Identification of Candidate Genes and Functional Pathways Associated with Body Size Traits in Hulunbuir Sheep Through GWAS Analysis
Highlights
- Our study identified three significant SNPs associated with body size traits (CG, CC, HW, BH, and BL) using MT-GWAS in Hulunbuir sheep.
- Three candidate genes (SLC9C1, VSTM2A, FRG1) associated with body size traits were identified through GWAS analysis and KEGG pathway enrichment.
- This study identified three candidate genes related to body size in Hulunbuir sheep, thus providing genetic targets for marker-assisted selection (MAS) in Hulunbuir sheep.
Abstract
1. Introduction
2. Materials and Methods
2.1. Experimental Animals
2.2. Determination of Body Size Traits
2.3. Genotyping and Quality Control
2.4. Principal Component Analysis
2.5. Genome-Wide Association Study
2.6. Gene Annotation
2.7. Enrichment Analysis of the Candidate Genes
3. Results
3.1. Descriptive Statistics
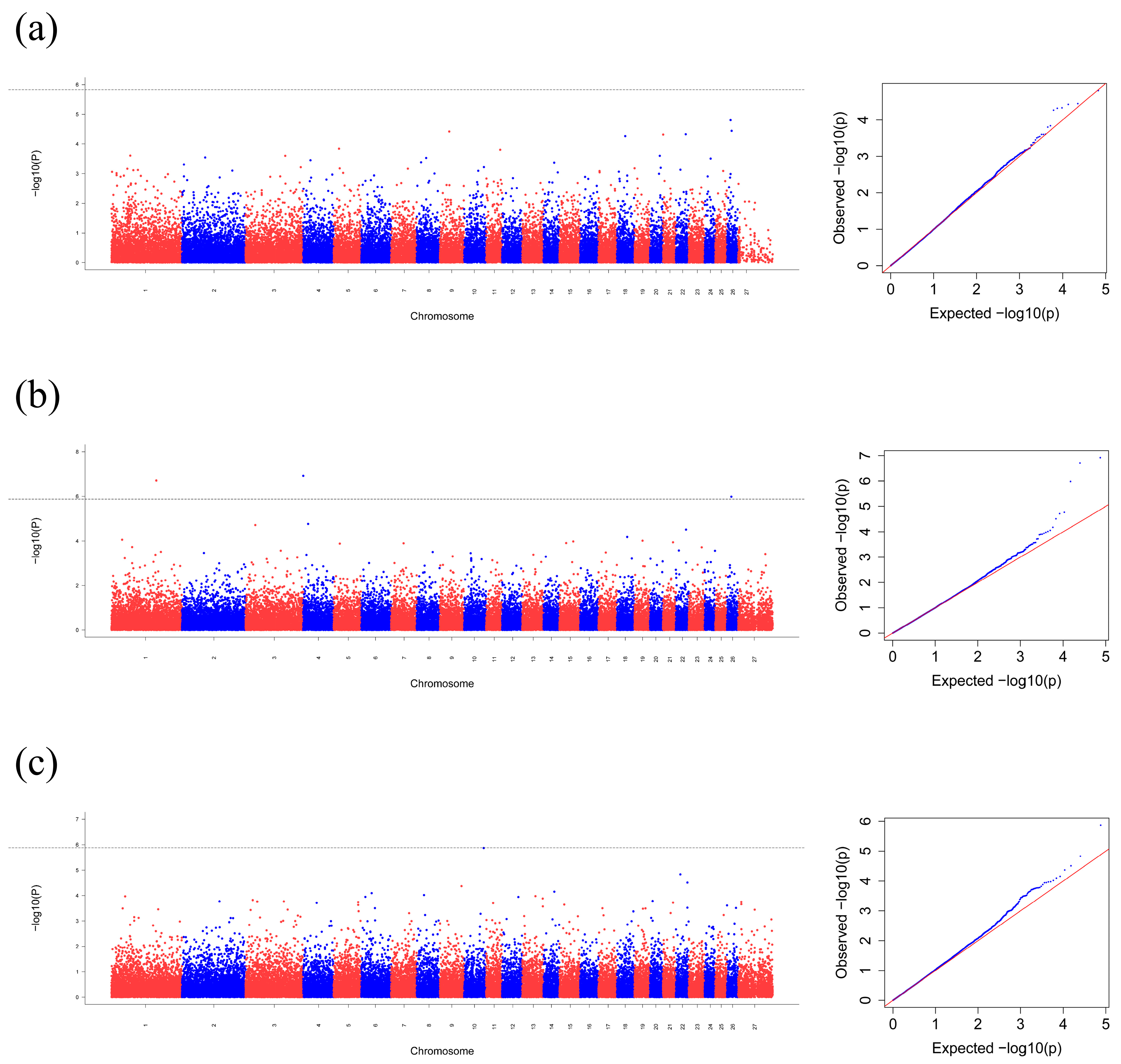
3.2. Genome-Wide Association Study
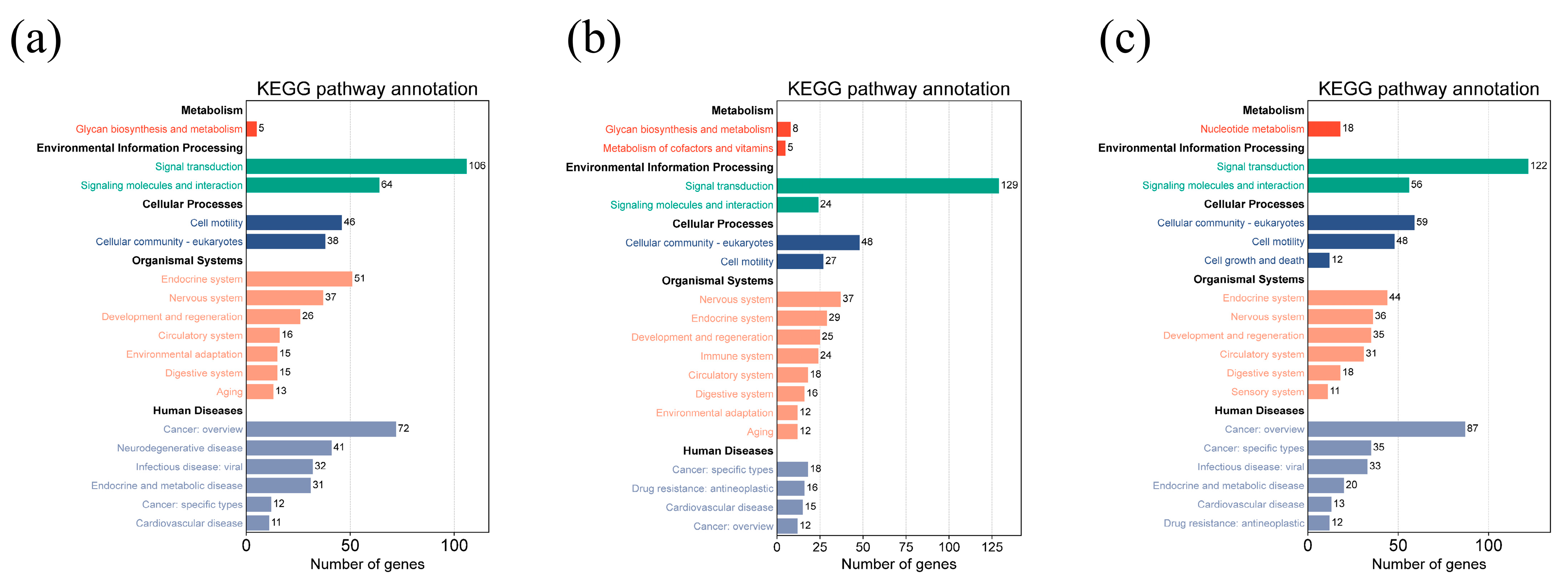
3.3. Gene-Set Enrichment and Analysis
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Shi, Y.; Qi, Y.; Liu, Y.; Rong, Y.; Ao, X.; Zhang, M.; Xia, Q.; Zhang, Y.; Wang, R. Study of the Influence of Non-Genetic Factors on the Growth and Development Traits and Cashmere Production Traits of Inner Mongolia White Cashmere Goats (Erlangshan Type). Vet. Sci. 2024, 11, 308. [Google Scholar] [CrossRef]
- Zhang, X.; Li, G.; Li, F.; Zhang, D.; Yuan, L.; Zhao, Y.; Zhang, Y.; Li, X.; Song, Q.; Wang, W. Effect of feed efficiency on growth performance, body composition, and fat deposition in growing Hu lambs. Anim. Biotechnol. 2023, 34, 183–198. [Google Scholar] [CrossRef] [PubMed]
- Kemper, K.E.; Visscher, P.M.; Goddard, M.E. Genetic architecture of body size in mammals. Genome Biol. 2012, 13, 244. [Google Scholar] [CrossRef]
- Ahmad, S.F.; Khan, N.N.; Ganai, N.A.; Shanaz, S.; Rather, M.A.; Alam, S. Multivariate quantitative genetic analysis of body weight traits in Corriedale sheep. Trop. Anim. Health Prod. 2021, 53, 197. [Google Scholar] [CrossRef]
- Erdenee, S.; Akhatayeva, Z.; Pan, C.; Cai, Y.; Xu, H.; Chen, H.; Lan, X. An insertion/deletion within the CREB1 gene identified using the RNA-sequencing is associated with sheep body morphometric traits. Gene 2021, 775, 145444. [Google Scholar] [CrossRef]
