Genetic Parameters, Prediction of Genotypic Values, and Forage Stability in Paspalum nicorae Parodi Ecotypes via REML/BLUP
Abstract
1. Introduction
2. Materials and Methods
2.1. Location and Research Period
2.2. Plant Material
2.3. Site Preparation and Soil Fertility
2.4. Seed Germination, Plant Establishment, and Experimental Design
2.5. Evaluated Traits
2.6. Statistical Analysis
2.6.1. REML/BLUP
2.6.2. Selection Index
2.6.3. Multi-Trait Genotype–Ideotype Distance Index
3. Results
3.1. Estimates of Variance Components via REML
3.2. Genetic Parameters
3.3. Genotypic Values and Genetic Gains
3.4. Adaptability, Stability, and Performance via BLUP-Based Indices
3.5. Multi-Trait Genotype–Ideotype Distance Index
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| UFRGS | Universidade Federal do Rio Grande do Sul |
| DPFA | Department of Forage Plants and Agrometeorology |
| Cfa | Humid subtropical climate, no dry season, hot summer |
| LD | Linear dichroism |
| RCBD | Randomized complete block design |
| OM | Organic matter |
| P | Phosphorus |
| K | Potassium |
| Al | Aluminum |
| Ca | Calcium |
| Mg | magnesium |
| H + Al | Potential acidity |
| CECeffective | Cation exchange capacity determined by soil pH |
| CTCpH7 | Estimated cation exchange capacity at pH 7 |
| V | Base saturation |
| m | Aluminum saturation |
| S | Sulfur |
| Cu | Copper |
| Zn | Zinc |
| Mn | Manganese |
| B | Boron |
| NT | Number of tillers |
| FM | Fresh mass |
| LDM | Leaf dry mass |
| SDM | Stem dry mass |
| IDM | Inflorescence dry mass |
| TDM | Total dry mass |
| LSR | Leaf-to-stem ratio |
| HI | Harvest index |
| CT | Cold tolerance |
| FP | Forage persistence |
| GH | Growth habit |
| PH | Plant height |
| ANADEV | Analysis of Deviance |
| BLUP | Best linear unbiased prediction |
| CVe (%) | Experimental coefficient of variation |
| CVg (%) | Genetic coefficient of variation |
| CVr | Relative coefficient of variation (CVe/CVg) |
| HMRPGV | Harmonic mean of relative performance of genotypic values |
| Environmental variance | |
| Genotype-by-measurement interaction variance | |
| Permanent environment variance | |
| Genotypic variance | |
| h2 | Broad-sense heritability of individual plots, considering total genotypic effects; ρ: repeatability at the plot level |
| Coefficient of determination of plot effects | |
| Coefficient for determining the effects of the genotypes x measurements interaction | |
