Global Transcriptome Analysis Reveals the Molecular Mechanism Underlying Seed Physical Dormancy Formation in Medicago sativa
Abstract
1. Introduction
2. Materials and Methods
2.1. Plant Material and Sample Collection
2.2. Seed Imbibition
2.3. Transcriptome Sequencing and Analysis of Seed Coat-Specific Expression Genes
2.4. Differential Gene Expression Analysis and Functional Enrichment
2.5. Transcription Factor Family Analysis
2.6. Quantitative Real-Time PCR Analysis
2.7. Gene Expression Analysis of M. truncatula Homologous Genes
3. Results
3.1. Transcriptomic Screening of Genes Specifically Expressed in Alfalfa Seed Coat
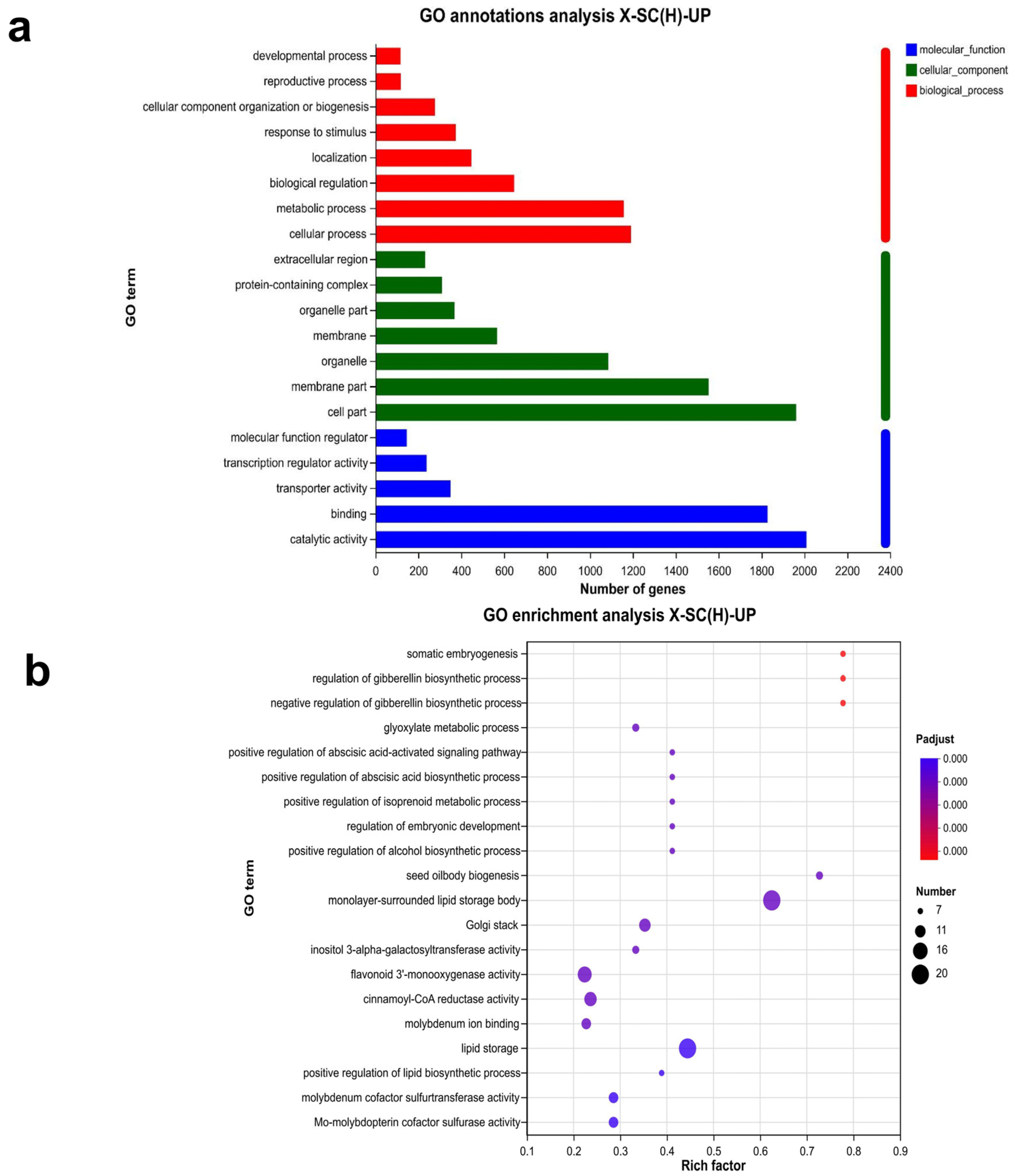
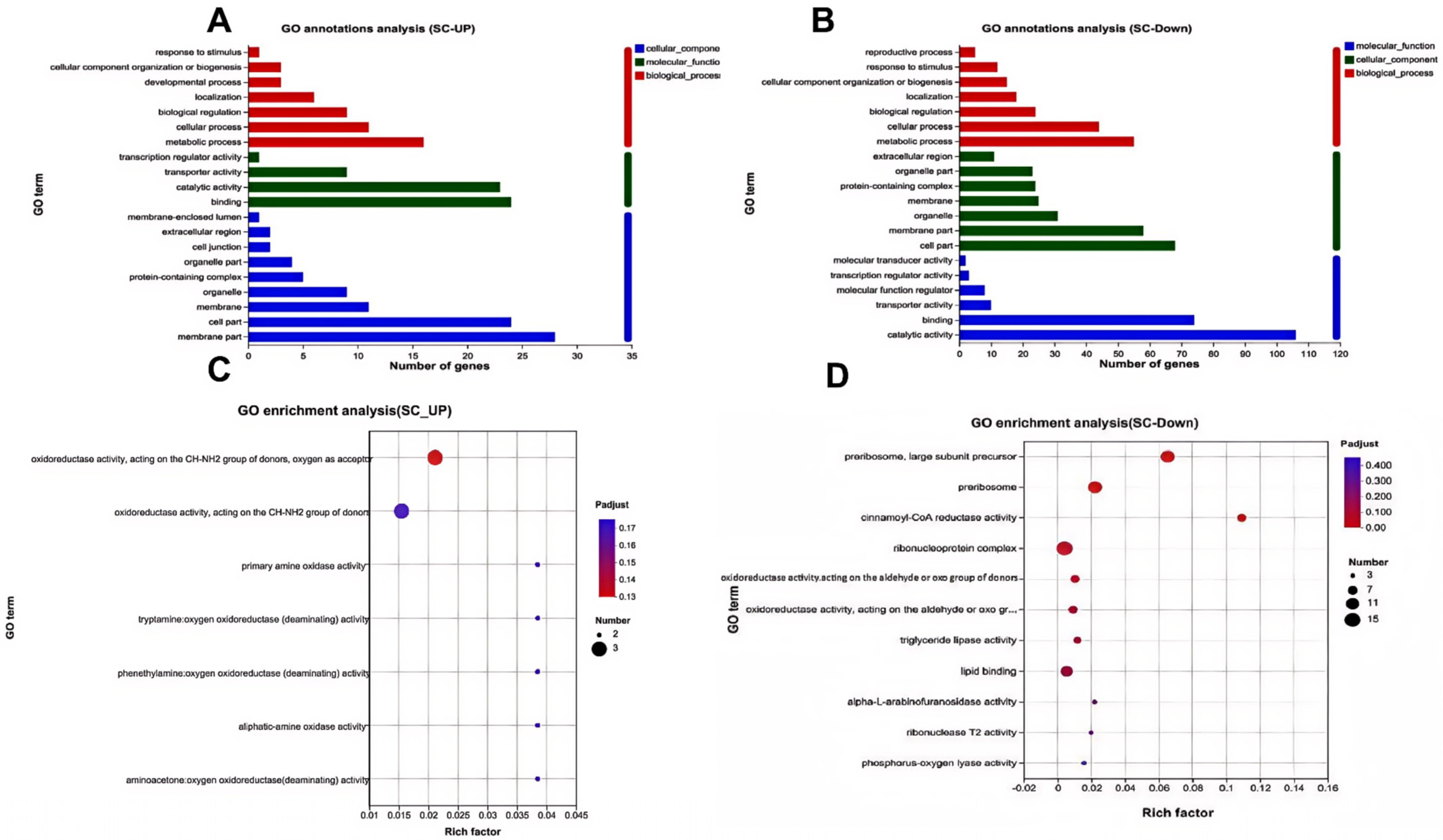
3.2. Gene Ontology Annotation and Enrichment of Genes Specifically Expressed in Seed Coat
3.3. Kyoto Encyclopedia of Genes and Genomes Annotation and Enrichment of Genes Specifically Expressed in Seed Coat
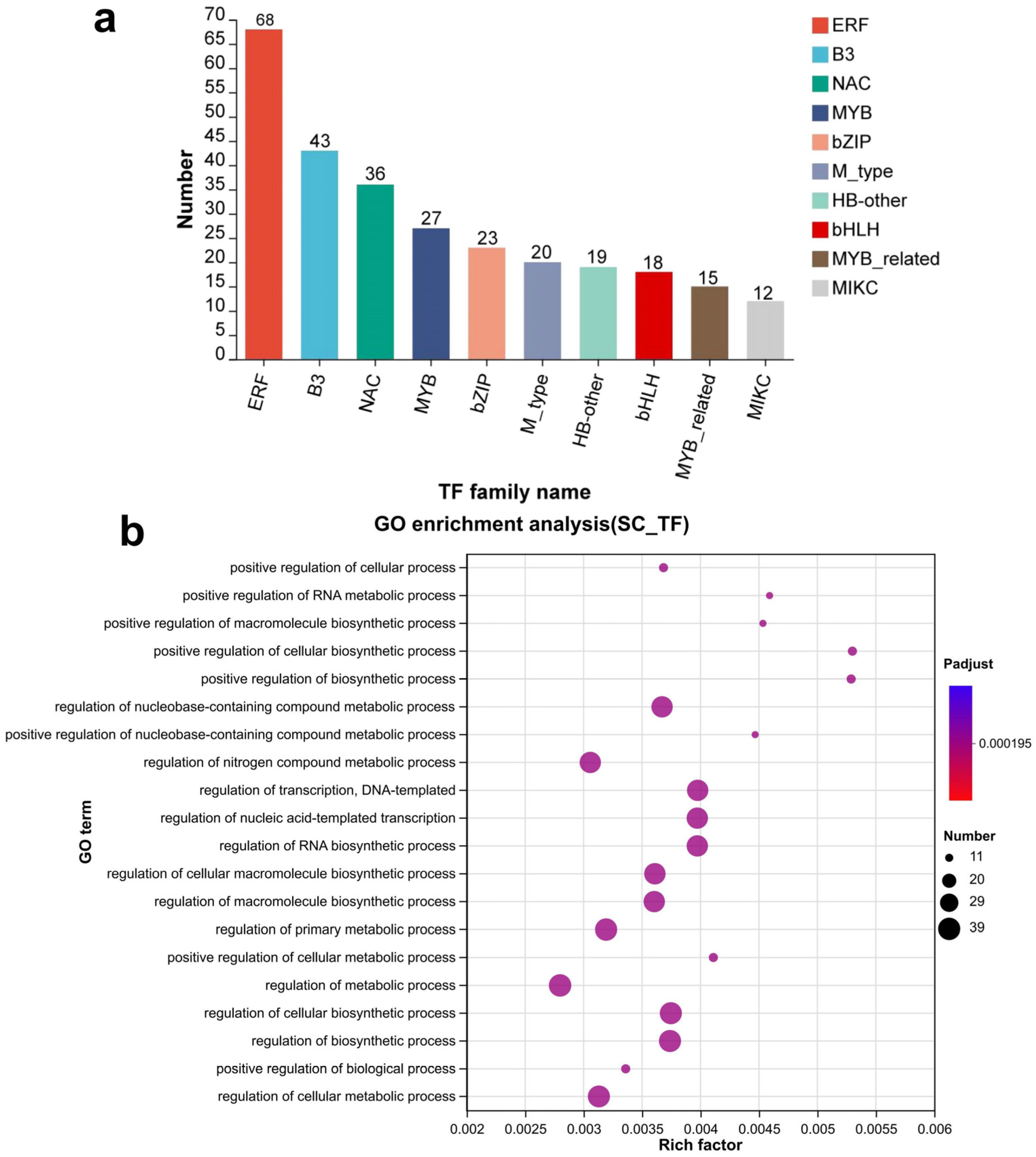
3.4. Transcription Factor Prediction of Genes Specifically Expressed in Seed Coat
3.5. Hard-Seededness Variation in Different Alfalfa Cultivars
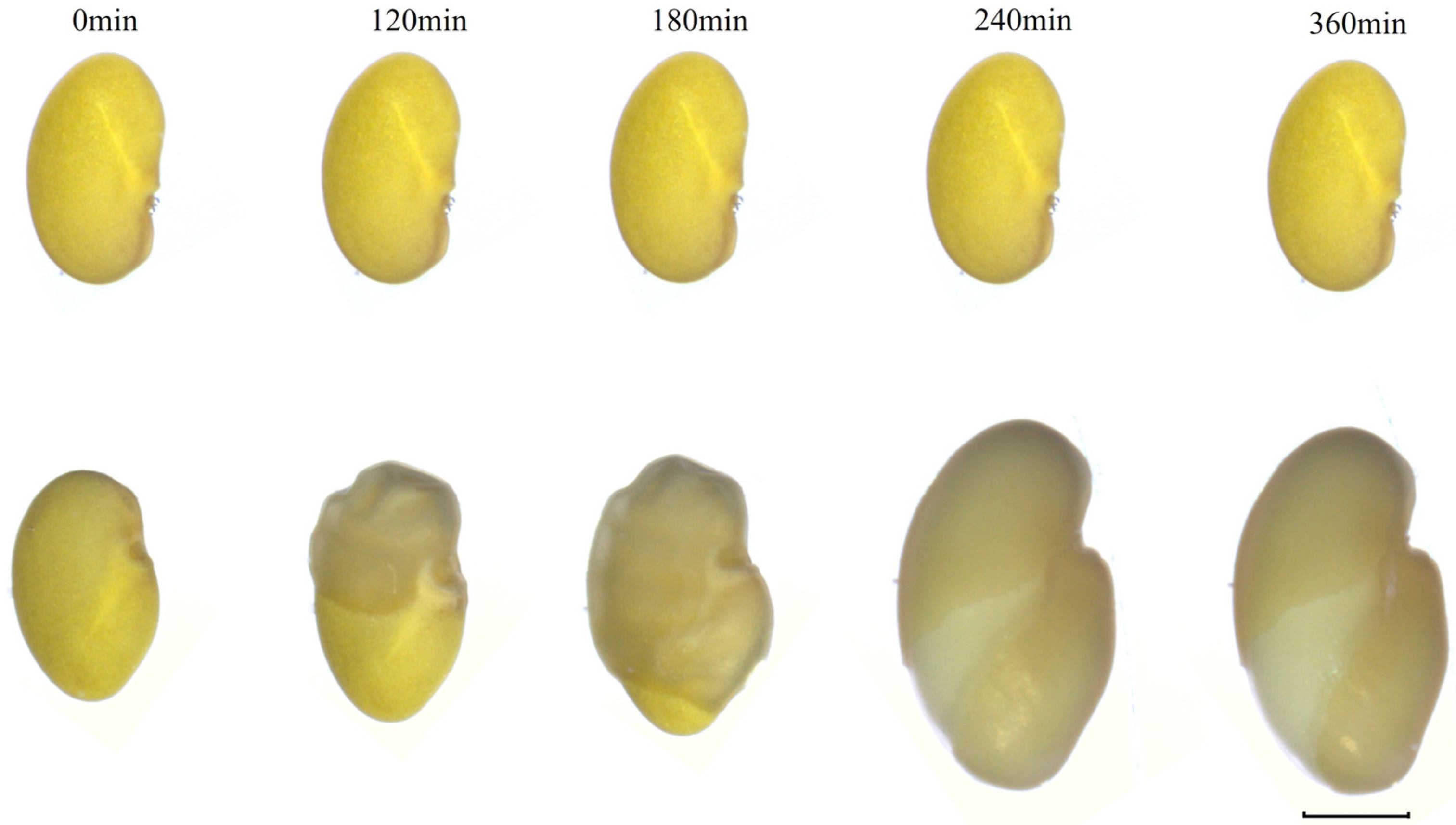
3.6. Phenotypic Changes of Seeds with and Without Physical Dormancy
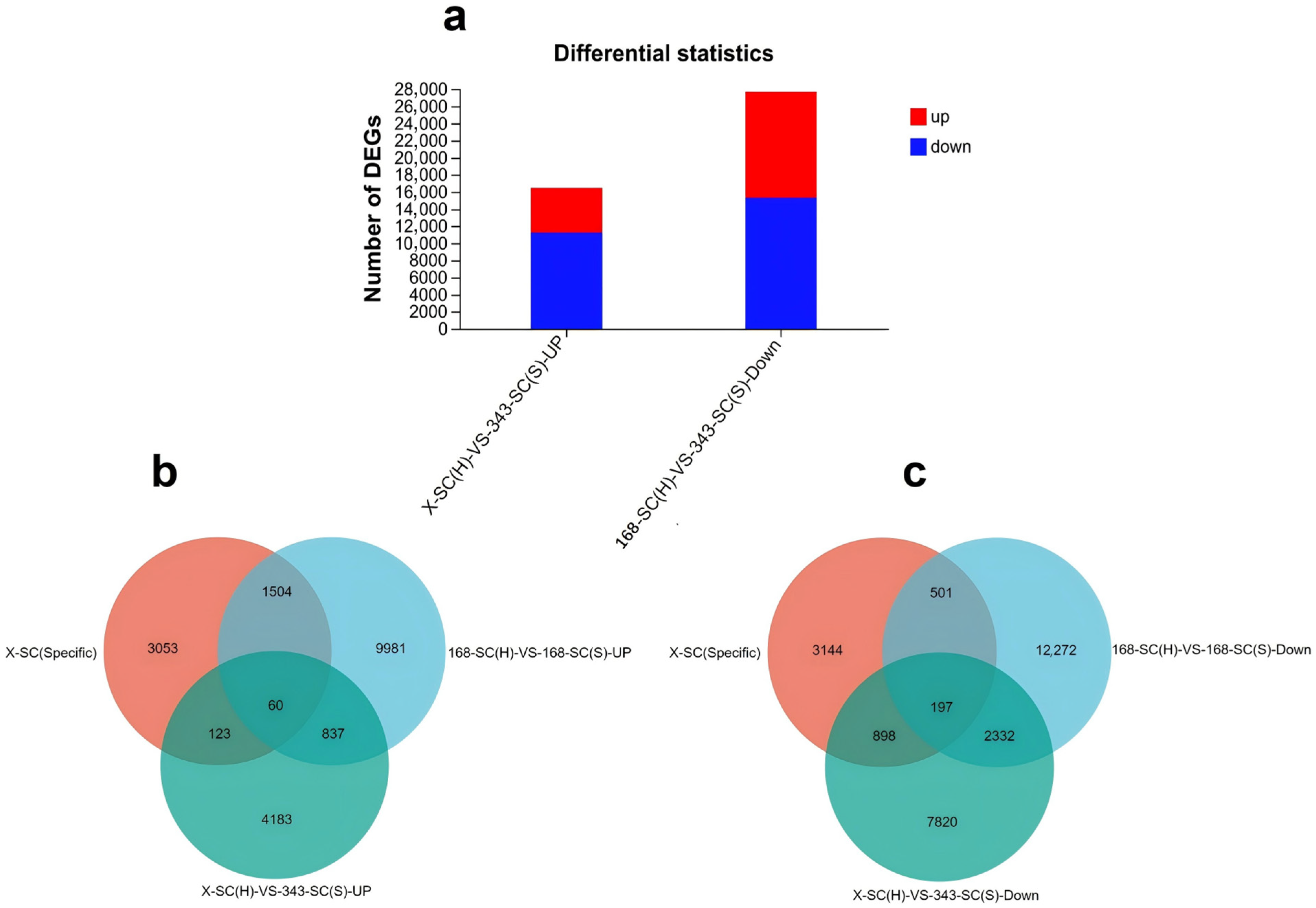
3.7. Transcription Analysis of Seed Coats from Seeds with and Without Physical Dormancy
3.8. GO Annotation and Enrichment Analysis of DEGs in Seed Coats with and Without Physical Dormancy
3.9. KEGG Annotation and Enrichment of DEGs in Hard and Non-Hard Seed Coats
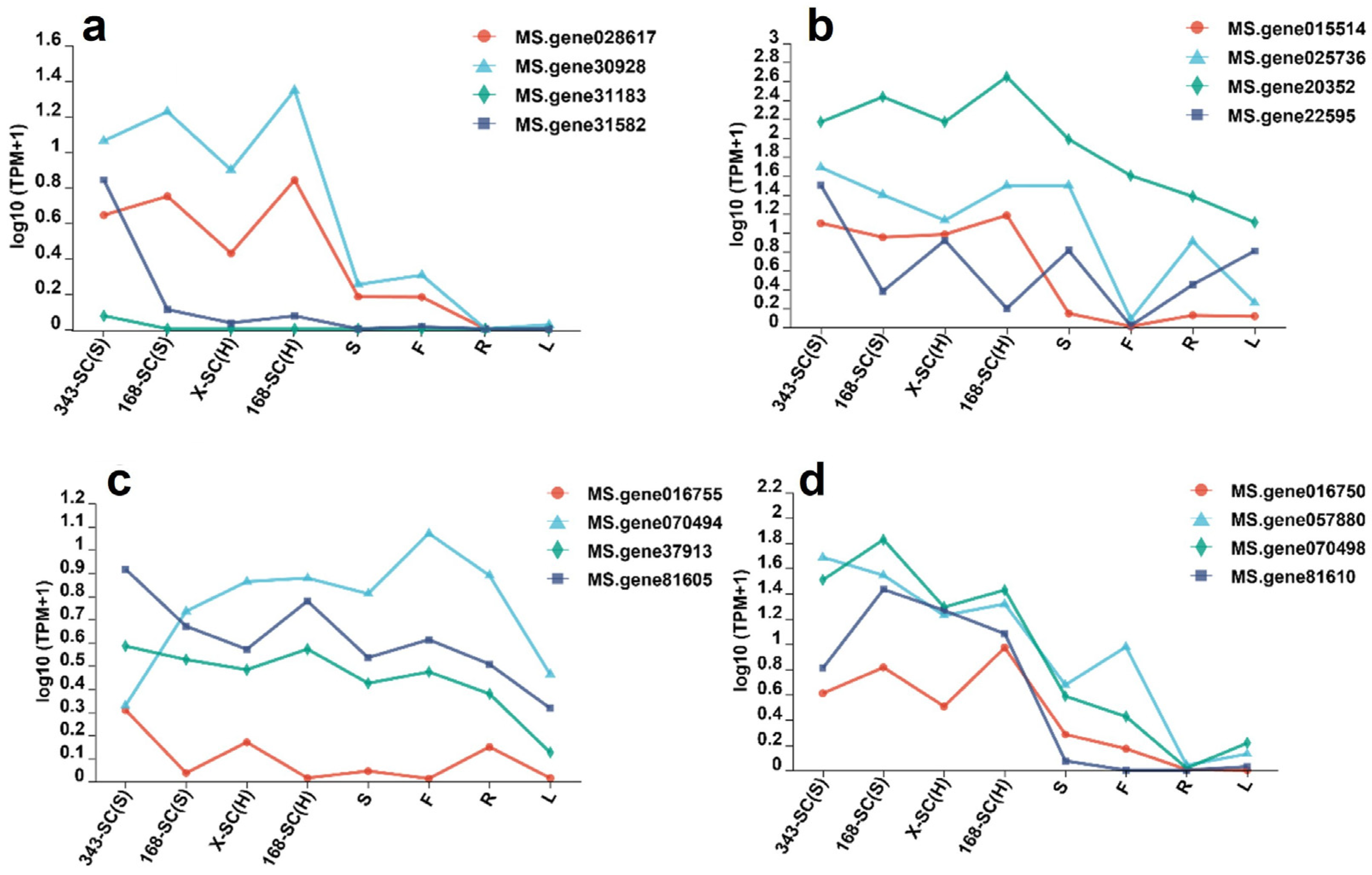
3.10. Screening Genes Associated with Seeds’ Physical Dormancy
3.11. Quantitative Real-Time PCR Verification of Screened DEGs in Seed Coats with Hard-Seededness
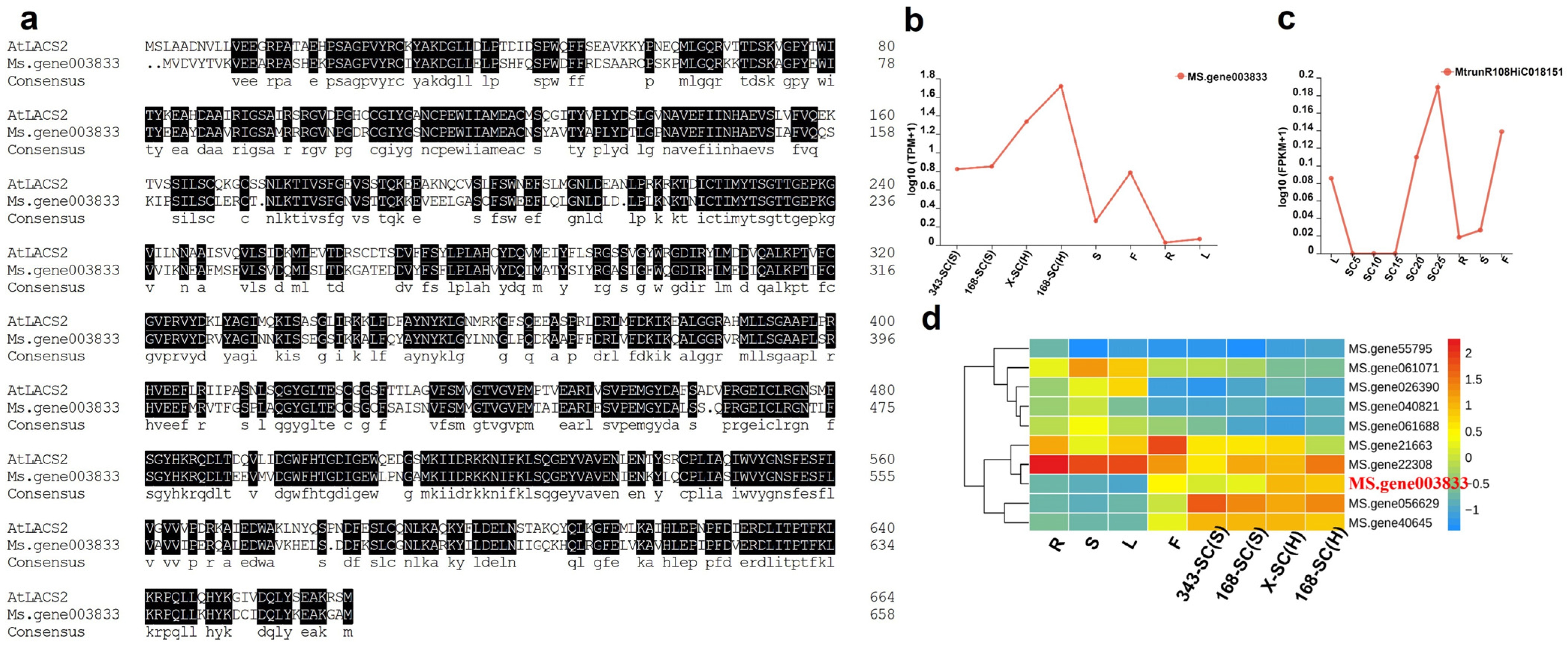
3.12. BLAST and Bioinformatics Analysis of Candidate Genes Associated with Hard-Seededness
3.13. Analysis of Homologous Genes of MtKCS12, MtKNOX4, GmHs1-1, and GmqHs1 in Alfalfa