- Kijas, J.W.; Townley, D.; Dalrymple, B.P.; Heaton, M.P.; Maddox, J.F.; McGrath, A.; Wilson, P.; Ingersoll, R.G.; McCulloch, R.; McWilliam, S.; et al. A genome wide survey of SNP variation reveals the genetic structure of sheep breeds. PLoS ONE 2009, 4, e4668. [Google Scholar] [CrossRef]
- Tosser-Klopp, G.; Bardou, P.; Bouchez, O.; Cabau, C.; Crooijmans, R.; Dong, Y.; Donnadieu-Tonon, C.; Eggen, A.; Heuven, H.C.; Jamli, S.; et al. Design and characterization of a 52K SNP chip for goats. PLoS ONE 2014, 9, e86227. [Google Scholar] [CrossRef]
- Zhu, C.; Fan, H.; Yuan, Z.; Hu, S.; Ma, X.; Xuan, J.; Wang, H.; Zhang, L.; Wei, C.; Zhang, Q.; et al. Genome-wide detection of CNVs in Chinese indigenous sheep with different types of tails using ovine high-density 600K SNP arrays. Sci. Rep. 2016, 6, 27822. [Google Scholar] [CrossRef]
- Becker, D.; Tetens, J.; Brunner, A.; Bürstel, D.; Ganter, M.; Kijas, J.; Drögemüller, C. Microphthalmia in Texel sheep is associated with a missense mutation in the paired-like homeodomain 3 (PITX3) gene. PLoS ONE 2010, 5, e8689. [Google Scholar] [CrossRef]
- Stephens, M. A unified framework for association analysis with multiple related phenotypes. PLoS ONE 2013, 8, e65245. [Google Scholar] [CrossRef]
- Bolormaa, S.; Pryce, J.E.; Reverter, A.; Zhang, Y.; Barendse, W.; Kemper, K.; Tier, B.; Savin, K.; Hayes, B.J.; Goddard, M.E. A multi-trait, meta-analysis for detecting pleiotropic polymorphisms for stature, fatness and reproduction in beef cattle. PLoS Genet. 2014, 10, e1004198. [Google Scholar] [CrossRef] [PubMed]
- Xu, H.; Zhang, X.; Zang, R.; Cai, Y.; Cao, X.; Yang, J.; Li, J.; Lan, X.; Wu, J. Genetic variations in the sheep SIRT7 gene and their correlation with body size traits. Arch. Anim. Breed. 2019, 62, 189–197. [Google Scholar] [CrossRef] [PubMed]
- He, S.; Di, J.; Han, B.; Chen, L.; Liu, M.; Li, W. Genome-Wide Scan for Runs of Homozygosity Identifies Candidate Genes Related to Economically Important Traits in Chinese Merino. Animals 2020, 10, 524. [Google Scholar] [CrossRef]
- Tao, L.; Liu, Y.F.; Zhang, H.; Li, H.Z.; Zhao, F.P.; Wang, F.Y.; Zhang, R.S.; Di, R.; Chu, M.X. Genome-wide association study and inbreeding depression on body size traits in Qira black sheep (Ovis aries). Anim. Genet. 2021, 52, 560–564. [Google Scholar] [CrossRef]
