Genetic Variant of the Canine FGF5 Gene for the Hair Length Trait in the Akita: Utility for Hair Coat Variations and Welfare in Conservation Breeding
Abstract
1. Introduction
2. Materials and Methods
2.1. Animals, Sample Collection, and DNA Extraction
2.2. Sanger Sequencing
2.3. Genotyping by Real-Time PCR for the c.578C>T Variant
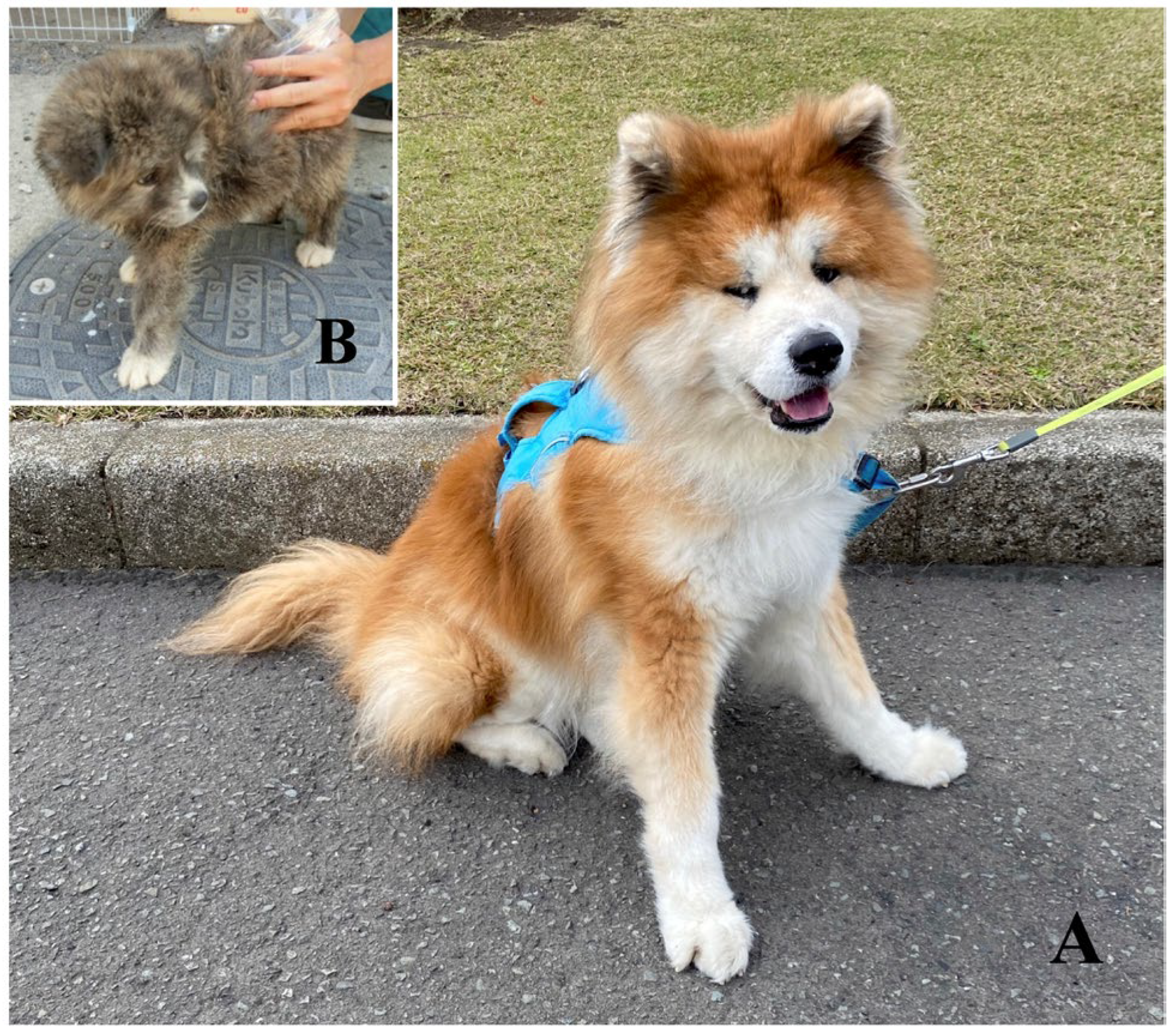
2.4. Scoring Method for Hair Length
2.5. Statistical Analyses
3. Results
3.1. Identification of a Variant Causing Long Hair in Akitas
3.2. Genotyping by Real-Time PCR
3.3. Scoring of Hair Length
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| FGF5 | fibroblast growth factor-5 |
| RSPO2 | R-spondin-2 |
| KRT71 | keratin-71 |
| CFA | dog chromosome |
| OMIA | Online Mendelian Inheritance in Animals |
| NIPPO | Nihonken Hozonkai |
| AKIHO | Akita Inu Hozonkai (Akita Inu Preservation Society) |
| PCR | polymerase chain reaction |
| HWE | Hardy–Weinberg equilibrium |
References
- Cadieu, E.; Neff, M.W.; Quignon, P.; Walsh, K.; Chase, K.; Parker, H.G.; Vonholdt, B.M.; Rhue, A.; Boyko, A.; Byers, A.; et al. Coat variation in the domestic dog is governed by variants in three genes. Science 2009, 326, 150–153. [Google Scholar] [CrossRef] [PubMed]
- Housley, D.J.; Venta, P.J. The long and the short of it: Evidence that FGF5 is a major determinant of canine ‘hair’-itability. Anim. Genet. 2006, 37, 309–315. [Google Scholar] [CrossRef] [PubMed]
- Whitaker, D.T.; Ostrander, E.A. Hair of the dog: Identification of a cis-regulatory module predicted to influence canine coat composition. Genes 2019, 10, 323. [Google Scholar] [CrossRef] [PubMed]
- Online Mendelian Inheritance in Animals [OMIA:000439-9615, Hair, Long in Canis Lupus Familiaris (Dog)]. Available online: https://www.omia.org/OMIA000439/9615/ (accessed on 30 July 2025).
