A Genetically-Informed Network Model of Myelodysplastic Syndrome: From Splicing Aberrations to Therapeutic Vulnerabilities
Abstract
1. Introduction
2. Interconnected Molecular Mechanisms in MDS Pathogenesis
2.1. Single-Cell Multi-Omics Reveals the Cellular Context of Splicing Aberrations
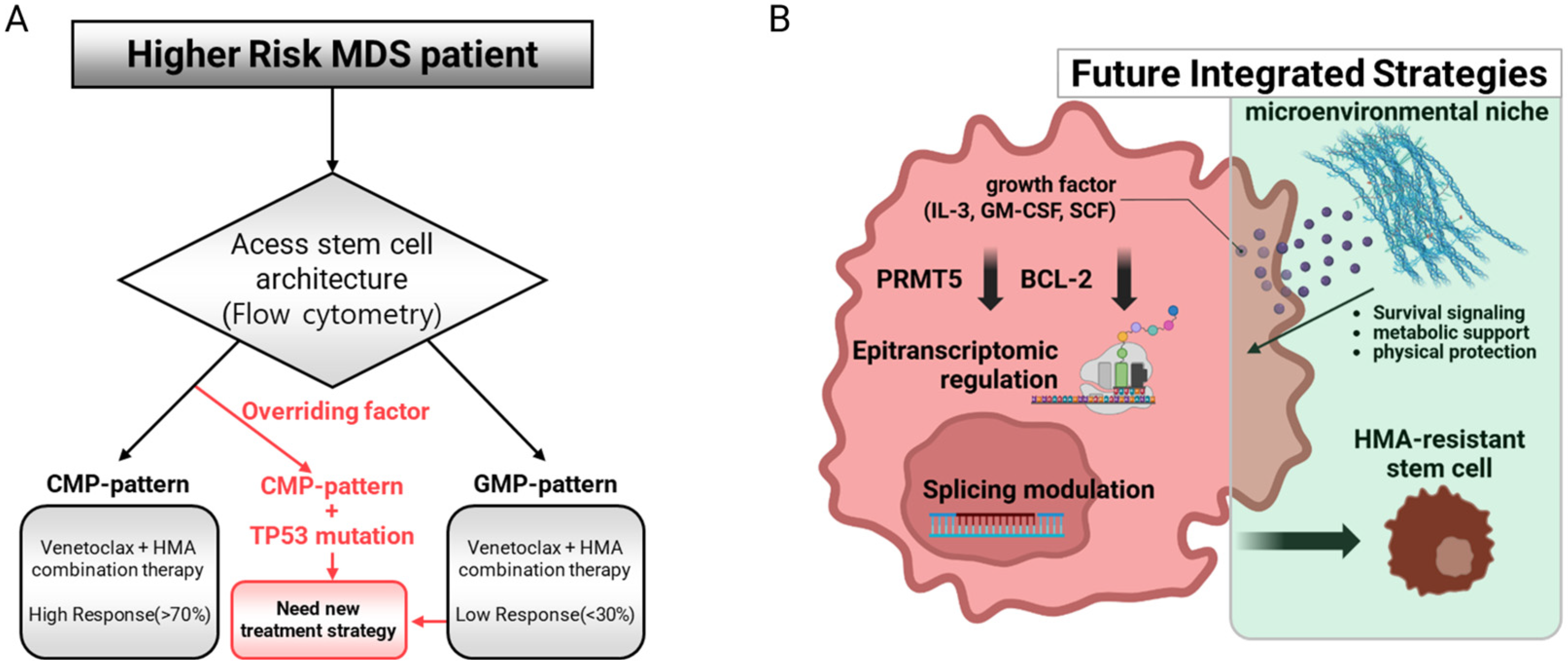
2.2. Stem Cell Architecture as a Unifying Framework for Disease Heterogeneity
2.3. Epitranscriptomic Regulation: A Novel Layer of Control
2.4. Microenvironmental Alterations: The Supporting Cast Becomes Central
2.5. Mechanistic Interconnections: A Systems-Level Perspective
3. Precision Therapeutic Strategies: From Mechanism to Medicine
3.1. Biomarker-Guided Treatment Selection: Venetoclax and Stem Cell Architecture
3.2. Venetoclax Combinations and Rational Drug Development
3.3. Understanding and Overcoming Treatment Resistance
3.4. Novel Targets and Emerging Therapeutic Approaches
4. Advanced Diagnostics and Clinical Implementation
4.1. Comprehensive Molecular Profiling for Precision Diagnosis
4.2. Molecular Risk Stratification and Prognostic Integration
4.3. Non-Invasive Monitoring and Circulating Biomarkers
5. Clinical Implementation and Future Perspectives
5.1. Translating Complexity into Clinical Practice
5.2. Addressing Current Limitations and Future Research Priorities
6. Outstanding Research Questions and Future Directions
6.1. Mechanistic Integration Questions
6.2. Therapeutic Development Priorities
6.3. Clinical Implementation Challenges
7. Conclusions
Author Contributions
Funding
Conflicts of Interest
Abbreviations
| MDS | Myelodysplastic syndrome |
| HSC | Hematopoietic stem cell |
| HMA | Hypomethylating agent |
| GoT-Splice | Genotyping of Transcriptomes with Splicing |
| tRF | Transfer RNA-derived fragment |
| MTOG | Mini tRFs containing a 5′ terminal oligoguanine |
| NK | Natural killer |
| OGM | Optical genome mapping |
References
- Arber, D.A.; Orazi, A.; Hasserjian, R.P.; Borowitz, M.J.; Calvo, K.R.; Kvasnicka, H.M.; Wang, S.A.; Bagg, A.; Barbui, T.; Branford, S.; et al. International Consensus Classification of Myeloid Neoplasms and Acute Leukemias: Integrating morphologic, clinical, and genomic data. Blood 2022, 140, 1200–1228. [Google Scholar] [CrossRef]
- Khoury, J.D.; Solary, E.; Abla, O.; Akkari, Y.; Alaggio, R.; Apperley, J.F.; Bejar, R.; Berti, E.; Busque, L.; Chan, J.K.C.; et al. The 5th edition of the World Health Organization Classification of Haematolymphoid Tumours: Myeloid and Histiocytic/Dendritic Neoplasms. Leukemia 2022, 36, 1703–1719. [Google Scholar] [CrossRef]
- Platzbecker, U. Treatment of MDS. Blood 2019, 133, 1096–1107. [Google Scholar] [CrossRef]
- Malcovati, L.; Hellström-Lindberg, E.; Bowen, D.; Adès, L.; Cermak, J.; Del Cañizo, C.; Della Porta, M.G.; Fenaux, P.; Gattermann, N.; Germing, U.; et al. European Leukemia Net Diagnosis and treatment of primary myelodysplastic syndromes in adults: Recommendations from the European Leukemia Net. Blood 2013, 122, 2943–2964. [Google Scholar] [CrossRef]
- Fenaux, P.; Mufti, G.J.; Hellstrom-Lindberg, E.; Santini, V.; Finelli, C.; Giagounidis, A.; Schoch, R.; Gattermann, N.; Sanz, G.; List, A.; et al. International Vidaza High-Risk MDS Survival Study Group. Efficacy of azacitidine compared with that of conventional care regimens in the treatment of higher-risk myelodysplastic syndromes: A randomised, open-label, phase III study. Lancet Oncol. 2009, 10, 223–232. [Google Scholar] [CrossRef]