- Jiang, J.; Cao, Y.; Shan, H.; Wu, J.; Song, X.; Jiang, Y. The GWAS Analysis of Body Size and Population Verification of Related SNPs in Hu Sheep. Front. Genet. 2021, 12, 642552. [Google Scholar] [CrossRef]
- Tuersuntuoheti, M.; Zhang, J.; Zhou, W.; Zhang, C.L.; Liu, C.; Chang, Q.; Liu, S. Exploring the growth trait molecular markers in two sheep breeds based on Genome-wide association analysis. PLoS ONE 2023, 18, e0283383. [Google Scholar] [CrossRef]
- Posbergh, C.J.; Huson, H.J. All sheeps and sizes: A genetic investigation of mature body size across sheep breeds reveals a polygenic nature. Anim. Genet. 2021, 52, 99–107. [Google Scholar] [CrossRef]
- Li, X.; He, S.G.; Li, W.R.; Luo, L.Y.; Yan, Z.; Mo, D.X.; Wan, X.; Lv, F.H.; Yang, J.; Xu, Y.X.; et al. Genomic analyses of wild argali, domestic sheep, and their hybrids provide insights into chromosome evolution, phenotypic variation, and germplasm innovation. Genome Res. 2022, 32, 1669–1684. [Google Scholar] [CrossRef]
- Li, J.; Zhang, N.; Tian, X.; Tian, P.; Yang, C.; Chen, J.; Yan, H.; Duan, C.; Guo, Y.; Liu, Y.; et al. Construction of Growth Model of Mutton Sheep and Prediction of Growth Performance. Chin. J. Anim. Nutr. 2021, 33, 6462–6473. [Google Scholar] [CrossRef]
- Guo, Y.; Bai, F.; Wang, J.; Fu, S.; Zhang, Y.; Liu, X.; Zhang, Z.; Shao, J.; Li, R.; Wang, F.; et al. Design and characterization of a high-resolution multiple-SNP capture array by target sequencing for sheep. J. Anim. Sci. 2023, 101, skac383. [Google Scholar] [CrossRef]
- Purcell, S.; Neale, B.; Todd-Brown, K.; Thomas, L.; Ferreira, M.A.; Bender, D.; Maller, J.; Sklar, P.; de Bakker, P.I.; Daly, M.J.; et al. PLINK: A tool set for whole-genome association and population-based linkage analyses. Am. J. Hum. Genet. 2007, 81, 559–575. [Google Scholar] [CrossRef] [PubMed]
- Zhou, X.; Stephens, M. Efficient multivariate linear mixed model algorithms for genome-wide association studies. Nat. Methods 2014, 11, 407–409. [Google Scholar] [CrossRef]
- Secco, B.; Camiré, É.; Brière, M.A.; Caron, A.; Billong, A.; Gélinas, Y.; Lemay, A.M.; Tharp, K.M.; Lee, P.L.; Gobeil, S.; et al. Amplification of Adipogenic Commitment by VSTM2A. Cell Rep. 2017, 18, 93–106. [Google Scholar] [CrossRef]
- Al Dow, M.; Secco, B.; Mouchiroud, M.; Rochette, M.; Gilio, G.R.; Massicard, M.; Hardy, M.; Gélinas, Y.; Festuccia, W.T.; Morissette, M.C.; et al. Loss of VSTM2A promotes adipocyte hypertrophy and disrupts metabolic homeostasis. Obesity 2025, 33, 522–536. [Google Scholar] [CrossRef] [PubMed]