- Dierks, C.; Mömke, S.; Philipp, U.; Distl, O. Allelic heterogeneity of FGF5 mutations causes the long-hair phenotype in dogs. Anim. Genet. 2013, 44, 425–431. [Google Scholar] [CrossRef]
- Online Mendelian Inheritance in Animals [OMIA:000439-9685, Hair, Long in Felis Catus (Domestic Cat)]. Available online: https://www.omia.org/OMIA000439/9685/ (accessed on 10 July 2025).
- Drögemüller, C.; Rüfenacht, S.; Wichert, B.; Leeb, T. Mutations within the FGF5 gene are associated with hair length in cats. Anim. Genet. 2007, 38, 218–221. [Google Scholar] [CrossRef]
- Kehler, J.S.; David, V.A.; Schäffer, A.A.; Bajema, K.; Eizirik, E.; Ryugo, D.K.; Hannah, S.S.; O’Brien, S.J.; Menotti-Raymond, M. Four independent mutations in the feline fibroblast growth factor 5 gene determine the long-haired phenotype in domestic cats. J. Hered. 2007, 98, 555–566. [Google Scholar] [CrossRef]
- Shaffer, G.D.; Ballif, B.C.; Meurs, K.; Shaffer, L.G.; Flores-Smith, H. Identification of a novel missense mutation in the fibroblast growth factor 5 gene associated with longhair in the Maine Coon Cat. Hum. Genet. 2021, 140, 1517–1523. [Google Scholar] [CrossRef]
- Fatima, N.; Jia, L.; Liu, B.; Li, L.; Bai, L.; Wang, W.; Zhao, S.; Wang, R.; Liu, E. A homozygous missense mutation in the fibroblast growth factor 5 gene is associated with the long-hair trait in Angora rabbits. BMC Genom. 2023, 24, 298. [Google Scholar] [CrossRef]
- Yu, F.; Liu, Z.; Jiao, S.; Zhang, X.; Bai, C.; Zhang, J.; Yan, S.; Yu, F.; Liu, Z.; Jiao, S.; et al. A nonsense mutation in the FGF5 gene is associated with the long-haired phenotype in domestic guinea pigs (Cavia porcellus). Anim. Genet. 2018, 49, 269. [Google Scholar] [CrossRef]
- Yoshizawa, Y.; Wada, K.; Shimoi, G.; Shiomi, G.; Kameyama, Y.; Wakabayashi, Y.; Fukuta, K.; Hashizume, R. A 1-bp deletion in Fgf5 causes male-dominant long hair in the Syrian hamster. Mamm. Genome 2015, 26, 630–637. [Google Scholar] [CrossRef]
- Li, Y.; Song, S.; Zhang, Z.; Liu, X.; Zhang, Y.; E, G.; Ma, Y.; Jiang, L. A deletion variant within the FGF5 gene in goats is associated with gene expression levels and cashmere growth. Anim. Genet. 2022, 53, 657–664. [Google Scholar] [CrossRef]
- Legrand, R.; Tiret, L.; Abitbol, M. Two recessive mutations in FGF5 are associated with the long-hair phenotype in donkeys. Genet. Sel. Evol. 2014, 46, 65. [Google Scholar] [CrossRef]
- Maraqa, T.; Alhajeri, B.H.; Alhaddad, H. FGF5 missense mutation is associated with dromedary hair length variation. Anim. Genet. 2021, 52, 848–856. [Google Scholar] [CrossRef]
- Daverio, M.S.; Vidal-Rioja, L.; Frank, E.N.; Di Rocco, F. Molecular characterization of the llama FGF5 gene and identification of putative loss of function mutations. Anim. Genet. 2017, 48, 716–719. [Google Scholar] [CrossRef] [PubMed]
- Pervin, S.; Islam, M.S.; Yorisada, Y.; Sakai, A.; Masamune, S.; Yabuki, A.; Rakib, T.M.; Maki, S.; Tacharina, M.R.; Yamato, O. Carrier rate and mutant allele frequency of GM1 gangliosidosis in miniature Shiba Inus (Mame Shiba): Population screening of breeding dogs in Japan. Animals 2022, 12, 1242. [Google Scholar] [CrossRef] [PubMed]
- Nihonken Hozonkai (NIPPO). Available online: https://www.nihonken-hozonkai.or.jp (accessed on 30 July 2025).
