Discovery of Novel APOC3 Isoforms in Hepatic and Intestinal Cell Models Using Long-Read RNA Sequencing
Abstract
1. Introduction
2. Materials and Methods
2.1. RNA Sequencing Data
2.2. Bioinformatics Analyses
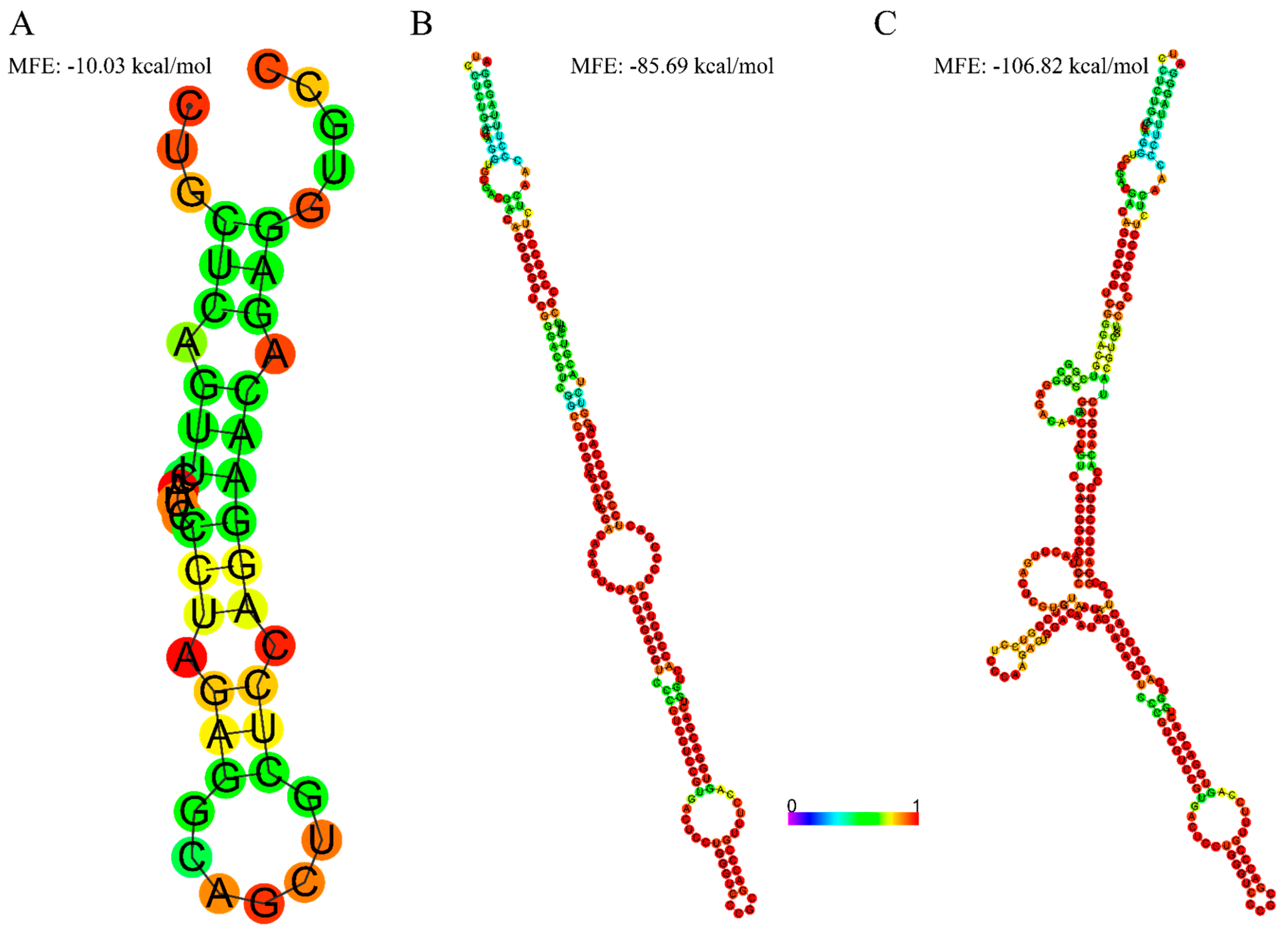
2.3. Secondary Structure Prediction
2.4. Phylogenetic Trees
2.5. Cell Lines and Culture
2.6. RNA Extraction
2.7. RT-PCR and Gel Electrophoresis
2.8. Band Stab PCR and Sanger Sequencing
2.9. Data and Code Availability
3. Results
3.1. Long-Read Transcriptome Annotation Reveals Novel APOC3 Isoforms
3.2. Extended 5′UTR in Novel-2 Is Associated with More Secondary Structures
3.3. Novel Isoforms Are Expressed in Caco-2 Cells
3.4. RT-PCR and Sanger Sequencing Confirm Novel APOC3 Isoforms
3.5. Novel Isoforms Cluster Closely with Known Human APOC3 Transcripts
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Sarwar, N.; Danesh, J.; Eiriksdottir, G.; Sigurdsson, G.; Wareham, N.; Bingham, S.; Boekholdt, S.M.; Khaw, K.-T.; Gudnason, V. Triglycerides and the risk of coronary heart disease: 10,158 incident cases among 262,525 participants in 29 Western prospective studies. Circulation 2007, 115, 450–458. [Google Scholar] [CrossRef] [PubMed]
- Austin, M.A.; Hokanson, J.E.; Edwards, K.L. Hypertriglyceridemia as a cardiovascular risk factor. Am. J. Cardiol. 1998, 81, 7b–12b. [Google Scholar] [CrossRef] [PubMed]
- Kersten, S. Physiological regulation of lipoprotein lipase. Biochim. Biophys. Acta 2014, 1841, 919–933. [Google Scholar] [CrossRef] [PubMed]
- Ding, Y.; Wang, Y.; Zhu, H.; Fan, J.; Yu, L.; Liu, G.; Liu, E. Hypertriglyceridemia and delayed clearance of fat load in transgenic rabbits expressing human apolipoprotein CIII. Transgenic Res. 2011, 20, 867–875. [Google Scholar] [CrossRef]
- Jørgensen, A.B.; Frikke-Schmidt, R.; Nordestgaard, B.G.; Tybjærg-Hansen, A. Loss-of-function mutations in APOC3 and risk of ischemic vascular disease. N. Engl. J. Med. 2014, 371, 32–41. [Google Scholar] [CrossRef]
- Crosby, J.; Peloso, G.M.; Auer, P.L.; Crosslin, D.R.; Stitziel, N.O.; Lange, L.A.; Lu, Y.; Tang, Z.Z.; Zhang, H.; Hindy, G.; et al. Loss-of-function mutations in APOC3, triglycerides, and coronary disease. N. Engl. J. Med. 2014, 371, 22–31. [Google Scholar] [CrossRef]
- Pollin, T.I.; Damcott, C.M.; Shen, H.; Ott, S.H.; Shelton, J.; Horenstein, R.B.; Post, W.; McLenithan, J.C.; Bielak, L.F.; Peyser, P.A.; et al. A null mutation in human APOC3 confers a favorable plasma lipid profile and apparent cardioprotection. Science 2008, 322, 1702–1705. [Google Scholar] [CrossRef]