- Tan, X.; Liu, L.; Dong, J.; Huang, M.; Zhang, J.; Li, Q.; Wang, H.; Bai, L.; Cui, M.; Zhou, Z.; et al. Genome-wide detections for runs of homozygosity and selective signatures reveal novel candidate genes under domestication in chickens. BMC Genom. 2024, 25, 485. [Google Scholar] [CrossRef]
- Cavarocchi, E.; Whitfield, M.; Chargui, A.; Stouvenel, L.; Lorès, P.; Coutton, C.; Arnoult, C.; Santulli, P.; Patrat, C.; Thierry-Mieg, N.; et al. The sodium/proton exchanger SLC9C1 (sNHE) is essential for human sperm motility and fertility. Clin. Genet. 2021, 99, 684–693. [Google Scholar] [CrossRef]
- Serrano, M.; Ramón, M.; Calvo, J.H.; Jiménez, M.; Freire, F.; Vázquez, J.M.; Arranz, J.J. Genome-wide association studies for sperm traits in Assaf sheep breed. Animal 2021, 15, 100065. [Google Scholar] [CrossRef]
- Hansda, A.K.; Tiwari, A.; Dixit, M. Current status and future prospect of FSHD region gene 1. J. Biosci. 2017, 42, 345–353. [Google Scholar] [CrossRef]
- Hanel, M.L.; Wuebbles, R.D.; Jones, P.L. Muscular dystrophy candidate gene FRG1 is critical for muscle development. Dev. Dyn. 2009, 238, 1502–1512. [Google Scholar] [CrossRef]
- Ding, N.; Tian, D.; Li, X.; Zhang, Z.; Tian, F.; Liu, S.; Han, B.; Liu, D.; Zhao, K. Genetic Polymorphisms of IGF1 and IGF1R Genes and Their Effects on Growth Traits in Hulun Buir Sheep. Genes 2022, 13, 666. [Google Scholar] [CrossRef]
- Li, X.; Ding, N.; Zhang, Z.; Tian, D.; Han, B.; Liu, S.; Liu, D.; Tian, F.; Zhao, K. Identification of Somatostatin Receptor Subtype 1 (SSTR1) Gene Polymorphism and Their Association with Growth Traits in Hulun Buir Sheep. Genes 2021, 13, 77. [Google Scholar] [CrossRef]
- Li, X.; Ding, N.; Zhang, Z.; Tian, D.; Han, B.; Liu, D.; Liu, S.; Tian, F.; Fu, D.; Song, X.; et al. Identification of SSTR5 Gene Polymorphisms and Their Association With Growth Traits in Hulun Buir Sheep. Front. Genet. 2022, 13, 831599. [Google Scholar] [CrossRef]
- Wang, X.; Shi, T.; Zhao, Z.; Hou, H.; Zhang, L. Proteomic analyses of sheep (Ovis aries) embryonic skeletal muscle. Sci. Rep. 2020, 10, 1750. [Google Scholar] [CrossRef]
- Lv, X.; Chen, W.; Wang, S.; Cao, X.; Yuan, Z.; Getachew, T.; Mwacharo, J.M.; Haile, A.; Sun, W. Whole-genome resequencing of Dorper and Hu sheep to reveal selection signatures associated with important traits. Anim. Biotechnol. 2023, 34, 3016–3026. [Google Scholar] [CrossRef]