- Silverman, L.R.; Demakos, E.P.; Peterson, B.L.; Kornblith, A.B.; Holland, J.C.; Odchimar-Reissig, R.; Stone, R.M.; Nelson, D.; Powell, B.L.; DeCastro, C.M.; et al. Randomized controlled trial of azacitidine in patients with the myelodysplastic syndrome: A study of the cancer and leukemia group B. J. Clin. Oncol. Off. J. Am. Soc. Clin. Oncol. 2002, 20, 2429–2440. [Google Scholar] [CrossRef] [PubMed]
- Cortés-López, M.; Chamely, P.; Hawkins, A.G.; Stanley, R.F.; Swett, A.D.; Ganesan, S.; Mouhieddine, T.H.; Dai, X.; Kluegel, L.; Chen, C.; et al. Single-cell multi-omics defines the cell-type-specific impact of splicing aberrations in human hematopoietic clonal outgrowths. Cell Stem Cell 2023, 30, 1262–1281.e8. [Google Scholar] [CrossRef] [PubMed]
- Alatawi, S.; Alzahrani, O.R.; Alatawi, F.A.; Almazni, I.A.; Almotiri, A.; Almsned, F.M. Identification of UBA7 expression downregulation in myelodysplastic neoplasm with SF3B1 mutations. Sci. Rep. 2025, 15, 10856. [Google Scholar] [CrossRef] [PubMed]
- Ganan-Gomez, I.; Yang, H.; Ma, F.; Montalban-Bravo, G.; Thongon, N.; Marchica, V.; Richard-Carpentier, G.; Chien, K.; Manyam, G.; Wang, F.; et al. Author Correction: Stem cell architecture drives myelodysplastic syndrome progression and predicts response to venetoclax-based therapy. Nat. Med. 2022, 28, 1097. [Google Scholar] [CrossRef]
- Guzzi, N.; Muthukumar, S.; Cieśla, M.; Todisco, G.; Ngoc, P.C.T.; Madej, M.; Munita, R.; Fazio, S.; Ekström, S.; Mortera-Blanco, T.; et al. Pseudouridine-modified tRNA fragments repress aberrant protein synthesis and predict leukaemic progression in myelodysplastic syndrome. Nat. Cell Biol. 2022, 24, 299–306. [Google Scholar] [CrossRef]
- Medyouf, H.; Mossner, M.; Jann, J.C.; Nolte, F.; Raffel, S.; Herrmann, C.; Lier, A.; Eisen, C.; Nowak, V.; Zens, B.; et al. Myelodysplastic cells in patients reprogram mesenchymal stromal cells to establish a transplantable stem cell niche disease unit. Cell Stem Cell 2014, 14, 824–837. [Google Scholar] [CrossRef]
- Braish, J.S.; Montalban-Bravo, G.; Ravandi, F.; Short, N.; Kadia, T.; Ohanian, M.; Chien, K.; Masarova, L.; Sasaki, K.; Yilmaz, M.; et al. Safety and efficacy of the combination of azacitidine with venetoclax after hypomethylating agent failure in higher-risk myelodysplastic syndrome. Leuk. Res. 2025, 153, 107692. [Google Scholar] [CrossRef] [PubMed]
- El-Cheikh, J.; Bidaoui, G.; Saleh, M.; Moukalled, N.; Abou Dalle, I.; Bazarbachi, A. Venetoclax: A New Partner in the Novel Treatment Era for Acute Myeloid Leukemia and Myelodysplastic Syndrome. Clin. Hematol. Int. 2023, 5, 143–154. [Google Scholar] [CrossRef]
- Shah, M.V.; Arber, D.A.; Hiwase, D.K. TP53 -Mutated Myeloid Neoplasms: 2024 Update on Diagnosis, Risk-Stratification, and Management. Am. J. Hematol. 2025, 100 (Suppl. S4), 88–115. [Google Scholar] [CrossRef] [PubMed]
- Baranwal, A.; Gurney, M.; Basmaci, R.; Katamesh, B.; He, R.; Viswanatha, D.S.; Greipp, P.; Foran, J.; Badar, T.; Murthy, H.; et al. Genetic landscape and clinical outcomes of patients with BCOR mutated myeloid neoplasms. Haematologica 2024, 109, 1779–1791. [Google Scholar] [CrossRef]
- Shivarov, V.; Gueorguieva, R.; Stoimenov, A.; Tiu, R. DNMT3A mutation is a poor prognosis biomarker in AML: Results of a meta-analysis of 4500 AML patients. Leuk. Res. 2013, 37, 1445–1450. [Google Scholar] [CrossRef]
- Moura, P.L.; Mortera-Blanco, T.; Hofman, I.J.; Todisco, G.; Kretzschmar, W.W.; Björklund, A.C.; Creignou, M.; Hagemann-Jensen, M.; Ziegenhain, C.; Cabrerizo Granados, D.; et al. Erythroid Differentiation Enhances RNA Mis-Splicing in SF3B1-Mutant Myelodysplastic Syndromes with Ring Sideroblasts. Cancer Res. 2024, 84, 211–225. [Google Scholar] [CrossRef] [PubMed]