| Genotypic correlation through measurements | |
| Heritability of the genotype mean | |
| u + g + gem | Average genotypic values |
| SG% | Selection gains |
| IVe (%) | Experimental variation index |
| IVg (%) | Genetic variation index |
| µ | Grand mean |
Appendix A
| NT (Tillers Plant−1) | FM (g Plant−1) | |||||||||||||
| Order | Ecotype | g | u + g | Gain | µn | u + g + gem | SG | Ecotypes | g | u + g | Gain | µn | u + g + gem | SG |
| 1 | N3.10 | 121.55 | 329.51 | 121.55 | 329.51 | 375.99 | 58.45 | N3.10 | 57.97 | 213.14 | 57.97 | 213.14 | 244.15 | 37.36 |
| 2 | N3.17 | 89.28 | 297.24 | 105.42 | 313.38 | 331.38 | 42.93 | N2.06 | 35.85 | 191.02 | 46.91 | 202.08 | 210.20 | 23.10 |
| 3 | N3.12 | 52.98 | 260.94 | 87.94 | 295.90 | 281.20 | 25.48 | N3.17 | 32.32 | 187.50 | 42.04 | 197.22 | 204.78 | 20.83 |
| 4 | N5.09 | 48.87 | 256.83 | 78.17 | 286.13 | 275.51 | 23.50 | N4.09 | 30.78 | 185.96 | 39.23 | 194.41 | 202.43 | 19.84 |
| 5 | N2.11 | 42.08 | 250.04 | 70.95 | 278.91 | 266.13 | 20.24 | N5.01 | 29.35 | 184.53 | 37.25 | 192.43 | 200.23 | 18.92 |
| 6 | N2.07 | 36.90 | 244.86 | 65.28 | 273.24 | 258.97 | 17.74 | N1.21 | 28.48 | 183.65 | 35.79 | 190.97 | 198.89 | 18.35 |
| 7 | N8.04 | 36.60 | 244.56 | 61.18 | 269.14 | 258.55 | 17.60 | N2.07 | 22.87 | 178.04 | 33.94 | 189.12 | 190.27 | 14.74 |
| 8 | N4.09 | 36.52 | 244.48 | 58.10 | 266.06 | 258.44 | 17.56 | N2.11 | 21.87 | 177.05 | 32.44 | 187.61 | 188.74 | 14.09 |
| 9 | N5.01 | 35.02 | 242.98 | 55.53 | 263.49 | 256.38 | 16.84 | N1.05 | 21.37 | 176.55 | 31.21 | 186.38 | 187.98 | 13.77 |
| 10 | N3.18 | 33.27 | 241.23 | 53.31 | 261.27 | 253.95 | 16.00 | N3.09 | 21.37 | 176.55 | 30.22 | 185.40 | 187.98 | 13.77 |
| 11 | N3.09 | 33.10 | 241.06 | 51.47 | 259.43 | 253.72 | 15.92 | N4.08 | 21.25 | 176.43 | 29.41 | 184.58 | 187.80 | 13.70 |
| 12 | N3.03 | 30.65 | 238.61 | 49.74 | 257.70 | 250.33 | 14.74 | N1.06 | 18.46 | 173.64 | 28.50 | 183.67 | 183.51 | 11.90 |
| 13 | N5.03 | 30.37 | 238.33 | 48.25 | 256.21 | 249.95 | 14.61 | N5.12 | 17.43 | 172.61 | 27.64 | 182.82 | 181.93 | 11.23 |
| 14 | N3.16 | 29.64 | 237.60 | 46.92 | 254.88 | 248.94 | 14.25 | N3.12 | 16.33 | 171.51 | 26.84 | 182.01 | 180.24 | 10.52 |
| 15 | N2.06 | 28.65 | 236.61 | 45.70 | 253.66 | 247.57 | 13.78 | N3.18 | 16.25 | 171.42 | 26.13 | 181.31 | 180.11 | 10.47 |
| 16 | N1.21 | 28.17 | 236.13 | 44.60 | 252.56 | 246.90 | 13.55 | N2.16 | 15.40 | 170.58 | 25.46 | 180.64 | 178.82 | 9.93 |