- Ito, Y.; Azrolan, N.; O’Connell, A.; Walsh, A.; Breslow, J.L. Hypertriglyceridemia as a Result of Human apo CII Gene Expression in Transgenic Mice. Science 1990, 249, 790–793. [Google Scholar] [CrossRef]
- Maeda, N.; Li, H.; Lee, D.; Oliver, P.; Quarfordt, S.; Osada, J. Targeted disruption of the apolipoprotein C-III gene in mice results in hypotriglyceridemia and protection from postprandial hypertriglyceridemia. J. Biol. Chem. 1994, 269, 23610–23616. [Google Scholar] [CrossRef]
- Qamar, A.; Khetarpal, S.A.; Khera, A.V.; Qasim, A.; Rader, D.J.; Reilly, M.P. Plasma Apolipoprotein C-III Levels, Triglycerides, and Coronary Artery Calcification in Type 2 Diabetics. Arterioscler. Thromb. Vasc. Biol. 2015, 35, 1880–1888. [Google Scholar] [CrossRef]
- Åvall, K.; Ali, Y.; Leibiger, I.B.; Leibiger, B.; Moede, T.; Paschen, M.; Dicker, A.; Daré, E.; Köhler, M.; Ilegems, E.; et al. Apolipoprotein CIII links islet insulin resistance to β-cell failure in diabetes. Proc. Natl. Acad. Sci. USA 2015, 112, E2611–E2619. [Google Scholar] [CrossRef] [PubMed]
- Bozzetto, L.; Berntzen, B.J.; Kaprio, J.; Rissanen, A.; Taskinen, M.-R.; Pietiläinen, K.H. A higher glycemic response to oral glucose is associated with higher plasma apolipoprotein C3 independently of BMI in healthy twins. Nutr. Metab. Cardiovasc. Dis. 2020, 30, 459–466. [Google Scholar] [CrossRef] [PubMed]
- Sol, E.-r.M.; Sundsten, T.; Bergsten, P. Role of MAPK in apolipoprotein CIII-induced apoptosis in INS-1E cells. Lipids Health Dis. 2009, 8, 3. [Google Scholar] [PubMed]
- Kawakami, A.; Aikawa, M.; Alcaide, P.; Luscinskas, F.W.; Libby, P.; Sacks, F.M. Apolipoprotein CIII Induces Expression of Vascular Cell Adhesion Molecule-1 in Vascular Endothelial Cells and Increases Adhesion of Monocytic Cells. Circulation 2006, 114, 681–687. [Google Scholar] [CrossRef]
- Harrison, P.W.; Amode, M.R.; Austine-Orimoloye, O.; Azov, A.G.; Barba, M.; Barnes, I.; Becker, A.; Bennett, R.; Berry, A.; Bhai, J.; et al. Ensembl 2024. Nucleic Acids Res. 2023, 52, D891–D899. [Google Scholar] [CrossRef]
- Wu, J.; Hu, W.; Li, S. Long-read transcriptome sequencing reveals allele-specific variants at high resolution. Trends Genet. 2023, 39, 31–33. [Google Scholar] [CrossRef]
- Glinos, D.A.; Garborcauskas, G.; Hoffman, P.; Ehsan, N.; Jiang, L.; Gokden, A.; Dai, X.; Aguet, F.; Brown, K.L.; Garimella, K.; et al. Transcriptome variation in human tissues revealed by long-read sequencing. Nature 2022, 608, 353–359. [Google Scholar] [CrossRef]
- Pyatnitskiy, M.A.; Arzumanian, V.A.; Radko, S.P.; Ptitsyn, K.G.; Vakhrushev, I.V.; Poverennaya, E.V.; Ponomarenko, E.A. Oxford Nanopore MinION Direct RNA-Seq for Systems Biology. Biology 2021, 10, 1131. [Google Scholar] [CrossRef]