- Mortuza, S.; Chin-Yee, B.; James, T.E.; Chin-Yee, I.H.; Hedley, B.D.; Ho, J.M.; Saini, L.; Lazo-Langner, A.; Schenkel, L.; Bhai, P.; et al. Myelodysplastic Neoplasms (MDS) with Ring Sideroblasts or SF3B1 Mutations: The Improved Clinical Utility of World Health Organization and International Consensus Classification 2022 Definitions, a Single-Centre Retrospective Chart Review. Curr. Oncol. (Tor. Ont.) 2024, 31, 1762–1773. [Google Scholar] [CrossRef]
- Herbert, A.; Hatfield, A.; Randazza, A.; Miyamoto, V.; Palmer, K.; Lackey, L. Precursor RNA structural patterns at SF3B1 mutation sensitive cryptic 3′ splice sites. Biorxiv Prepr. Serv. Biol. 2025, 02.19/638873. [Google Scholar] [CrossRef]
- Damianov, A.; Lin, C.H.; Zhang, J.; Manley, J.L.; Black, D.L. Cancer-associated SF3B1 mutation K700E causes widespread changes in U2/branchpoint recognition without altering splicing. Proc. Natl. Acad. Sci. USA 2025, 122, e2423776122. [Google Scholar] [CrossRef]
- Boussi, L.; Biswas, J.; Abdel-Wahab, O.; Stein, E. Therapeutic strategies targeting aberrant RNA splicing in myeloid malignancies. Br. J. Haematol. 2024, 205, 2153–2162. [Google Scholar] [CrossRef]
- Liu, X.; Devadiga, S.A.; Stanley, R.F.; Morrow, R.M.; Janssen, K.A.; Quesnel-Vallières, M.; Pomp, O.; Moverley, A.A.; Li, C.; Skuli, N.; et al. A mitochondrial surveillance mechanism activated by SRSF2 mutations in hematologic malignancies. J. Clin. Investig. 2024, 134, e175619. [Google Scholar] [CrossRef]
- Nian, Q.; Li, Y.; Li, J.; Zhao, L.; Rodrigues Lima, F.; Zeng, J.; Liu, R.; Ye, Z. U2AF1 in various neoplastic diseases and relevant targeted therapies for malignant cancers with complex mutations (Review). Oncol. Rep. 2024, 51, 5. [Google Scholar] [CrossRef] [PubMed]
- Cockey, S.G.; Zhang, H.; Hussaini, M.; Zhang, L.; Moscinski, L.; Yang, E.; Li, J.; Wang, L.; Song, J. Molecular landscape and clinical outcome of SRSF2/TET2 Co-mutated myeloid neoplasms. Leuk. Lymphoma 2025, 66, 469–478. [Google Scholar] [CrossRef]
- Senapati, J.; Loghavi, S.; Garcia-Manero, G.; Tang, G.; Kadia, T.; Short, N.J.; Abbas, H.A.; Arani, N.; DiNardo, C.D.; Borthakur, G.; et al. Clinical interrogation of TP53 aberrations and its impact on survival in patients with myeloid neoplasms. Haematologica 2025, 110, 1304–1315. [Google Scholar] [CrossRef]
- Mimura, N.; Hideshima, T.; Anderson, K.C. Novel therapeutic strategies for multiple myeloma. Exp. Hematol. 2015, 43, 732–741. [Google Scholar] [CrossRef]
- Guzzi, N.; Cieśla, M.; Ngoc, P.C.T.; Lang, S.; Arora, S.; Dimitriou, M.; Pimková, K.; Sommarin, M.N.E.; Munita, R.; Lubas, M.; et al. Pseudouridylation of tRNA-Derived Fragments Steers Translational Control in Stem Cells. Cell 2018, 173, 1204–1216.e26. [Google Scholar] [CrossRef]
- Flores-Figueroa, E.; Arana-Trejo, R.M.; Gutiérrez-Espíndola, G.; Pérez-Cabrera, A.; Mayani, H. Mesenchymal stem cells in myelodysplastic syndromes: Phenotypic and cytogenetic characterization. Leuk. Res. 2005, 29, 215–224. [Google Scholar] [CrossRef]
- Boy, M.; Bisio, V.; Zhao, L.P.; Guidez, F.; Schell, B.; Lereclus, E.; Henry, G.; Villemonteix, J.; Rodrigues-Lima, F.; Gagne, K.; et al. Myelodysplastic Syndrome associated TET2 mutations affect NK cell function and genome methylation. Nat. Commun. 2023, 14, 588. [Google Scholar] [CrossRef] [PubMed]
- Itzykson, R.; Kosmider, O.; Cluzeau, T.; Mansat-De Mas, V.; Dreyfus, F.; Beyne-Rauzy, O.; Quesnel, B.; Vey, N.; Gelsi-Boyer, V.; Raynaud, S.; et al. Groupe Francophone des Myelodysplasies (GFM). Impact of TET2 mutations on response rate to azacitidine in myelodysplastic syndromes and low blast count acute myeloid leukemias. Leukemia 2011, 25, 1147–1152. [Google Scholar] [CrossRef] [PubMed]