| 17 | N2.16 | 25.01 | 232.97 | 43.45 | 251.41 | 242.53 | 12.02 | N3.16 | 15.24 | 170.42 | 24.86 | 180.04 | 178.57 | 9.82 |
| 18 | N5.20 | 22.07 | 230.03 | 42.26 | 250.22 | 238.47 | 10.61 | N1.19 | 15.24 | 170.42 | 24.32 | 179.50 | 178.57 | 9.82 |
| 19 | N3.05 | 21.56 | 229.52 | 41.17 | 249.13 | 237.76 | 10.37 | N3.05 | 11.58 | 166.76 | 23.65 | 178.83 | 172.95 | 7.46 |
| 20 | N4.15 | 20.29 | 228.25 | 40.13 | 248.09 | 236.01 | 9.76 | N3.04 | 11.09 | 166.26 | 23.03 | 178.20 | 172.19 | 7.15 |
| 21 | N4.08 | 19.59 | 227.55 | 39.15 | 247.11 | 235.04 | 9.42 | N2.02 | 10.12 | 165.29 | 22.41 | 177.59 | 170.70 | 6.52 |
| 22 | N4.07 | 19.48 | 227.44 | 38.26 | 246.22 | 234.88 | 9.37 | N5.09 | 9.14 | 164.32 | 21.81 | 176.98 | 169.21 | 5.89 |
| 23 | N5.18 | 16.73 | 224.69 | 37.32 | 245.28 | 231.09 | 8.05 | N8.04 | 9.13 | 164.30 | 21.26 | 176.43 | 169.19 | 5.88 |
| 24 | N3.04 | 16.64 | 224.60 | 36.46 | 244.42 | 230.96 | 8.00 | N4.07 | 8.91 | 164.08 | 20.74 | 175.92 | 168.85 | 5.74 |
| 25 | N6.12 | 15.65 | 223.61 | 35.63 | 243.59 | 229.59 | 7.52 | N1.22 | 8.37 | 163.54 | 20.25 | 175.42 | 168.02 | 5.39 |
| 26 | N2.22 | 14.35 | 222.31 | 34.81 | 242.77 | 227.79 | 6.90 | N1.18 | 7.94 | 163.11 | 19.77 | 174.95 | 167.36 | 5.11 |
| 27 | N1.10 | 12.84 | 220.80 | 34.00 | 241.95 | 225.72 | 6.18 | N5.18 | 7.80 | 162.98 | 19.33 | 174.51 | 167.16 | 5.03 |
| 28 | N1.05 | 12.58 | 220.54 | 33.23 | 241.19 | 225.34 | 6.05 | N5.03 | 7.75 | 162.92 | 18.92 | 174.09 | 167.07 | 4.99 |
| 29 | N4.13 | 11.53 | 219.49 | 32.48 | 240.44 | 223.89 | 5.54 | N6.22 | 6.75 | 161.93 | 18.50 | 173.67 | 165.54 | 4.35 |
| 30 | N4.20 | 11.18 | 219.14 | 31.77 | 239.73 | 223.42 | 5.38 | N4.06 | 6.72 | 161.89 | 18.10 | 173.28 | 165.49 | 4.33 |
| 31 | N5.12 | 7.95 | 215.91 | 31.00 | 238.96 | 218.94 | 3.82 | N5.20 | 6.36 | 161.54 | 17.73 | 172.90 | 164.94 | 4.10 |
| 32 | N1.18 | 7.18 | 215.14 | 30.26 | 238.22 | 217.89 | 3.45 | N5.19 | 6.26 | 161.43 | 17.37 | 172.54 | 164.78 | 4.03 |
| 33 | N2.10 | 6.99 | 214.95 | 29.55 | 237.51 | 217.62 | 3.36 | N3.15 | 6.01 | 161.18 | 17.02 | 172.20 | 164.40 | 3.87 |
| 34 | N5.22 | 6.88 | 214.84 | 28.89 | 236.85 | 217.47 | 3.31 | N3.19 | 5.38 | 160.56 | 16.68 | 171.86 | 163.44 | 3.47 |
| 35 | N5.19 | 6.59 | 214.55 | 28.25 | 236.21 | 217.07 | 3.17 | N6.08 | 4.22 | 159.40 | 16.32 | 171.50 | 161.66 | 2.72 |