- Japan Kennel Club (JKC). Available online: https://www.jkc.or.jp (accessed on 10 July 2025).
- Akita Inu Preservation Society. Available online: https://akitainu-hozonkai.com/?lang=en (accessed on 30 July 2025).
- Mizukami, K.; Chang, H.-S.; Yabuki, A.; Kawamichi, T.; Kawahara, N.; Hayashi, D.; Hossain, M.A.; Rahman, M.M.; Uddin, M.M.; Yamato, O. Novel rapid genotyping assays for neuronal ceroid lipofuscinosis in Border Collie dogs and high frequency of the mutant allele in Japan. J. Vet. Diagn. Investig. 2011, 23, 1131–1139. [Google Scholar] [CrossRef] [PubMed]
- Greco, D.S. Hyperadrenocorticism. Associated with sex steroid excess. Clin. Tech. Small Anim. Pract. 2007, 22, 12–17. [Google Scholar] [CrossRef]
- Woo, J.; Suh, W.; Sung, J.H. Hair growth regulation by fibroblast growth factor 12 (FGF12). Int. J. Mol. Sci. 2022, 23, 9467. [Google Scholar] [CrossRef]
- Hébert, J.M.; Rosenquist, T.; Götz, J.; Martin, G.R. FGF5 as a regulator of the hair growth cycle: Evidence from targeted and spontaneous mutations. Cell 1994, 78, 1017–1025. [Google Scholar] [CrossRef]
- OMIM 165190, Fibroblast Growth Factor 5; FGF5. Available online: https://omim.org/entry/165190 (accessed on 30 July 2025).
- Higgins, C.A.; Petukhova, L.; Harel, S.; Ho, Y.Y.; Drill, E.; Shapiro, L.; Wajid, M.; Christiano, A.M. FGF5 is a crucial regulator of hair length in humans. Proc. Nat. Acad. Sci. USA 2014, 111, 10648–10653. [Google Scholar] [CrossRef]
- Kim, Y.J.; Jung, N.; Kim, N.; Ha, J.C.; Park, J.H.; Han, K.; Chang, M.; Lee, J.; Kim, C.H. Effect of cysteine-free human fibroblast growth factor-5s mutant (FGF5sC93S) on hair growth. Dermatol. Ther. 2020, 33, e14530. [Google Scholar] [CrossRef]
- Cui, S.; Li, Y.; Zhang, X.; Wu, B.; Li, M.; Gao, J.; Xia, H.; Xu, L. FGF5 protects heart from sepsis injury by attenuating cardiomyocyte pyroptosis through inhibiting CaMKII/NFκB signaling. Biochem. Biophys. Res. Commun. 2022, 636, 104–112. [Google Scholar] [CrossRef]
- Liu, C.; Li, H.; Qi, Q.; Lu, L.; Gan, W.; Loos, R.J.; Lin, X. Common variants in or near FGF5, CYP17A1 and MTHFR genes are associated with blood pressure and hypertension in Chinese Hans. J. Hypertens. 2011, 29, 70–75. [Google Scholar] [CrossRef] [PubMed]
- Maki, S.; Islam, M.S.; Itoh, T.; Nurimoto, M.; Yabuki, A.; Furusawa, Y.; Kamishina, H.; Kobatake, Y.; Rakib, T.M.; Tacharina, M.R.; et al. Molecular epidemiological survey for degenerative myelopathy in German Shepherd Dogs in Japan: Allele frequency and clinical progression rate. Animals 2022, 12, 1647. [Google Scholar] [CrossRef]
- Pervin, S.; Islam, M.S.; Tada, N.; Tsutsui, T.; Rahman, M.M.; Yabuki, A.; Tacharina, M.R.; Rakib, T.M.; Maki, S.; Yamato, O. Screening and carrier rate of neuronal ceroid lipofuscinosis in Chihuahua dogs in Japan. Animals 2022, 12, 1210. [Google Scholar] [CrossRef]




| Primer/Probe | Sequence 5′ to 3′ (mer) | Position (ROS_Cfam_1.0) | Tm (°C) * | Ta (°C) ** | Amplicon Size (bp) |
|---|---|---|---|---|---|
| Sanger sequencing: | |||||
| E1(3)-F | TGGAAGAATGAGCTTGTCCCT (21) | g.4533325_4533345 | 64.9 | 58.0 | 389 |