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Hadidi, M.; Orellana Palacios, J.C.; McClements, D.J.; Mahfouzi, M.; Moreno, A. Alfalfa as a sustainable source of plant-based food proteins. Trends Food Sci. Technol. 2023, 135, 202–214. [Google Scholar] [CrossRef]
- Singer, S.D.; Hannoufa, A.; Acharya, S. Molecular improvement of alfalfa for enhanced productivity and adaptability in a changing environment. Plant Cell Environ. 2018, 41, 1955–1971. [Google Scholar] [CrossRef]
- Lorenzo, C.D.; García-Gagliardi, P.; Antonietti, M.S.; Sánchez-Lamas, M.; Mancini, E.; Dezar, C.A.; Vazquez, M.; Watson, G.; Yanovsky, M.J.; Cerdán, P.D. Improvement of alfalfa forage quality and management through the down-regulation of MsFTa1. Plant Biotechnol. J. 2020, 18, 944–954. [Google Scholar] [CrossRef]
- Kulkarni, K.P.; Tayade, R.; Asekova, S.; Song, J.T.; Shannon, J.G.; Lee, J.D. Harnessing the Potential of Forage Legumes, Alfalfa, Soybean, and Cowpea for Sustainable Agriculture and Global Food Security. Front. Plant Sci. 2018, 9, 1314. [Google Scholar] [CrossRef]
- Bouton, J. The economic benefits of forage improvement in the United States. Euphytica 2007, 154, 263–270. [Google Scholar] [CrossRef]
- Zhou, Q.; Mao, P.; Luo, D.; Chai, X.; Deng, H.; Fang, Q.; Fang, L.; Nan, Z.; Wen, J.; Liu, Z. Comparative transcriptome analyses reveal that the MsNST1 gene affects lignin synthesis in alfalfa (Medicago sativa L.). Crop J. 2022, 10, 1059–1072. [Google Scholar] [CrossRef]
- Diamond, J. Evolution, consequences and future of plant and animal domestication. Nature 2002, 418, 700–707. [Google Scholar] [CrossRef] [PubMed]
- Larson, G.; Piperno, D.R.; Allaby, R.G.; Purugganan, M.D.; Andersson, L.; Arroyo-Kalin, M.; Barton, L.; Climer Vigueira, C.; Denham, T.; Dobney, K.; et al. Current perspectives and the future of domestication studies. Proc. Natl. Acad. Sci. USA 2014, 111, 6139–6146. [Google Scholar] [CrossRef] [PubMed]
- Nonogaki, H. Seed dormancy and germination-emerging mechanisms and new hypotheses. Front. Plant Sci. 2014, 5, 233. [Google Scholar] [CrossRef]