- Bejar, R.; Lord, A.; Stevenson, K.; Bar-Natan, M.; Pérez-Ladaga, A.; Zaneveld, J.; Wang, H.; Caughey, B.; Stojanov, P.; Getz, G.; et al. TET2 mutations predict response to hypomethylating agents in myelodysplastic syndrome patients. Blood 2014, 124, 2705–2712. [Google Scholar] [CrossRef]
- Krevvata, M.; Silva, B.C.; Manavalan, J.S.; Galan-Diez, M.; Kode, A.; Matthews, B.G.; Park, D.; Zhang, C.A.; Galili, N.; Nickolas, T.L.; et al. Inhibition of leukemia cell engraftment and disease progression in mice by osteoblasts. Blood 2014, 124, 2834–2846. [Google Scholar] [CrossRef] [PubMed]
- Pollyea, D.A.; Stevens, B.M.; Jones, C.L.; Winters, A.; Pei, S.; Minhajuddin, M.; D’Alessandro, A.; Culp-Hill, R.; Riemondy, K.A.; Gillen, A.E.; et al. Venetoclax with azacitidine disrupts energy metabolism and targets leukemia stem cells in patients with acute myeloid leukemia. Nat. Med. 2018, 24, 1859–1866. [Google Scholar] [CrossRef] [PubMed]
- Nanaa, A.; He, R.; Foran, J.M.; Badar, T.; Gangat, N.; Pardanani, A.; Hogan, W.J.; Litzow, M.R.; Patnaik, M.; Al-Kali, A.; et al. Venetoclax plus hypomethylating agents in DDX41-mutated acute myeloid leukaemia and myelodysplastic syndrome: Mayo Clinic series on 12 patients. Br. J. Haematol. 2024, 204, 171–176. [Google Scholar] [CrossRef] [PubMed]
- Desai, S.R.; Chakraborty, S.; Shastri, A. Mechanisms of resistance to hypomethylating agents and BCL-2 inhibitors. Best. Pract. Res. Clin. Haematol. 2023, 36, 101521. [Google Scholar] [CrossRef]
- Ramanathan, R.; Xie, Y.; Badar, T.; Zeidan, A.M.; Patel, S.A. Contemporary management paradigms and emerging therapeutics for myelodysplastic syndromes/neoplasms. Br. J. Haematol. 2025, 206, 1571–1581. [Google Scholar] [CrossRef]
- Bazinet, A.; Desikan, S.P.; Li, Z.; Rodriguez-Sevilla, J.J.; Venugopal, S.; Urrutia, S.; Montalban-Bravo, G.; Sasaki, K.; Chien, K.S.; Hammond, D.; et al. Cytogenetic and Molecular Associations with Outcomes in Higher-Risk Myelodysplastic Syndromes Treated with Hypomethylating Agents plus Venetoclax. Clin. Cancer Res. An. Off. J. Am. Assoc. Cancer Res. 2024, 30, 1319–1326. [Google Scholar] [CrossRef]
- Wang, X.; Yuan, L.; Lu, B.; Lin, D.; Xu, X. Glutathione promotes the synergistic effects of venetoclax and azacytidine against myelodysplastic syndrome-refractory anemia by regulating the cell cycle. Exp. Ther. Med. 2023, 26, 574. [Google Scholar] [CrossRef]
- DiNardo, C.D.; Jonas, B.A.; Pullarkat, V.; Thirman, M.J.; Garcia, J.S.; Wei, A.H.; Konopleva, M.; Döhner, H.; Letai, A.; Fenaux, P.; et al. Azacitidine and Venetoclax in Previously Untreated Acute Myeloid Leukemia. N. Engl. J. Med. 2020, 383, 617–629. [Google Scholar] [CrossRef]
- Kantarjian, H.M.; DiNardo, C.D.; Kadia, T.M.; Daver, N.G.; Altman, J.K.; Stein, E.M.; Jabbour, E.; Schiffer, C.A.; Lang, A.; Ravandi, F. Acute myeloid leukemia management and research in 2025. CA A Cancer J. Clin. 2025, 75, 46–67. [Google Scholar] [CrossRef]
- Kantarjian, H.; Borthakur, G.; Daver, N.; DiNardo, C.D.; Issa, G.; Jabbour, E.; Kadia, T.; Sasaki, K.; Short, N.J.; Yilmaz, M.; et al. Current status and research directions in acute myeloid leukemia. Blood Cancer J. 2024, 14, 163. [Google Scholar] [CrossRef]
- Mei, C.; Ye, L.; Ren, Y.; Zhou, X.; Ma, L.; Xu, G.; Xu, W.; Lu, C.; Yang, H.; Luo, Y.; et al. 15-days duration of venetoclax combined with azacitidine in the treatment of relapsed/refractory high-risk myelodysplastic syndromes: A retrospective single-center study. Hematol. Oncol. 2023, 41, 546–554. [Google Scholar] [CrossRef]