| 36 | N2.02 | 4.91 | 212.87 | 27.60 | 235.56 | 214.75 | 2.36 | N6.12 | 3.89 | 159.07 | 15.98 | 171.16 | 161.15 | 2.51 |
| 37 | N4.05 | 3.52 | 211.48 | 26.95 | 234.91 | 212.82 | 1.69 | N3.03 | 3.70 | 158.88 | 15.65 | 170.82 | 160.86 | 2.38 |
| 38 | N4.06 | 1.32 | 209.28 | 26.28 | 234.24 | 209.78 | 0.63 | N2.10 | 2.59 | 157.77 | 15.30 | 170.48 | 159.15 | 1.67 |
| 39 | N1.17 | −0.36 | 207.60 | 25.59 | 233.55 | 207.46 | −0.17 | N3.02 | 2.11 | 157.29 | 14.97 | 170.14 | 158.42 | 1.36 |
| 40 | N2.13 | −1.30 | 206.66 | 24.92 | 232.88 | 206.17 | −0.62 | N2.13 | 1.29 | 156.46 | 14.62 | 169.80 | 157.15 | 0.83 |
| 41 | N3.02 | −3.90 | 204.06 | 24.22 | 232.18 | 202.57 | −1.87 | N6.19 | −0.25 | 154.93 | 14.26 | 169.44 | 154.79 | −0.16 |
| 42 | N7.20 | −4.10 | 203.86 | 23.54 | 231.50 | 202.29 | −1.97 | N1.17 | −0.57 | 154.61 | 13.91 | 169.08 | 154.31 | −0.37 |
| 43 | N1.06 | −4.16 | 203.80 | 22.90 | 230.86 | 202.21 | −2.00 | N5.15 | −1.03 | 154.14 | 13.56 | 168.74 | 153.59 | −0.67 |
| 44 | N5.05 | −4.23 | 203.73 | 22.28 | 230.24 | 202.11 | −2.04 | N6.05 | −1.67 | 153.51 | 13.21 | 168.39 | 152.61 | −1.08 |
| 45 | N6.01 | −5.11 | 202.85 | 21.67 | 229.63 | 200.89 | −2.46 | N4.21 | −1.70 | 153.47 | 12.88 | 168.06 | 152.56 | −1.10 |
| 46 | N3.15 | −5.66 | 202.30 | 21.08 | 229.04 | 200.14 | −2.72 | N4.13 | −1.90 | 153.28 | 12.56 | 167.74 | 152.26 | −1.22 |
| 47 | N2.17 | −5.80 | 202.16 | 20.51 | 228.47 | 199.94 | −2.79 | N5.21 | −3.60 | 151.57 | 12.22 | 167.39 | 149.65 | −2.32 |
| 48 | N5.15 | −7.02 | 200.94 | 19.93 | 227.89 | 198.26 | −3.37 | N5.22 | −3.94 | 151.24 | 11.88 | 167.06 | 149.13 | −2.54 |
| 49 | N3.22 | −7.10 | 200.86 | 19.38 | 227.34 | 198.15 | −3.41 | N4.05 | −4.42 | 150.76 | 11.55 | 166.72 | 148.39 | −2.85 |
| 50 | N5.16 | −9.04 | 198.92 | 18.81 | 226.77 | 195.47 | −4.35 | N4.15 | −4.53 | 150.65 | 11.23 | 166.40 | 148.22 | −2.92 |
| 51 | N5.21 | −9.28 | 198.68 | 18.26 | 226.22 | 195.13 | −4.46 | N2.01 | −5.88 | 149.30 | 10.89 | 166.07 | 146.16 | −3.79 |
| 52 | N6.08 | −10.00 | 197.96 | 17.72 | 225.68 | 194.14 | −4.81 | N2.22 | −6.90 | 148.28 | 10.55 | 165.73 | 144.59 | −4.44 |
| 53 | N3.07 | −10.75 | 197.21 | 17.18 | 225.14 | 193.09 | −5.17 | N1.10 | −7.58 | 147.60 | 10.21 | 165.38 | 143.55 | −4.88 |
| 54 | N2.01 | −14.48 | 193.48 | 16.60 | 224.56 | 187.94 | −6.96 | N3.08 | −7.89 | 147.28 | 9.87 | 165.05 | 143.06 | −5.09 |
| 55 | N3.06 | −14.92 | 193.04 | 16.02 | 223.98 | 187.33 | −7.18 | N5.05 | −8.20 | 146.98 | 9.54 | 164.72 | 142.59 | −5.28 |