- Lou, M.; Zhang, S.; Yang, W.; Li, S.; Cao, H.; Zhang, Z.; Ling, Y. Transcriptome analysis revealed the mechanism of skeletal muscle growth and development in different hybrid sheep. Anim. Biosci. 2025, 38, 408–418. [Google Scholar] [CrossRef]
- Verbrugge, S.A.J.; Schönfelder, M.; Becker, L.; Yaghoob Nezhad, F.; Hrabě de Angelis, M.; Wackerhage, H. Genes Whose Gain or Loss-Of-Function Increases Skeletal Muscle Mass in Mice: A Systematic Literature Review. Front. Physiol. 2018, 9, 553. [Google Scholar] [CrossRef]
- Samereh, S.; Hajarian, H.; Karamishabankareh, H.; Soltani, L.; Foroutanifar, S. Effects of different concentrations of Chir98014 as an activator of Wnt/beta-catenin signaling pathway on oocyte in-vitro maturation and subsequent embryonic development in Sanjabi ewes. Reprod. Domest. Anim. 2021, 56, 965–971. [Google Scholar] [CrossRef]
- Chen, Q.; Bao, J.J.; Zhang, H.C.; Huang, C.; Zhao, Q.; Pu, Y.B.; Jiang, L.; Hosseiny, A.; Ibrahim, M.; Hussain, T.; et al. LncRNA GTL2 regulates myoblast proliferation and differentiation via the PKA-CREB pathway in Duolang sheep. Zool. Res. 2024, 45, 1261–1275. [Google Scholar] [CrossRef]
- Yengo, L.; Sidorenko, J.; Kemper, K.E.; Zheng, Z.; Wood, A.R.; Weedon, M.N.; Frayling, T.M.; Hirschhorn, J.; Yang, J.; Visscher, P.M. Meta-analysis of genome-wide association studies for height and body mass index in ∼700000 individuals of European ancestry. Hum. Mol. Genet. 2018, 27, 3641–3649. [Google Scholar] [CrossRef]
- Li, M.; Zhu, L.; Li, X.; Shuai, S.; Teng, X.; Xiao, H.; Li, Q.; Chen, L.; Guo, Y.; Wang, J. Expression profiling analysis for genes related to meat quality and carcass traits during postnatal development of backfat in two pig breeds. Sci. China C Life Sci. 2008, 51, 718–733. [Google Scholar] [CrossRef]
- Schaid, D.J.; Chen, W.; Larson, N.B. From genome-wide associations to candidate causal variants by statistical fine-mapping. Nat. Rev. Genet. 2018, 19, 491–504. [Google Scholar] [CrossRef] [PubMed]
- Yang, T.; Wang, M.; Liu, Y.; Li, Y.; Feng, M.; Zhao, C. A mutation in POLR2A gene associated with body size traits in Dezhou donkeys revealed with GWAS. J. Anim. Sci. 2024, 102, skae217. [Google Scholar] [CrossRef] [PubMed]


| Group | Age | The Number of Ewes | The Number of Rams |
|---|---|---|---|