- Hu, X.; Li, L.; Nkwocha, J.; Sharma, K.; Zhou, L.; Grant, S. Synergistic Interactions between the Hypomethylating Agent Thio-Deoxycytidine and Venetoclax in Myelodysplastic Syndrome Cells. Hematol. Rep. 2023, 15, 91–100. [Google Scholar] [CrossRef]
- Sumiyoshi, R.; Tashiro, H.; Shirasaki, R.; Matsuo, T.; Yamamoto, T.; Matsumoto, K.; Ooi, J.; Shirafuji, N. The FLT3 internal tandem duplication mutation at disease diagnosis is a negative prognostic factor in myelodysplastic syndrome patients. Leuk. Res. 2022, 113, 106790. [Google Scholar] [CrossRef] [PubMed]
- Lane, A.A.; Garcia, J.S.; Raulston, E.G.; Garzon, J.L.; Galinsky, I.; Baxter, E.W.; Leonard, R.; DeAngelo, D.J.; Luskin, M.R.; Reilly, C.R.; et al. Phase 1b trial of tagraxofusp in combination with azacitidine with or without venetoclax in acute myeloid leukemia. Blood Adv. 2024, 8, 591–602. [Google Scholar] [CrossRef]
- Senapati, J.; Fiskus, W.C.; Daver, N.; Wilson, N.R.; Ravandi, F.; Garcia-Manero, G.; Kadia, T.; DiNardo, C.D.; Jabbour, E.; Burger, J.; et al. Phase I Results of Bromodomain and Extra-Terminal Inhibitor PLX51107 in Combination with Azacitidine in Patients with Relapsed/Refractory Myeloid Malignancies. Clin. Cancer Res. An. Off. J. Am. Assoc. Cancer Res. 2023, 29, 4352–4360. [Google Scholar] [CrossRef]
- Diesch, J.; Zwick, A.; Garz, A.K.; Palau, A.; Buschbeck, M.; Götze, K.S. A clinical-molecular update on azanucleoside-based therapy for the treatment of hematologic cancers. Clin. Epigenetics 2016, 8, 71. [Google Scholar] [CrossRef]
- Unnikrishnan, A.; Papaemmanuil, E.; Beck, D.; Deshpande, N.P.; Verma, A.; Kumari, A.; Woll, P.S.; Richards, L.A.; Knezevic, K.; Chandrakanthan, V.; et al. Integrative Genomics Identifies the Molecular Basis of Resistance to Azacitidine Therapy in Myelodysplastic Syndromes. Cell Rep. 2017, 20, 572–585. [Google Scholar] [CrossRef]
- He, M.Y.; Kridel, R. Treatment resistance in diffuse large B-cell lymphoma. Leukemia 2021, 35, 2151–2165. [Google Scholar] [CrossRef] [PubMed]
- Sallman, D.A.; DeZern, A.E.; Garcia-Manero, G.; Steensma, D.P.; Roboz, G.J.; Sekeres, M.A.; Cluzeau, T.; Sweet, K.L.; McLemore, A.; McGraw, K.L.; et al. Eprenetapopt (APR-246) and Azacitidine in TP53-Mutant Myelodysplastic Syndromes. J. Clin. Oncol. Off. J. Am. Soc. Clin. Oncol. 2021, 39, 1584–1594. [Google Scholar] [CrossRef] [PubMed]
- Walter, M.J.; Shen, D.; Ding, L.; Shao, J.; Koboldt, D.C.; Chen, K.; Larson, D.E.; McLellan, M.D.; Dooling, D.; Abbott, R.; et al. Clonal architecture of secondary acute myeloid leukemia. N. Engl. J. Med. 2012, 366, 1090–1098. [Google Scholar] [CrossRef]
- Ding, L.; Ley, T.J.; Larson, D.E.; Miller, C.A.; Koboldt, D.C.; Welch, J.S.; Ritchey, J.K.; Young, M.A.; Lamprecht, T.; McLellan, M.D.; et al. Clonal evolution in relapsed acute myeloid leukaemia revealed by whole-genome sequencing. Nature 2012, 481, 506–510. [Google Scholar] [CrossRef]
- Haferlach, T.; Nagata, Y.; Grossmann, V.; Okuno, Y.; Bacher, U.; Nagae, G.; Schnittger, S.; Sanada, M.; Kon, A.; Alpermann, T.; et al. Landscape of genetic lesions in 944 patients with myelodysplastic syndromes. Leukemia 2014, 28, 241–247. [Google Scholar] [CrossRef]
- Lindsley, R.C.; Mar, B.G.; Mazzola, E.; Grauman, P.V.; Shareef, S.; Allen, S.L.; Pigneux, A.; Wetzler, M.; Stuart, R.K.; Erba, H.P.; et al. Acute myeloid leukemia ontogeny is defined by distinct somatic mutations. Blood 2015, 125, 1367–1376. [Google Scholar] [CrossRef] [PubMed]