| 56 | N6.22 | −15.32 | 192.64 | 15.46 | 223.42 | 186.78 | −7.37 | N5.16 | −8.25 | 146.93 | 9.23 | 164.40 | 142.52 | −5.32 |
| 57 | N1.22 | −16.60 | 191.36 | 14.90 | 222.86 | 185.02 | −7.98 | N6.18 | −8.47 | 146.71 | 8.92 | 164.09 | 142.18 | −5.46 |
| 58 | N6.19 | −17.94 | 190.02 | 14.33 | 222.29 | 183.16 | −8.63 | N3.07 | −9.22 | 145.96 | 8.60 | 163.78 | 141.03 | −5.94 |
| 59 | N3.19 | −19.29 | 188.67 | 13.76 | 221.72 | 181.29 | −9.28 | N4.20 | −9.25 | 145.93 | 8.30 | 163.48 | 140.98 | −5.96 |
| 60 | N4.10 | −19.96 | 188.00 | 13.20 | 221.16 | 180.36 | −9.60 | N6.01 | −9.84 | 145.34 | 8.00 | 163.17 | 140.07 | −6.34 |
| 61 | N4.21 | −20.80 | 187.16 | 12.65 | 220.60 | 179.21 | −10.00 | N5.10 | −9.95 | 145.23 | 7.70 | 162.88 | 139.91 | −6.41 |
| 62 | N1.09 | −21.64 | 186.32 | 12.09 | 220.05 | 178.05 | −10.41 | N3.22 | −11.29 | 143.89 | 7.40 | 162.57 | 137.85 | −7.28 |
| 63 | N1.07 | −22.79 | 185.17 | 11.54 | 219.50 | 176.45 | −10.96 | N1.09 | −11.39 | 143.78 | 7.10 | 162.28 | 137.69 | −7.34 |
| 64 | N5.02 | −22.97 | 184.99 | 11.00 | 218.96 | 176.21 | −11.05 | N5.02 | −12.61 | 142.57 | 6.79 | 161.97 | 135.82 | −8.13 |
| 65 | N4.14 | −23.51 | 184.45 | 10.47 | 218.43 | 175.46 | −11.31 | N5.14 | −12.79 | 142.39 | 6.49 | 161.67 | 135.55 | −8.24 |
| 66 | N5.10 | −24.13 | 183.83 | 9.94 | 217.90 | 174.61 | −11.60 | N2.17 | −14.33 | 140.84 | 6.17 | 161.35 | 133.18 | −9.24 |
| 67 | N1.19 | −24.65 | 183.31 | 9.43 | 217.39 | 173.88 | −11.85 | N4.14 | −14.67 | 140.51 | 5.86 | 161.04 | 132.66 | −9.45 |
| 68 | N5.11 | −25.43 | 182.53 | 8.92 | 216.87 | 172.81 | −12.23 | N4.10 | −15.06 | 140.11 | 5.56 | 160.73 | 132.06 | −9.71 |
| 69 | N2.18 | −25.69 | 182.27 | 8.41 | 216.37 | 172.45 | −12.35 | N7.20 | −15.18 | 139.99 | 5.25 | 160.43 | 131.87 | −9.78 |
| 70 | N1.13 | −26.63 | 181.33 | 7.91 | 215.87 | 171.14 | −12.81 | N8.03 | −15.23 | 139.94 | 4.96 | 160.14 | 131.79 | −9.82 |
| 71 | N5.06 | −29.14 | 178.82 | 7.39 | 215.35 | 167.67 | −14.01 | N5.06 | −15.26 | 139.92 | 4.68 | 159.85 | 131.76 | −9.83 |
| 72 | N4.19 | −29.17 | 178.79 | 6.88 | 214.84 | 167.63 | −14.03 | N2.18 | −17.83 | 137.34 | 4.36 | 159.54 | 127.80 | −11.49 |
| 73 | N8.03 | −30.05 | 177.91 | 6.38 | 214.34 | 166.42 | −14.45 | N1.16 | −18.10 | 137.08 | 4.06 | 159.23 | 127.40 | −11.66 |
| 74 | N4.12 | −30.42 | 177.54 | 5.88 | 213.84 | 165.91 | −14.63 | N4.19 | −19.80 | 135.37 | 3.73 | 158.91 | 124.78 | −12.76 |