| Group 1 | 4 months old | 2 | 0 |
| 6 months old | 122 | 125 | |
| Group 2 | 12 months old | 2 | 3 |
| 24 months old | 233 | 26 | |
| Group 3 | 36 months old | 142 | 5 |
| 48 months old | 106 | 1 | |
| 60 months old | 30 | 0 | |
| 70 months old | 2 | 0 |
| Trait | Group | Mean | SD |
|---|---|---|---|
| Chest girth, cm | 1 | 70.74 | 6.08 |
| 2 | 85.89 | 8.25 | |
| 3 | 93.06 | 6.91 | |
| Cannon circumference, cm | 1 | 7.25 | 0.53 |
| 2 | 7.7 | 0.48 | |
| 3 | 7.71 | 0.46 | |
| Hip width, cm | 1 | 15.91 | 1.92 |
| 2 | 18.92 | 1.85 | |
| 3 | 19.59 | 1.63 | |
| Body height, cm | 1 | 58.46 | 3.97 |
| 2 | 61.96 | 6.87 | |
| 3 | 65.20 | 5.58 | |
| Body length, cm | 1 | 62.47 | 5.41 |
| 2 | 63.33 | 9.03 | |
| 3 | 63.82 | 7.53 |
| Group | Chr. | Ref/Mut | Position (bp) | p-Value | Nearest Gene | Location |
|---|---|---|---|---|---|---|
| 2 | 4 | G/C | 512,090 | 1.21 × 10−7 | VSTM2A; LOC101105741 | intergenic |
| 2 | 1 | G/A | 175,232,922 | 1.95 × 10−7 | SLC9C1 | intron |
| 2 | 26 | G/T | 17,254,807 | 1.05 × 10−6 | LOC105605168; FRG1 | intergenic |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yang, H.; Li, T.; Zhang, N.; Chen, J.; Zhang, Y.; Peng, S.; Zhou, L.; Ma, R.; Zhang, Z.; Liu, Q.; et al. Identification of Candidate Genes and Functional Pathways Associated with Body Size Traits in Hulunbuir Sheep Through GWAS Analysis. Genes 2025, 16, 410. https://doi.org/10.3390/genes16040410
Yang H, Li T, Zhang N, Chen J, Zhang Y, Peng S, Zhou L, Ma R, Zhang Z, Liu Q, et al. Identification of Candidate Genes and Functional Pathways Associated with Body Size Traits in Hulunbuir Sheep Through GWAS Analysis. Genes. 2025; 16(4):410. https://doi.org/10.3390/genes16040410
Chicago/Turabian StyleYang, Hengqian, Tingting Li, Na Zhang, Jieran Chen, Yuting Zhang, Shiyu Peng, Lisheng Zhou, Runlin Ma, Zhichao Zhang, Qiuyue Liu, and et al. 2025. "Identification of Candidate Genes and Functional Pathways Associated with Body Size Traits in Hulunbuir Sheep Through GWAS Analysis" Genes 16, no. 4: 410. https://doi.org/10.3390/genes16040410
APA StyleYang, H., Li, T., Zhang, N., Chen, J., Zhang, Y., Peng, S., Zhou, L., Ma, R., Zhang, Z., Liu, Q., Wang, H., & He, J. (2025). Identification of Candidate Genes and Functional Pathways Associated with Body Size Traits in Hulunbuir Sheep Through GWAS Analysis. Genes, 16(4), 410. https://doi.org/10.3390/genes16040410