- Watts, J.; Minden, M.D.; Bachiashvili, K.; Brunner, A.M.; Abedin, S.; Crossman, T.; Zajac, M.; Moroz, V.; Egger, J.L.; Tarkar, A.; et al. Phase I/II study of the clinical activity and safety of GSK3326595 in patients with myeloid neoplasms. Ther. Adv. Hematol. 2024, 15, 20406207241275376. [Google Scholar] [CrossRef]
- Okabe, S.; Moriyama, M.; Arai, Y.; Gotoh, A. Glutaminase 1 plays critical roles in myelodysplastic syndrome and acute myeloid leukemia cells. Cancer Biomark. Sect. A Dis. Markers 2024, 41, 55–68. [Google Scholar] [CrossRef] [PubMed]
- Yuan, X.; Sabzvar, M.K.; Patil, A.D.; Chinnaswamy, K.; Howie, K.L.; Andhavaram, R.; Wang, B.; Siegler, M.A.; Dumaz, A.; Stuckey, J.A.; et al. Comprehensive Analyses of the Effects of the Small-Molecule Inhibitor of the UHM Domain in the Splicing Factor U2AF1 in Leukemia Cells. Res. Sq. 2024, rs-3, rs-4477663. [Google Scholar] [CrossRef]
- Haque, T.; Cadenas, F.L.; Xicoy, B.; Alfonso-Pierola, A.; Platzbecker, U.; Avivi, I.; Brunner, A.M.; Chromik, J.; Morillo, D.; Patel, M.R.; et al. Phase 1 study of JNJ-64619178, a protein arginine methyltransferase 5 inhibitor, in patients with lower-risk myelodysplastic syndromes. Leuk. Res. 2023, 134, 107390. [Google Scholar] [CrossRef] [PubMed]
- Konturek-Ciesla, A.; Zhang, Q.; Kharazi, S.; Bryder, D. A non-genotoxic stem cell therapy boosts lymphopoiesis and averts age-related blood diseases in mice. Nat. Commun. 2025, 16, 5129. [Google Scholar] [CrossRef]
- Fei, F.; Jariwala, A.; Pullarkat, S.; Loo, E.; Liu, Y.; Tizro, P.; Ali, H.; Otoukesh, S.; Amanam, I.; Artz, A.; et al. Genomic Landscape of Myelodysplastic/Myeloproliferative Neoplasms: A Multi-Central Study. Int. J. Mol. Sci. 2024, 25, 10214. [Google Scholar] [CrossRef]
- Tiu, R.V.; Gondek, L.P.; O’Keefe, C.L.; Elson, P.; Huh, J.; Mohamedali, A.; Kulasekararaj, A.; Advani, A.S.; Paquette, R.; List, A.F.; et al. Prognostic impact of SNP array karyotyping in myelodysplastic syndromes and related myeloid malignancies. Blood 2011, 117, 4552–4560. [Google Scholar] [CrossRef] [PubMed]
- Edelman Saul, E.; Urrutia, S.; Yang, H.; Montalban-Bravo, G.; Tang, G.; Toruner, G.; Patel, K.; Luthra, R.; Bueso-Ramos, C.; Wang, S.A.; et al. A Case of Cryptic CBFB::MYH11 Acute Myeloid Leukemia With Noncanonical Breakpoints Detected by Optical Genome Mapping. J. Natl. Compr. Cancer Netw. JNCCN 2025, 23, e257015. [Google Scholar] [CrossRef] [PubMed]
- Wang, M.; Chen, P.; Li, D.; Zhao, M. Prognostic impact of DTA mutation and co-occurring mutations in patients with myelodysplastic syndrome. Mol. Biol. Rep. 2024, 51, 985. [Google Scholar] [CrossRef]
- Wang, H.; Li, X.; Qi, J.; Liu, H.; Chu, T.; Xu, X.; Qiu, H.; Fu, C.; Tang, X.; Ruan, C.; et al. Prognostic mutations identified by whole-exome sequencing and validation of the Molecular International Prognostic Scoring System in myelodysplastic syndromes after allogeneic haematopoietic stem cell transplantation. Br. J. Haematol. 2024, 205, 1899–1909. [Google Scholar] [CrossRef]
- Papaemmanuil, E.; Gerstung, M.; Bullinger, L.; Gaidzik, V.I.; Paschka, P.; Roberts, N.D.; Potter, N.E.; Heuser, M.; Thol, F.; Bolli, N.; et al. Genomic classification and prognosis in acute myeloid leukemia. N. Engl. J. Med. 2016, 374, 2209–2221. [Google Scholar] [CrossRef]
- Bowen, D.; Groves, M.J.; Burnett, A.K.; Patel, Y.; Allen, C.; Green, C.; Gale, R.E.; Hills, R.; Linch, D.C. TP53 gene mutation is frequent in patients with acute myeloid leukemia and complex karyotype, and is associated with very poor prognosis. Leukemia 2009, 23, 203–206. [Google Scholar] [CrossRef]