- Smýkal, P.; Jovanović, Ž.; Stanisavljević, N.; Zlatković, B.; Ćupina, B.; Đorđević, V.; Mikić, A.; Medović, A. A comparative study of ancient DNA isolated from charred pea (Pisum sativum L.) seeds from an Early Iron Age settlement in southeast Serbia: Inference for pea domestication. Genet. Resour. Crop Evol. 2014, 61, 1533–1544. [Google Scholar] [CrossRef]
- Chen, K.; Li, G.J.; Bressan, R.A.; Song, C.P.; Zhu, J.K.; Zhao, Y. Abscisic acid dynamics, signaling, and functions in plants. J. Integr. Plant Biol. 2020, 62, 25–54. [Google Scholar] [CrossRef]
- Hradilová, I.; Duchoslav, M.; Brus, J.; Pechanec, V.; Hýbl, M.; Kopecký, P.; Smržová, L.; Štefelová, N.; Vaclávek, T.; Bariotakis, M.; et al. Variation in wild pea (Pisum sativum subsp. elatius) seed dormancy and its relationship to the environment and seed coat traits. PeerJ 2019, 7, e6263. [Google Scholar] [CrossRef]
- Rubio de Casas, R.; Willis, C.G.; Pearse, W.D.; Baskin, C.C.; Baskin, J.M.; Cavender-Bares, J. Global biogeography of seed dormancy is determined by seasonality and seed size: A case study in the legumes. New Phytol. 2017, 214, 1527–1536. [Google Scholar] [CrossRef]
- Smýkal, P.; Vernoud, V.; Blair, M.W.; Soukup, A.; Thompson, R.D. The role of the testa during development and in establishment of dormancy of the legume seed. Front. Plant Sci. 2014, 5, 351. [Google Scholar] [CrossRef]
- Stone, E.C.; Juhren, G. The effect of fire on the germination of the seed of Rhus ovata Wats. Am. J. Bot. 1951, 38, 368–372. [Google Scholar] [CrossRef]
- Graeber, K.; Nakabayashi, K.; Miatton, E.; Leubner-Metzger, G.; Soppe, W.J. Molecular mechanisms of seed dormancy. Plant Cell Environ. 2012, 35, 1769–1786. [Google Scholar] [CrossRef] [PubMed]
- Baskin, J.M.; Baskin, C.C. A classification system for seed dormancy. Seed Sci. Res. 2004, 14, 1–16. [Google Scholar] [CrossRef]
- Meyer, C.J.; Steudle, E.; Peterson, C.A. Patterns and kinetics of water uptake by soybean seeds. J. Exp. Bot. 2007, 58, 717–732. [Google Scholar] [CrossRef]