| E1(3)-R | GCGCGAGCAACTTACTTAAC (20) | g.4533694_4533713 | 61.5 | ||
| E1(4)-F | AGAACCGGCCCTACAAGATG (20) | g.4533286_4533305 | 65.1 | 60.0 | 495 |
| E1(4)-R | AGGGTGCAAAACAACCGCGGTC (22) | g.4533759_4533780 | 74.6 | ||
| E2-F | GCTATAAAGAATGAAAAGAATCTATG (26) | g.4541429_4541454 | 57.4 | 60.0 | 302 |
| E2-R | TCTGAGCCAATTGTTCATCTAAC (23) | g.4541708_4541730 | 61.9 | ||
| E3-F | AGGCCAAGTTTACAGATGACTG (22) | g.4552791_4552812 | 62.0 | 61.3 | 285 |
| E3-R | GAACCTTTGGCTTGACGTGG (20) | g.4553056_4553075 | 66.7 | ||
| E3(2)-F | CCTTTTACCGCAGAAGACCTC (21) | g.4552714_4552734 | 63.8 | 60.0 | 502 |
| E3(2)-R | CTCTTCTGGGAGCTGTAAAG (20) | g.4553196_4553215 | 58.2 | ||
| Real-time polymerase chain reaction: | |||||
| Forward primer | CTCCGCAATACACCGAAGTGA (21) | g.4552858_4552878 | 67.1 | 60.0 | 83 |
| Reverse primer | TGCAGCCCCGCTTAGC (16) | g.4552925_4552940 | 66.5 | ||
| Probe for C allele | CTTGTTGAGCGCCACGTA (18) | g.4552915_4552898 | 63.7 | ||
| Probe for T allele | CTTGTTGAGCACCACGTA (18) | g.4552915_4552898 | 58.2 | ||
| Generational Population | Number of Dogs Examined | Number of Dogs with Each Genotype (%) | Mutant T Allele Frequency | Chi-Square Test | ||
|---|---|---|---|---|---|---|
| C/C | C/T | T/T | ||||
| Modern Akitas (2021): | ||||||
| Actual measured data | 60 | 21 (35.0%) | 36 (60.0%) | 3 (5.0%) | 0.350 ** | p = 0.0475 † |
| Expected data * | 25.4 (42.3%) | 27.3 (45.5%) | 7.4 (12.3%) | |||
| Classic Akitas (1970s and 1980s): | ||||||
| Actual measured data | 73 | 47 (64.4%) | 21 (28.8%) | 5 (6.8%) | 0.212 ** | p = 0.489 †† |
| Expected data * | 45.3 (62.0%) | 24.4 (33.5%) | 3.3 (4.5%) | |||
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Maki, S.; Islam, M.S.; Kansaku, N.; Arakawa, N.; Yabuki, A.; Rakib, T.M.; Faruq, A.A.; Yamato, O. Genetic Variant of the Canine FGF5 Gene for the Hair Length Trait in the Akita: Utility for Hair Coat Variations and Welfare in Conservation Breeding. Genes 2025, 16, 927. https://doi.org/10.3390/genes16080927
Maki S, Islam MS, Kansaku N, Arakawa N, Yabuki A, Rakib TM, Faruq AA, Yamato O. Genetic Variant of the Canine FGF5 Gene for the Hair Length Trait in the Akita: Utility for Hair Coat Variations and Welfare in Conservation Breeding. Genes. 2025; 16(8):927. https://doi.org/10.3390/genes16080927
Chicago/Turabian StyleMaki, Shinichiro, Md Shafiqul Islam, Norio Kansaku, Nanami Arakawa, Akira Yabuki, Tofazzal Md Rakib, Abdullah Al Faruq, and Osamu Yamato. 2025. "Genetic Variant of the Canine FGF5 Gene for the Hair Length Trait in the Akita: Utility for Hair Coat Variations and Welfare in Conservation Breeding" Genes 16, no. 8: 927. https://doi.org/10.3390/genes16080927
APA StyleMaki, S., Islam, M. S., Kansaku, N., Arakawa, N., Yabuki, A., Rakib, T. M., Faruq, A. A., & Yamato, O. (2025). Genetic Variant of the Canine FGF5 Gene for the Hair Length Trait in the Akita: Utility for Hair Coat Variations and Welfare in Conservation Breeding. Genes, 16(8), 927. https://doi.org/10.3390/genes16080927