| 75 | N3.08 | −30.48 | 177.48 | 5.40 | 213.35 | 165.83 | −14.66 | N1.12 | −19.85 | 135.32 | 3.42 | 158.60 | 124.70 | −12.79 |
| 76 | N1.16 | −31.75 | 176.21 | 4.91 | 212.87 | 164.07 | −15.27 | N3.06 | −20.02 | 135.16 | 3.11 | 158.29 | 124.45 | −12.90 |
| 77 | N1.15 | −32.22 | 175.74 | 4.42 | 212.38 | 163.42 | −15.49 | N1.14 | −22.86 | 132.32 | 2.77 | 157.95 | 120.09 | −14.73 |
| 78 | N5.14 | −32.27 | 175.69 | 3.95 | 211.91 | 163.36 | −15.52 | N1.07 | −25.00 | 130.18 | 2.42 | 157.60 | 116.81 | −16.11 |
| 79 | N6.05 | −32.54 | 175.42 | 3.49 | 211.45 | 162.97 | −15.65 | N5.11 | −25.01 | 130.17 | 2.07 | 157.25 | 116.80 | −16.11 |
| 80 | N1.12 | −46.03 | 161.93 | 2.87 | 210.83 | 144.33 | −22.13 | N1.13 | −25.72 | 129.46 | 1.72 | 156.90 | 115.70 | −16.57 |
| 81 | N1.14 | −47.95 | 160.01 | 2.25 | 210.21 | 141.68 | −23.06 | N7.15 | −28.29 | 126.89 | 1.35 | 156.53 | 111.76 | −18.23 |
| 82 | N6.18 | −48.90 | 159.06 | 1.62 | 209.58 | 140.36 | −23.51 | N4.12 | −31.70 | 123.47 | 0.95 | 156.13 | 106.51 | −20.43 |
| 83 | N4.17 | −61.89 | 146.07 | 0.86 | 208.82 | 122.41 | −29.76 | N1.15 | −32.52 | 122.65 | 0.55 | 155.72 | 105.26 | −20.96 |
| 84 | N7.15 | −71.11 | 136.85 | 0.00 | 207.96 | 109.66 | −34.19 | N4.17 | −45.40 | 109.78 | 0.00 | 155.18 | 85.49 | −29.26 |
| LDM (g plant−1) | SDM (g plant−1) | |||||||||||||
| Order | Ecotypes | g | u + g | Gain | µn | u + g + gem | SG | Ecotypes | g | u + g | Gain | µn | u + g + gem | SG |
| 1 | N3.10 | 18.67 | 80.98 | 18.67 | 80.98 | 96.00 | 29.96 | N3.10 | 1.94 | 8.49 | 1.94 | 8.49 | 10.69 | 29.65 |
| 2 | N2.06 | 10.46 | 72.77 | 14.56 | 76.88 | 81.19 | 16.78 | N3.17 | 1.40 | 7.94 | 1.67 | 8.21 | 9.53 | 21.31 |
| 3 | N1.05 | 9.67 | 71.99 | 12.93 | 75.25 | 79.77 | 15.52 | N2.06 | 1.23 | 7.77 | 1.52 | 8.07 | 9.17 | 18.77 |
| 4 | N3.17 | 8.57 | 70.88 | 11.84 | 74.16 | 77.77 | 13.75 | N3.05 | 0.62 | 7.17 | 1.30 | 7.84 | 7.88 | 9.52 |
| 5 | N2.11 | 7.20 | 69.51 | 10.91 | 73.23 | 75.30 | 11.55 | N6.22 | 0.62 | 7.16 | 1.16 | 7.71 | 7.86 | 9.39 |
| 6 | N5.01 | 6.46 | 68.77 | 10.17 | 72.49 | 73.97 | 10.37 | N1.06 | 0.61 | 7.15 | 1.07 | 7.61 | 7.84 | 9.26 |
| 7 | N3.12 | 6.18 | 68.50 | 9.60 | 71.92 | 73.47 | 9.92 | N1.21 | 0.60 | 7.15 | 1.00 | 7.55 | 7.84 | 9.22 |
| 8 | N3.18 | 6.05 | 68.37 | 9.16 | 71.47 | 73.24 | 9.71 | N1.19 | 0.54 | 7.09 | 0.94 | 7.49 | 7.70 | 8.29 |
| 9 | N8.04 | 5.63 | 67.94 | 8.76 | 71.08 | 72.47 | 9.03 | N3.04 | 0.51 | 7.05 | 0.90 | 7.44 | 7.63 | 7.76 |