- Sarygina, E.; Kozlova, A.; Deinichenko, K.; Radko, S.; Ptitsyn, K.; Khmeleva, S.; Kurbatov, L.K.; Spirin, P.; Prassolov, V.S.; Ilgisonis, E.; et al. Principal Component Analysis of Alternative Splicing Profiles Revealed by Long-Read ONT Sequencing in Human Liver Tissue and Hepatocyte-Derived HepG2 and Huh7 Cell Lines. Int. J. Mol. Sci. 2023, 24, 15502. [Google Scholar] [CrossRef]
- Kaur, G.; Perteghella, T.; Carbonell-Sala, S.; Gonzalez-Martinez, J.; Hunt, T.; Mądry, T.; Jungreis, I.; Arnan, C.; Lagarde, J.; Borsari, B.; et al. GENCODE: Massively expanding the lncRNA catalog through capture long-read RNA sequencing. bioRxiv 2024. [Google Scholar] [CrossRef]
- Yang, J.; Hirai, Y.; Iida, K.; Ito, S.; Trumm, M.; Terada, S.; Sakai, R.; Tsuchiya, T.; Tabata, O.; Kamei, K.-I. Integrated-gut-liver-on-a-chip platform as an in vitro human model of non-alcoholic fatty liver disease. Commun. Biol. 2023, 6, 310. [Google Scholar] [CrossRef]
- Pardo-Palacios, F.J.; Arzalluz-Luque, A.; Kondratova, L.; Salguero, P.; Mestre-Tomás, J.; Amorín, R.; Estevan-Morió, E.; Liu, T.; Nanni, A.; McIntyre, L.; et al. SQANTI3: Curation of long-read transcriptomes for accurate identification of known and novel isoforms. Nat. Methods 2024, 21, 793–797. [Google Scholar] [CrossRef]
- Gruber, A.R.; Lorenz, R.; Bernhart, S.H.; Neuböck, R.; Hofacker, I.L. The Vienna RNA websuite. Nucleic Acids Res. 2008, 36, W70–W74. [Google Scholar] [CrossRef] [PubMed]
- Wilton, S.D.; Lim, L.; Dye, D.; Laing, N. Bandstab: A PCR-based alternative to cloning PCR products. Biotechniques 1997, 22, 642–645. [Google Scholar] [CrossRef] [PubMed]
- Ebert, P.; Audano, P.A.; Zhu, Q.; Rodriguez-Martin, B.; Porubsky, D.; Bonder, M.J.; Sulovari, A.; Ebler, J.; Zhou, W.; Mari, R.S.; et al. Haplotype-resolved diverse human genomes and integrated analysis of structural variation. Science 2021, 372, eabf7117. [Google Scholar] [CrossRef] [PubMed]
- Steijger, T.; Abril, J.F.; Engström, P.G.; Kokocinski, F.; The RGASP Consortium; Hubbard, T.J.; Guigó, R.; Harrow, J.; Bertone, P. Assessment of transcript reconstruction methods for RNA-seq. Nat. Methods 2013, 10, 1177–1184. [Google Scholar] [CrossRef]
- Carninci, P.; Kasukawa, T.; Katayama, S.; Gough, J.; Frith, M.C.; Maeda, N.; Oyama, R.; Ravasi, T.; Lenhard, B.; Wells, C.; et al. The Transcriptional Landscape of the Mammalian Genome. Science 2005, 309, 1559–1563. [Google Scholar] [CrossRef]
- Eden, E.; Brunak, S. Analysis and recognition of 5′ UTR intron splice sites in human pre-mRNA. Nucleic Acids Res. 2004, 32, 1131–1142. [Google Scholar] [CrossRef]
