. Integr. Agric. 2020, 19, 1–10. [Google Scholar] [CrossRef]
- Zhang, Q.; Maroof, M.S.; Lu, T.Y.; Shen, B.J.T.; Genetics, A. Genetic diversity and differentiation of indica and japonica rice detected by RFLP analysis. Theor. Appl. Genet. 1992, 83, 495–499. [Google Scholar] [CrossRef]
- Gough, N.R. Rice that tolerates a chill. Sci. Signal. 2015, 8, ec76. [Google Scholar] [CrossRef]
- Maleki, C.; Oliver, P.; Lewin, S.; Liem, G.; Keast, R. Preference mapping of different water-to-rice ratios in cooked aromatic white jasmine rice. J. Food Sci. 2020, 85, 1576–1585. [Google Scholar] [CrossRef]
- Man, J.; Yang, Y.; Zhang, C.; Zhou, X.; Dong, Y.; Zhang, F.; Liu, Q.; Wei, C. Structural changes of high-amylose rice starch residues following in vitro and in vivo digestion. J. Agric. Food Chem. 2012, 60, 9332–9341. [Google Scholar] [CrossRef]
- GB/T 17891-2017; High Quality Paddy. China Standards Press: Beijing, China, 2017.
- Yamamoto, T.; Horisue, N.; Ikeda, R. Rice Breeding Manual; Yokendo Ltd.: Tokyo, Japan, 1996; pp. 74–124. [Google Scholar]
- GB/T 5492-2008; General Administration of Quality Supervision, Inspection and Quarantine of the People’s Republic of China; Stand-ardization Administration of the People’s Republic of China. Inspection of Grain and Oils—Identification of Colour, Odour and Taste of Grain and Oilseeds. Standards Press of China: Beijing, China, 2008.
- Rio, D.C.; Ares, M.; Hannon, G.J.; Nilsen, T.W. Purification of RNA using TRIzol (TRI reagent). Cold Spring Harb. Protoc. 2010, 2010, pdb.prot5439. [Google Scholar] [CrossRef]
- Chen, S.; Zhou, Y.; Chen, Y.; Gu, J. fastp: An ultra-fast all-in-one FASTQ preprocessor. Bioinformatics 2018, 34, i884–i890. [Google Scholar] [CrossRef]
- Kim, D.; Langmead, B.; Salzberg, S.L. HISAT: A fast spliced aligner with low memory requirements. Nat. Methods 2015, 12, 357–360. [Google Scholar] [CrossRef]
- Pertea, M.; Pertea, G.M.; Antonescu, C.M.; Chang, T.C.; Mendell, J.T.; Salzberg, S.L. StringTie enables improved reconstruction of a transcriptome from RNA-seq reads. Nat. Biotechnol. 2015, 33, 290–295. [Google Scholar] [CrossRef]
- Dewey, C.N.; Bo, L. RSEM: Accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinform. 2011, 12, 323. [Google Scholar] [CrossRef]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef]
- Shannon, P.; Markiel, A.; Ozier, O.; Baliga, N.S.; Wang, J.T.; Ramage, D.; Amin, N.; Schwikowski, B.; Ideker, T. Cytoscape: A Software Environment for Integrated Models of Biomolecular Interaction Networks. Genome Res. 2003, 13, 2498–2504. [Google Scholar] [CrossRef]
- Zhang, W. Responses of indica rice yield and quality to extreme high and low temperatures during the reproductive period. Eur. J. Agron. 2019, 106, 30–38. [Google Scholar] [CrossRef]
- Ai, X.; Xiong, R.; Tan, X.; Wang, H.; Zeng, Y.; Huang, S.; Shang, Q.; Pan, X.; Shi, Q.; Zhang, J.; et al. Low temperature and light combined stress after heading on starch fine structure and physicochemical properties of late-season indica rice with different grain quality in southern China. Food Res. Int. 2023, 164, 112320. [Google Scholar] [CrossRef]
- Srinivas, J. Effects of ripening temperature on starch structure and gelatinization, pasting, and cooking properties in rice (Oryza sativa). J. Agric. Food Chem. 2015, 63, 3085–3093. [Google Scholar] [CrossRef]
- Zhang, H. The effects of chilling stress after anthesis on the physicochemical properties of rice (Oryza sativa L) starch. Food Chem. 2017, 237, 936–941. [Google Scholar] [CrossRef]
- Shi, W. The deterioration of starch physiochemical and minerals in high-quality indica rice under low-temperature stress during grain filling. Front. Plant Sci. 2024, 14, 1295003. [Google Scholar]