- Normanno, N.; Rachiglio, A.M.; Lambiase, M.; Martinelli, E.; Fenizia, F.; Esposito, C.; Roma, C.; Troiani, T.; Rizzi, D.; Tatangelo, F.; et al. CAPRI-GOIM investigators Heterogeneity of KRAS, NRAS, BRAF and PIK3CA mutations in metastatic colorectal cancer and potential effects on therapy in the CAPRI GOIM trial. Ann. Oncol. Off. J. Eur. Soc. Med. Oncol. 2015, 26, 1710–1714. [Google Scholar] [CrossRef] [PubMed]
- Han, X.; Liu, W.; Kang, Z.; Li, D. Prognostic significance of GATA2 in patients with MDS/AML: A systematic review and meta-analysis. Ann. Hematol. 2024, 103, 4943–4952. [Google Scholar] [CrossRef]
- Schreiber, F.; Piontek, G.; Schneider-Kimoto, Y.; Schwarz-Furlan, S.; De Vito, R.; Locatelli, F.; Gengler, C.; Yoshimi, A.; Jung, A.; Klauschen, F.; et al. Development of MDS in Pediatric Patients with GATA2 Deficiency: Increased Histone Trimethylation and Deregulated Apoptosis as Potential Drivers of Transformation. Cancers 2023, 15, 5594. [Google Scholar] [CrossRef]
- Miao, L.; Wang, X.; Yao, M.; Tao, Y.; Han, Y. Clinicopathological and prognostic significance of DDX41 mutation in myeloid neoplasms: A systematic review and meta-analysis. Ann. Hematol. 2025, 104, 2581–2591. [Google Scholar] [CrossRef]
- Woerner, J.; Huang, Y.; Hutter, S.; Gurnari, C.; Sánchez, J.M.H.; Wang, J.; Huang, Y.; Schnabel, D.; Aaby, M.; Xu, W.; et al. Circulating microbial content in myeloid malignancy patients is associated with disease subtypes and patient outcomes. Nat. Commun. 2022, 13, 1038. [Google Scholar] [CrossRef] [PubMed]
- Mo, A.; Poynton, M.; Wood, E.; Shortt, J.; Brunskill, S.J.; Doree, C.; Sandercock, J.; Saadah, N.; Luk, E.; Stanworth, S.J.; et al. Do anemia treatments improve quality of life and physical function in patients with myelodysplastic syndromes (MDS)? A systematic review. Blood Rev. 2023, 61, 101114. [Google Scholar] [CrossRef]
- Grimwade, D.; Freeman, S.D. Defining minimal residual disease in acute myeloid leukemia: Which platforms are ready for prime time? Blood 2014, 124, 3345–3355. [Google Scholar] [CrossRef]
- Lee, J.; Cho, H.J.; Kim, J.; Baek, D.W.; Choi, H.; Ham, J.Y.; Chang, S.H.; Kim, J.G.; Sohn, S.K.; Moon, J.H. Prognostic Impact of Transfusion Dependency in Patients with Lower-Risk Myelodysplastic Syndrome. Clin. Lab. 2025, 71, 272–280. [Google Scholar] [CrossRef]
- Potenza, L.; Giusti, D.; Borelli, E.; Colaci, E.; Banchelli, F.; Cuoghi Costantini, R.; Forghieri, F.; Morselli, M.; Bettelli, F.; Pioli, V.; et al. Early Palliative Care, Goals of Care Conversations and Quality EOL Care in Acute Leukemia and HR-MDS. J. Pain Symptom Manag. 2025. S0885-3924(25)00583-4, advance online publication. [Google Scholar] [CrossRef] [PubMed]
- Marin, D.; Gabriel, I.H.; Ahmad, S.; Foroni, L.; de Lavallade, H.; Clark, R.; O’Brien, S.; Sergeant, R.; Hedgley, C.; Milojkovic, D.; et al. KIR2DS1 genotype predicts for complete cytogenetic response and survival in newly diagnosed chronic myeloid leukemia patients treated with imatinib. Leukemia 2012, 26, 296–302. [Google Scholar] [CrossRef] [PubMed]

| Molecular Mechanism | Cell-Type Specificity | Key Clinical Manifestations | Therapeutic Implications | Ref |
|---|---|---|---|---|
| Splicing Factor Mutations (SF3B1, SRSF2, U2AF1) | SF3B1: Ring sideroblast formation megakaryocytic-erythroid preference [7,17] SRSF2: Mitochondrial dysfunction, multiple lineages [20,22] U2AF1: Distinct 3′ splice site patterns [23] | Ring sideroblasts (>15%) [7] macrocytic anemia [17] lineage-specific dysplasia [8] iron overload [19] | PRMT5 inhibition synthetic lethality [21] H3B-8800 splicing modulators in trials [22] personalized mutation-based therapy [23] | [7,8,17,19,21,22,23] |