- Corley, M.; Solem, A.; Phillips, G.; Lackey, L.; Ziehr, B.; Vincent, H.A.; Mustoe, A.M.; Ramos, S.B.V.; Weeks, K.M.; Moorman, N.J.; et al. An RNA structure-mediated, posttranscriptional model of human α-1-antitrypsin expression. Proc. Natl. Acad. Sci. USA 2017, 114, E10244–E10253. [Google Scholar] [CrossRef]
- Sobczak, K.; Krzyzosiak, W.J. Structural Determinants of BRCA1 Translational Regulation. J. Biol. Chem. 2002, 277, 17349–17358. [Google Scholar] [CrossRef]
- Pelletier, J.; Sonenberg, N. Insertion mutagenesis to increase secondary structure within the 5′ noncoding region of a eukaryotic mRNA reduces translational efficiency. Cell 1985, 40, 515–526. [Google Scholar] [CrossRef] [PubMed]
- Zhan, Y.; Hu, Z.; Lu, Z.; Lin, Z. The evolution of 5′ UTR length was shaped by natural selection for its functional impact on gene expression. bioRxiv 2024. [Google Scholar] [CrossRef]
- Wieder, N.; D’souza, E.N.; Martin-Geary, A.C.; Lassen, F.H.; Talbot-Martin, J.; Fernandes, M.; Chothani, S.P.; Rackham, O.J.L.; Schafer, S.; Aspden, J.L.; et al. Differences in 5′untranslated regions highlight the importance of translational regulation of dosage sensitive genes. Genome Biol. 2024, 25, 111. [Google Scholar] [CrossRef]
- Nitschke, L.; Tewari, A.; Coffin, S.L.; Xhako, E.; Pang, K.; Gennarino, V.A.; Johnson, J.L.; Blanco, F.A.; Liu, Z.; Zoghbi, H.Y. miR760 regulates ATXN1 levels via interaction with its 5′ untranslated region. Genes Dev. 2020, 34, 1147–1160. [Google Scholar] [CrossRef]
- Sureau, A.; Gattoni, R.; Dooghe, Y.; Stévenin, J.; Soret, J. SC35 autoregulates its expression by promoting splicing events that destabilize its mRNAs. EMBO J. 2001, 20, 1785–1796. [Google Scholar] [CrossRef]
- Zhou, Y.; Liu, S.; Liu, G.; Oztürk, A.; Hicks, G.G. ALS-associated FUS mutations result in compromised FUS alternative splicing and autoregulation. PLoS Genet. 2013, 9, e1003895. [Google Scholar] [CrossRef]
- Wollerton, M.C.; Gooding, C.; Wagner, E.J.; Garcia-Blanco, M.A.; Smith, C.W. Autoregulation of polypyrimidine tract binding protein by alternative splicing leading to nonsense-mediated decay. Mol. Cell 2004, 13, 91–100. [Google Scholar] [CrossRef]
- Sun, Y.; Bao, Y.; Han, W.; Song, F.; Shen, X.; Zhao, J.; Zuo, J.; Saffen, D.; Chen, W.; Wang, Z.; et al. Autoregulation of RBM10 and cross-regulation of RBM10/RBM5 via alternative splicing-coupled nonsense-mediated decay. Nucleic Acids Res. 2017, 45, 8524–8540. [Google Scholar] [CrossRef]
- Jong, M.C.; Hofker, M.H.; Havekes, L.M. Role of ApoCs in Lipoprotein Metabolism. Arterioscler. Thromb. Vasc. Biol. 1999, 19, 472–484. [Google Scholar] [CrossRef]