- Penfield, S. Seed dormancy and germination. Curr. Biol. 2017, 27, R874–R878. [Google Scholar] [CrossRef]
- Hradilová, I.; Trněný, O.; Válková, M.; Cechová, M.; Janská, A.; Prokešová, L.; Aamir, K.; Krezdorn, N.; Rotter, B.; Winter, P.; et al. A Combined Comparative Transcriptomic, Metabolomic, and Anatomical Analyses of Two Key Domestication Traits: Pod Dehiscence and Seed Dormancy in Pea (Pisum sp.). Front. Plant Sci. 2017, 8, 542. [Google Scholar] [CrossRef]
- Mullin, W.J.; Xu, W. Study of soybean seed coat components and their relationship to water absorption. J. Agric. Food Chem. 2001, 49, 5331–5335. [Google Scholar] [CrossRef] [PubMed]
- Sun, L.; Miao, Z.; Cai, C.; Zhang, D.; Zhao, M.; Wu, Y.; Zhang, X.; Swarm, S.A.; Zhou, L.; Zhang, Z.J.; et al. GmHs1-1, encoding a calcineurin-like protein, controls hard-seededness in soybean. Nat. Genet. 2015, 47, 939–943. [Google Scholar] [CrossRef] [PubMed]
- Bakker, J. Seeds, Ecology, Biogeography and Evolution of Dormancy, and Germination. C.C. Baskin & J.M. Baskin. Plant Ecol. 2001, 152, 204–205. [Google Scholar] [CrossRef]
- Ma, F.; Cholewa, E.; Mohamed, T.; Peterson, C.A.; Gijzen, M. Cracks in the palisade cuticle of soybean seed coats correlate with their permeability to water. Ann. Bot. 2004, 94, 213–228. [Google Scholar] [CrossRef]
- Chai, M.; Zhou, C.; Molina, I.; Fu, C.; Nakashima, J.; Li, G.; Zhang, W.; Park, J.; Tang, Y.; Jiang, Q.; et al. A class II KNOX gene, KNOX4, controls seed physical dormancy. Proc. Natl. Acad. Sci. USA 2016, 113, 6997–7002. [Google Scholar] [CrossRef]
- Wen, Z.; Du, J.; Chai, M.; Lu, X.; Liu, H.; Bao, Q.; Zhang, Y.; Huo, H.; Wang, Z.; Wen, J.; et al. Anthocyanidin Reductase promotes physical dormancy in Medicago truncatula seeds. Plant Physiol. 2025, 199, kiaf525. [Google Scholar] [CrossRef]