| Stem Cell Architecture Alterations | CMP-pattern: Increased CD34+CD38+CD123+ progenitors [9] GMP-pattern: Expanded CD34+CD38+CD123- progenitors [9] | CMP-pattern: TP53 mutations [14,25] GMP-pattern: RUNX1/DNMT3A Mutations [15,16] | Venetoclax response prediction >70% (CMP) vs. <30% (GMP) [9] guides combination therapy selection [12,33] | [9,12,14,15,16,25,33] |
| Epitranscriptomic Dysregulation (Pseudouridine modifications) | CD34+ HSPCs through mTOG-mediated translational control [10,27] ribosome dysfunction [27] | Aberrant protein synthesis [10] impaired differentiation [27] increased transformation risk [10] | PUS7 inhibition [27] mTOG pathway targeting [10] proteostasis modulators [27] | [10,27] |
| Microenvironmental Alterations | TET2 mutations affect NK cells [29] stromal reprogramming [11,28] cytokine dysregulation [32] | Immune evasion [29] impaired normal hematopoiesis [11] chronic inflammation [32] | Immune checkpoint inhibitors + HMA [29] NK cell therapy [29] stromal targeting [11,32] | [11,28,29,32] |
| Resistance Mechanism | Primitive Stem Cell Persistence | Microenvironment-Mediated Protection | Dominant Genetic Alterations |
|---|---|---|---|
| Molecular/Cellular Basis | Maintained self-renewal in disease-initiating cells [9,11] | Stromal cell survival signals [11,28] drug metabolism barriers [32] | TP53 biallelic mutations [14,25] |
| Clinical Manifestation | Initial response followed by relapse [12,33] | Variable response rates [35] | Refractory disease regardless of biomarkers [14,50] |
| Current Therapeutic Approaches | Intensified HMA dosing extended treatment [3,5] | Standard combination regimens [12,13] | Early transplant consideration [14] |
| Emerging Strategies | Stem cell architecture-guided therapy [9] | Niche-disrupting agents [11,32] | p53 pathway restoration [50] |
| Ref | [3,5,9,12,33] | [11,12,13,28,32,35] | [14,25,50] |
| Resistance Mechanism | Adaptive Clonal Evolution | Multi-pathway Dysfunction | Epitranscriptomic Resistance |
| Molecular/Cellular Basis | Selection pressure-driven mutation acquisition [48,51,52] | Complex genetic landscapes [53,54] | Dysregulated protein synthesis machinery [10,27] |
| Clinical Manifestation | Progressive treatment failure [48] | Poor single-agent response [35,48] | Intrinsic treatment resistance [10] |
| Current Therapeutic Approaches | Sequential therapy changes [35] | Empirical combination approaches [12,37] | No specific targeting available |
| Emerging Strategies | Dynamic treatment algorithms [48] | Personalized multi-target strategies [36] | PUS7-mTOG pathway modulators [10,27] |
| Ref | [35,48,51,52] | [12,35,36,37,48,53,54] | [10,27] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yu, S.; Kim, J.; Kim, M.S. A Genetically-Informed Network Model of Myelodysplastic Syndrome: From Splicing Aberrations to Therapeutic Vulnerabilities. Genes 2025, 16, 928. https://doi.org/10.3390/genes16080928
Yu S, Kim J, Kim MS. A Genetically-Informed Network Model of Myelodysplastic Syndrome: From Splicing Aberrations to Therapeutic Vulnerabilities. Genes. 2025; 16(8):928. https://doi.org/10.3390/genes16080928
Chicago/Turabian StyleYu, Sanghyeon, Junghyun Kim, and Man S. Kim. 2025. "A Genetically-Informed Network Model of Myelodysplastic Syndrome: From Splicing Aberrations to Therapeutic Vulnerabilities" Genes 16, no. 8: 928. https://doi.org/10.3390/genes16080928
APA StyleYu, S., Kim, J., & Kim, M. S. (2025). A Genetically-Informed Network Model of Myelodysplastic Syndrome: From Splicing Aberrations to Therapeutic Vulnerabilities. Genes, 16(8), 928. https://doi.org/10.3390/genes16080928