- Graham, M.J.; Lee, R.G.; Bell, T.A., III; Fu, W.; Mullick, A.E.; Alexander, V.J.; Singleton, W.; Viney, N.; Geary, R.; Su, J.; et al. Antisense oligonucleotide inhibition of apolipoprotein C-III reduces plasma triglycerides in rodents, nonhuman primates, and humans. Circ. Res. 2013, 112, 1479–1490. [Google Scholar] [CrossRef]
- Gordts, P.L.; Nock, R.; Son, N.-H.; Ramms, B.; Lew, I.; Gonzales, J.C.; Thacker, B.E.; Basu, D.; Lee, R.G.; Mullick, A.E.; et al. ApoC-III inhibits clearance of triglyceride-rich lipoproteins through LDL family receptors. J. Clin. Investig. 2016, 126, 2855–2866. [Google Scholar] [CrossRef] [PubMed]
- Ramms, B.; Patel, S.; Sun, X.; Pessentheiner, A.R.; Ducasa, G.M.; Mullick, A.E.; Lee, R.G.; Crooke, R.M.; Tsimikas, S.; Witztum, J.L.; et al. Interventional hepatic apoC-III knockdown improves atherosclerotic plaque stability and remodeling by triglyceride lowering. JCI Insight 2022, 7, e158414. [Google Scholar] [CrossRef] [PubMed]
- Wu, A.L.; Windmueller, H.G. Identification of circulating apolipoproteins synthesized by rat small intestine in vivo. J. Biol. Chem. 1978, 253, 2525–2528. [Google Scholar] [PubMed]


| Isoform | Structural_Category | Associated_Transcript | Coding | Subcategory |
|---|---|---|---|---|
| Novel-1 | novel_in_catalog | novel | coding | combination_of_known_junctions |
| Novel-2 | full-splice_match | ENST00000227667.8 | coding | alternative_5end |
| Novel-3 | novel_in_catalog | novel | non_coding | combination_of_known_splicesites |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Farstad-O’Halloran, K.; Sooda, A.; Iqbal, T.; Wilton, S.; Aung-Htut, M.T. Discovery of Novel APOC3 Isoforms in Hepatic and Intestinal Cell Models Using Long-Read RNA Sequencing. Genes 2025, 16, 412. https://doi.org/10.3390/genes16040412
Farstad-O’Halloran K, Sooda A, Iqbal T, Wilton S, Aung-Htut MT. Discovery of Novel APOC3 Isoforms in Hepatic and Intestinal Cell Models Using Long-Read RNA Sequencing. Genes. 2025; 16(4):412. https://doi.org/10.3390/genes16040412
Chicago/Turabian StyleFarstad-O’Halloran, Kara, Anuradha Sooda, Tooba Iqbal, Steve Wilton, and May T. Aung-Htut. 2025. "Discovery of Novel APOC3 Isoforms in Hepatic and Intestinal Cell Models Using Long-Read RNA Sequencing" Genes 16, no. 4: 412. https://doi.org/10.3390/genes16040412
APA StyleFarstad-O’Halloran, K., Sooda, A., Iqbal, T., Wilton, S., & Aung-Htut, M. T. (2025). Discovery of Novel APOC3 Isoforms in Hepatic and Intestinal Cell Models Using Long-Read RNA Sequencing. Genes, 16(4), 412. https://doi.org/10.3390/genes16040412