- Chai, M.; Queralta Castillo, I.; Sonntag, A.; Wang, S.; Zhao, Z.; Liu, W.; Du, J.; Xie, H.; Liao, F.; Yun, J.; et al. A seed coat-specific β-ketoacyl-CoA synthase, KCS12, is critical for preserving seed physical dormancy. Plant Physiol. 2021, 186, 1606–1615. [Google Scholar] [CrossRef]
- Ali, S.; Kucek, L.K.; Riday, H.; Krom, N.; Krogman, S.; Cooper, K.; Jacobs, L.; Mehta, P.; Trammell, M.; Bhamidimarri, S.; et al. Transcript profiling of hairy vetch (Vicia villosa Roth) identified interesting genes for seed dormancy. Plant Genome 2023, 16, e20330. [Google Scholar] [CrossRef]
- Kucek, L.K.; Azevedo, M.D.; Eagen, S.S.; Ehlke, N.J.; Hayes, R.J.; Mirsky, S.; Reberg-Horton, C.; Ryan, M.; Wayman, S.; Wiering, N.P.; et al. Seed Dormancy in Hairy Vetch (Vicia villosa Roth) Is Influenced by Genotype and Environment. Agronomy 2020, 10, 1804. [Google Scholar] [CrossRef]
- Lei, Y.; Hannoufa, A.; Yu, P. The Use of Gene Modification and Advanced Molecular Structure Analyses towards Improving Alfalfa Forage. Int. J. Mol. Sci. 2017, 18, 298. [Google Scholar] [CrossRef]
- Jiresova, J.; Sera, B.; Scholtz, V.; Khun, J.; Sery, M. The Dormancy Overcoming and Affection of Early Growth of Alfalfa (Medicago sativa L.) Seeds by Non-Thermal Plasma and Plasma Activated Water. Rom. Rep. Phys. 2021, 73, 711. [Google Scholar]
- Wang, C.; Lin, J.; Bu, Y.; Sun, R.; Lu, Y.; Gai, J.; Xing, H.; Guo, N.; Zhao, J. Genome-wide transcriptome analysis reveals key regulatory networks and genes involved in the determination of seed hardness in vegetable soybean. Hortic. Res. 2024, 11, uhae084. [Google Scholar] [CrossRef]
- Wang, X.; Zhang, H.; Song, R.; Sun, M.; Liu, P.; Tian, P.; Mao, P.; Jia, S. Multiple omics datasets reveal significant physical and physiological dormancy in alfalfa hard seeds identified by multispectral imaging analysis. Crop J. 2023, 11, 1458–1468. [Google Scholar] [CrossRef]
- Laosatit, K.; Amkul, K.; Yimram, T.; Chen, J.; Lin, Y.; Yuan, X.; Wang, L.; Chen, X.; Somta, P. A Class II KNOX Gene, KNAT7-1, Regulates Physical Seed Dormancy in Mungbean [Vigna radiata (L.) Wilczek]. Front. Plant Sci. 2022, 13, 852373. [Google Scholar] [CrossRef] [PubMed]