- Huabing, D. Grain Quality Characterization of Hybrid Rice Restorer Lines with Resilience to Suboptimal Temperatures during Filling Stage. Foods 2022, 11, 3513. [Google Scholar] [CrossRef]
- Gilbert, R.G. The biosynthesis, structure and gelatinization properties of starches from wild and cultivated African rice species (Oryza barthii and Oryza glaberrima). Carbohydr. Polym. 2015, 129, 92–100. [Google Scholar] [CrossRef]
- Xu, Q. Effect of indica pedigree on eating and cooking quality in rice backcross inbred lines of indica and japonica crosses. Breed. Sci. 2017, 67, 450–458. [Google Scholar] [CrossRef][Green Version]
- Hu, Y.; Li, L.; Tian, J.; Zhang, C.; Wang, J.; Yu, E.; Xing, Z.; Guo, B.; Wei, H.; Huo, Z.; et al. Effects of dynamic low temperature during the grain filling stage on starch morphological structure, physicochemical properties, and eating quality of soft japonica rice. Cereal Chem. 2020, 97, 540–550. [Google Scholar] [CrossRef]
- Sun, C.; Wang, Y.; Yang, X.; Tang, L.; Wan, C.; Liu, J.; Chen, C.; Zhang, H.; He, C.; Liu, C.; et al. MATE transporter GFD1 cooperates with sugar transporters, mediates carbohydrate partitioning, and controls grain filling duration, grain size and number in rice. Plant Biotechnol. J. 2022, 21, 621–634. [Google Scholar] [CrossRef]
- Zhang, W. An integrated physiology and proteomics analysis reveals the response of wheat grain to low temperature stress during booting. J. Integr. Agric. 2025, 24, 114–131. [Google Scholar] [CrossRef]
- Zhang, W.; Zhao, Y.; Li, L.; Xu, X.; Yang, L.; Luo, Z.; Wang, B.; Ma, S.; Fan, Y.; Huang, Z. The Effects of Short-Term Exposure to Low Temperatures During the Booting Stage on Starch Synthesis and Yields in Wheat Grain. Front. Plant Sci. 2021, 12, 684784. [Google Scholar] [CrossRef] [PubMed]
- Qu, Z.; Jia, Y.; Duan, Y.; Chen, H.; Wang, X.; Zheng, H.; Liu, H.; Wang, J.; Zou, D.; Zhao, H. Integrated Isoform Sequencing and Dynamic Transcriptome Analysis Reveals Diverse Transcripts Responsible for Low Temperature Stress at Anther Meiosis Stage in Rice. Front. Plant Sci. 2021, 12, 795834. [Google Scholar] [CrossRef] [PubMed]
- Huang, S. Molecular mechanisms underlying low temperature inhibition of grain filling in maize (Zea mays L.): Coordination of growth and cold responses. Plant J. Cell Mol. Biol. 2024, 119, 982–997. [Google Scholar]
- Chen, Y. Integrated physiological, transcriptomic and metabolomic analyses reveal the mechanism of peanut kernel weight reduction under waterlogging stress. Plant Cell Environ. 2024, 47, 3198–3214. [Google Scholar]
- Angelika, M. Localization of sucrose synthase in wheat roots: Increased in situ activity of sucrose synthase correlates with cell wall thickening by cellulose deposition under hypoxia. Planta 2003, 217, 252–260. [Google Scholar] [CrossRef]
- Pilar, C. Differential expression of two types of sucrose synthase-encoding genes in wheat in response to anaerobiosis, cold shock and light. Gene 1990, 88, 167–172. [Google Scholar] [CrossRef]
- Varshney, R.K. Trehalose: A sugar molecule involved in temperature stress management in plants. Crop J. 2024, 12, 1–16. [Google Scholar]
- Ge, L.F.; Chao, D.Y.; Shi, M.; Zhu, M.Z.; Gao, J.P.; Lin, H.X. Overexpression of the trehalose-6-phosphate phosphatase gene OsTPP1 confers stress tolerance in rice and results in the activation of stress responsive genes. Planta 2008, 228, 191–201. [Google Scholar] [CrossRef]
- Li, J. Natural variation of indels in the CTB3 promoter confers cold tolerance in japonica rice. Nat. Commun. 2025, 16, 1613. [Google Scholar] [CrossRef]