- Chen, S. fastp 1.0: An ultra-fast all-round tool for FASTQ data quality control and preprocessing. iMeta 2025, 4, e70078. [Google Scholar] [CrossRef] [PubMed]
- Kim, D.; Langmead, B.; Salzberg, S.L. HISAT: A fast spliced aligner with low memory requirements. Nat. Methods 2015, 12, 357–360. [Google Scholar] [CrossRef]
- Li, B.; Dewey, C.N. RSEM: Accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinform. 2011, 12, 323. [Google Scholar] [CrossRef]
- Wagner, G.P.; Kin, K.; Lynch, V.J. Measurement of mRNA abundance using RNA-seq data: RPKM measure is inconsistent among samples. Theory Biosci. 2012, 131, 281–285. [Google Scholar] [CrossRef]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef]
- Moïse, J.A.; Han, S.; Gudynaitę-Savitch, L.; Johnson, D.A.; Miki, B.L.A. Seed coats: Structure, development, composition, and biotechnology. Vitr. Cell. Dev. Biol.-Plant 2005, 41, 620–644. [Google Scholar] [CrossRef]
- Koizumi, M.; Kikuchi, K.; Isobe, S.; Ishida, N.; Naito, S.; Kano, H. Role of seed coat in imbibing soybean seeds observed by micro-magnetic resonance imaging. Ann. Bot. 2008, 102, 343–352. [Google Scholar] [CrossRef] [PubMed]
- Chen, H.; Zeng, Y.; Yang, Y.; Huang, L.; Tang, B.; Zhang, H.; Hao, F.; Liu, W.; Li, Y.; Liu, Y.; et al. Allele-aware chromosome-level genome assembly and efficient transgene-free genome editing for the autotetraploid cultivated alfalfa. Nat. Commun. 2020, 11, 2494. [Google Scholar] [CrossRef] [PubMed]
- Dong, S.; Sang, L.; Xie, H.; Chai, M.; Wang, Z.Y. Comparative Transcriptome Analysis of Salt Stress-Induced Leaf Senescence in Medicago truncatula. Front. Plant Sci. 2021, 12, 666660. [Google Scholar] [CrossRef] [PubMed]
- Jiang, X.; Yang, X.; Zhang, F.; Yang, T.; Yang, C.; He, F.; Gao, T.; Wang, C.; Yang, Q.; Wang, Z.; et al. Combining QTL mapping and RNA-Seq Unravels candidate genes for Alfalfa (Medicago sativa L.) leaf development. BMC Plant Biol. 2022, 22, 485. [Google Scholar] [CrossRef]
- Decsi, K.; Kutasy, B.; Kiniczky, M.; Hegedűs, G.; Virág, E. RNA-seq datasets of field soybean cultures conditioned by Elice16Indures® biostimulator. Data Brief 2022, 42, 108182. [Google Scholar] [CrossRef]
- Hou, W.; Zhang, X.; Liu, Y.; Liu, Y.; Feng, B.L. RNA-Seq and genetic diversity analysis of faba bean (Vicia faba L.) varieties in China. PeerJ 2023, 11, e14259. [Google Scholar] [CrossRef]
- Mahdavi Mashaki, K.; Garg, V.; Nasrollahnezhad Ghomi, A.A.; Kudapa, H.; Chitikineni, A.; Zaynali Nezhad, K.; Yamchi, A.; Soltanloo, H.; Varshney, R.K.; Thudi, M. RNA-Seq analysis revealed genes associated with drought stress response in kabuli chickpea (Cicer arietinum L.). PLoS ONE 2018, 13, e0199774. [Google Scholar] [CrossRef]
- Finch-Savage, W.E.; Leubner-Metzger, G. Seed dormancy and the control of germination. New Phytol. 2006, 171, 501–523. [Google Scholar] [CrossRef]
- Schnurr, J.; Shockey, J.; Browse, J. The acyl-CoA synthetase encoded by LACS2 is essential for normal cuticle development in Arabidopsis. Plant Cell 2004, 16, 629–642. [Google Scholar] [CrossRef]
- Xie, L.J.; Tan, W.J.; Yang, Y.C.; Tan, Y.F.; Zhou, Y.; Zhou, D.M.; Xiao, S.; Chen, Q.F. Long-Chain acyl-CoA Synthetase LACS2 Contributes to Submergence Tolerance by Modulating Cuticle Permeability in Arabidopsis. Plants 2020, 9, 262. [Google Scholar] [CrossRef]

| Gene ID | Pathway | Non-Redundant Protein Sequence Database |
|---|---|---|
| MS.gene071857 | lipid binding | major allergen Pru ar 1-like protein [Trifolium pratense] |
| MS.gene04019 | lipid binding | pleckstrin homology domain-containing protein 1 [Medicago truncatula] |
| MS.gene72825 | lipid binding | acyl-CoA-binding protein [Medicago truncatula] |
| MS.gene87096 | lipid binding | non-specific lipid-transfer protein 1 [Medicago truncatula] |
| MS.gene27472 | lipid binding | lipid transfer protein [Medicago truncatula] |
| MS.gene27474 | lipid binding | lipid transfer protein [Medicago truncatula] |