- Liu, H.; Si, X.; Wang, Z.; Cao, L.; Gao, L.; Wang, K.; Jiao, C.; Zhuang, L.; Liu, Y.; Hou, J.; et al. TaTPP-7A positively feedback regulates grain filling and wheat grain yield through T6P-SnRK1 signaling pathway and sugar-ABA interaction. Plant Biotechnol. J. 2023, 21, 1159–1175. [Google Scholar] [CrossRef] [PubMed]
- Kazuko, Y. Transcriptional regulatory network of plant cold-stress responses. Trends Plant Sci. 2022, 27, 922–935. [Google Scholar] [CrossRef]
- Shi, Y.; Ding, Y.; Yang, S. Molecular Regulation of CBF Signaling in Cold Acclimation. Trends Plant Sci. 2018, 23, 623–637. [Google Scholar] [CrossRef]
- Zhang, Z. Changes in rice disasters across China in recent decades and the meteorological and agronomic causes. Reg. Environ. Change 2013, 13, 743–759. [Google Scholar]
- Wang, J.; Ren, Y.; Liu, X.; Luo, S.; Zhang, X.; Liu, X.; Lin, Q.; Zhu, S.; Wan, H.; Yang, Y.; et al. Transcriptional Activation and Phosphorylation of OsCNGC9 Confer Enhanced Chilling Tolerance in Rice. Mol. Plant 2020, 14, 315–329. [Google Scholar] [CrossRef]
- Tang, J.; Tian, X.; Mei, E.; He, M.; Gao, J.; Yu, J.; Xu, M.; Liu, J.; Song, L.; Li, X.; et al. WRKY53 negatively regulates rice cold tolerance at the booting stage by fine-tuning anther gibberellin levels. Plant Cell 2022, 34, 4495–4515. [Google Scholar] [CrossRef]
- Immacolata, C. Overexpression of the rice Osmyb4 gene increases chilling and freezing tolerance of Arabidopsis thaliana plants. Plant J. Cell Mol. Biol. 2004, 37, 115–127. [Google Scholar]
- Wang, Y. OsJRL negatively regulates rice cold tolerance via interfering phenylalanine metabolism and flavonoid biosynthesis. Plant Cell Environ. 2024, 47, 4071–4085. [Google Scholar]
- Zhou, Y. CPK27 enhances cold tolerance by promoting flavonoid biosynthesis through phosphorylating HY5 in tomato. New Phytol. 2025, 246, 2174–2191. [Google Scholar] [CrossRef] [PubMed]
- Jiang, B.; Shi, Y.; Peng, Y.; Jia, Y.; Yan, Y.; Dong, X.; Li, H.; Dong, J.; Li, J.; Gong, Z.; et al. Cold-Induced CBF–PIF3 Interaction Enhances Freezing Tolerance by Stabilizing the phyB Thermosensor in Arabidopsis. Mol. Plant 2020, 13, 894–906. [Google Scholar] [CrossRef] [PubMed]
- Saijo, Y.; Hata, S.; Kyozuka, J.; Shimamoto, K.; Izui, K. Over-expression of a single Ca2+-dependent protein kinase confers both cold and salt/drought tolerance on rice plants. Plant J. Cell Mol. Biol. 2000, 23, 319–327. [Google Scholar] [CrossRef] [PubMed]
- Setsuko, K. OsCDPK13, a calcium-dependent protein kinase gene from rice, is induced by cold and gibberellin in rice leaf sheath. Plant Mol. Biol. 2004, 55, 541–552. [Google Scholar]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Cao, L.; Wang, X.; Liu, Y.; Ding, G.; Zhou, J.; Lei, L.; Bai, L.; Luo, Y.; Sun, S. Transcriptomic Dynamics of Rice Varieties with Differential Cold Tolerance Under Low-Temperature Stress During Grain-Filling Stage. Genes 2025, 16, 950. https://doi.org/10.3390/genes16080950
Cao L, Wang X, Liu Y, Ding G, Zhou J, Lei L, Bai L, Luo Y, Sun S. Transcriptomic Dynamics of Rice Varieties with Differential Cold Tolerance Under Low-Temperature Stress During Grain-Filling Stage. Genes. 2025; 16(8):950. https://doi.org/10.3390/genes16080950
Chicago/Turabian StyleCao, Liangzi, Xueyang Wang, Yingying Liu, Guohua Ding, Jinsong Zhou, Lei Lei, Liangming Bai, Yu Luo, and Shichen Sun. 2025. "Transcriptomic Dynamics of Rice Varieties with Differential Cold Tolerance Under Low-Temperature Stress During Grain-Filling Stage" Genes 16, no. 8: 950. https://doi.org/10.3390/genes16080950
APA StyleCao, L., Wang, X., Liu, Y., Ding, G., Zhou, J., Lei, L., Bai, L., Luo, Y., & Sun, S. (2025). Transcriptomic Dynamics of Rice Varieties with Differential Cold Tolerance Under Low-Temperature Stress During Grain-Filling Stage. Genes, 16(8), 950. https://doi.org/10.3390/genes16080950