| MS.gene00236 | lipid binding | major allergen Pru ar 1-like protein [Trifolium pratense] |
| MS.gene89266 | lipid binding | putative lipid-transfer protein DIR1 isoform X1 [Medicago truncatula] |
| MS.gene27869 | lipase activity | GDSL esterase/lipase At5g45670 [Medicago truncatula] |
| MS.gene019909 | lipase activity | triacylglycerol lipase 2 [Medicago truncatula] |
| MS.gene36001 | lipase activity | triacylglycerol lipase 2 [Medicago truncatula] |
| MS.gene74700 | lipase activity | triacylglycerol lipase 2 [Medicago truncatula] |
| MS.gene72397 | lipase activity | triacylglycerol lipase-like protein [Medicago truncatula] |
| MS.gene003833 | lipid metabolism | long chain acyl-CoA synthetase 2 [Medicago truncatula] |
| MS.gene003796 | lipid metabolism | probable 1-acyl-sn-glycerol-3-phosphate acyltransferase 5 [Medicago truncatula] |
| MS.gene87717 | lipid transport | non-specific lipid-transfer protein 2 [Medicago truncatula] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, H.; Kang, X.; Li, X.; Yuan, F.; Wang, Z.-Y.; Chai, M. Global Transcriptome Analysis Reveals the Molecular Mechanism Underlying Seed Physical Dormancy Formation in Medicago sativa. Genes 2025, 16, 1438. https://doi.org/10.3390/genes16121438
Li H, Kang X, Li X, Yuan F, Wang Z-Y, Chai M. Global Transcriptome Analysis Reveals the Molecular Mechanism Underlying Seed Physical Dormancy Formation in Medicago sativa. Genes. 2025; 16(12):1438. https://doi.org/10.3390/genes16121438
Chicago/Turabian StyleLi, He, Xiaoying Kang, Xu Li, Feng Yuan, Zeng-Yu Wang, and Maofeng Chai. 2025. "Global Transcriptome Analysis Reveals the Molecular Mechanism Underlying Seed Physical Dormancy Formation in Medicago sativa" Genes 16, no. 12: 1438. https://doi.org/10.3390/genes16121438
APA StyleLi, H., Kang, X., Li, X., Yuan, F., Wang, Z.-Y., & Chai, M. (2025). Global Transcriptome Analysis Reveals the Molecular Mechanism Underlying Seed Physical Dormancy Formation in Medicago sativa. Genes, 16(12), 1438. https://doi.org/10.3390/genes16121438