| 10 | N1.06 | 5.56 | 67.87 | 8.44 | 70.76 | 72.34 | 8.92 | N3.09 | 0.47 | 7.02 | 0.85 | 7.40 | 7.56 | 7.23 |
| 11 | N3.03 | 5.29 | 67.60 | 8.16 | 70.47 | 71.85 | 8.48 | N5.12 | 0.47 | 7.01 | 0.82 | 7.36 | 7.54 | 7.13 |
| 12 | N4.09 | 4.80 | 67.12 | 7.88 | 70.19 | 70.98 | 7.71 | N8.04 | 0.45 | 7.00 | 0.79 | 7.33 | 7.51 | 6.89 |
| 13 | N6.22 | 4.46 | 66.78 | 7.61 | 69.93 | 70.37 | 7.16 | N4.09 | 0.43 | 6.98 | 0.76 | 7.31 | 7.47 | 6.60 |
| 14 | N5.15 | 4.31 | 66.62 | 7.38 | 69.69 | 70.09 | 6.91 | N3.15 | 0.40 | 6.94 | 0.73 | 7.28 | 7.39 | 6.05 |
| 15 | N3.08 | 4.27 | 66.58 | 7.17 | 69.49 | 70.02 | 6.85 | N5.21 | 0.39 | 6.94 | 0.71 | 7.26 | 7.38 | 5.95 |
| 16 | N4.08 | 4.07 | 66.39 | 6.98 | 69.29 | 69.66 | 6.53 | N5.01 | 0.39 | 6.94 | 0.69 | 7.24 | 7.38 | 5.94 |
| 17 | N5.14 | 3.99 | 66.31 | 6.80 | 69.12 | 69.52 | 6.41 | N4.06 | 0.38 | 6.92 | 0.67 | 7.22 | 7.35 | 5.76 |
| 18 | N3.09 | 3.62 | 65.93 | 6.63 | 68.94 | 68.84 | 5.80 | N5.15 | 0.36 | 6.91 | 0.66 | 7.20 | 7.32 | 5.55 |
| 19 | N1.21 | 3.42 | 65.74 | 6.46 | 68.77 | 68.49 | 5.49 | N1.18 | 0.32 | 6.87 | 0.64 | 7.18 | 7.23 | 4.89 |
| 20 | N2.07 | 3.38 | 65.70 | 6.30 | 68.62 | 68.42 | 5.43 | N3.16 | 0.32 | 6.87 | 0.62 | 7.17 | 7.23 | 4.88 |
| 21 | N1.18 | 3.12 | 65.44 | 6.15 | 68.47 | 67.95 | 5.01 | N2.01 | 0.25 | 6.80 | 0.60 | 7.15 | 7.08 | 3.83 |
| 22 | N3.15 | 2.41 | 64.73 | 5.98 | 68.30 | 66.66 | 3.87 | N3.02 | 0.24 | 6.79 | 0.59 | 7.13 | 7.06 | 3.70 |
| 23 | N3.04 | 2.29 | 64.61 | 5.82 | 68.14 | 66.46 | 3.68 | N4.20 | 0.22 | 6.76 | 0.57 | 7.12 | 7.01 | 3.32 |
| 24 | N2.02 | 2.16 | 64.47 | 5.67 | 67.98 | 66.21 | 3.46 | N5.03 | 0.21 | 6.76 | 0.56 | 7.10 | 7.00 | 3.27 |
| 25 | N5.12 | 2.13 | 64.44 | 5.53 | 67.84 | 66.16 | 3.42 | N1.09 | 0.20 | 6.74 | 0.54 | 7.09 | 6.97 | 3.01 |
| 26 | N8.03 | 1.93 | 64.24 | 5.39 | 67.70 | 65.80 | 3.09 | N3.12 | 0.19 | 6.73 | 0.53 | 7.07 | 6.94 | 2.83 |
| 27 | N1.22 | 1.87 | 64.19 | 5.26 | 67.57 | 65.70 | 3.01 | N5.19 | 0.18 | 6.73 | 0.52 | 7.06 | 6.94 | 2.81 |
| 28 | N4.15 | 1.79 | 64.10 | 5.13 | 67.45 | 65.54 | 2.87 | N5.14 | 0.18 | 6.72 | 0.50 | 7.05 | 6.92 | 2.68 |
| 29 | N4.06 | 1.77 | 64.08 | 5.02 | 67.33 | 65.50 | 2.84 | N5.10 | 0.17 | 6.72 | 0.49 | 7.04 | 6.91 | 2.63 |
| 30 | N3.02 | 1.28 | 63.59 | 4.89 | 67.21 | 64.62 | 2.05 | N3.19 | 0.17 | 6.72 | 0.48 | 7.03 | 6.91 | 2.59 |
| 31 | N2.16 | 1.19 | 63.50 | 4.77 | 67.09 | 64.46 | 1.91 | N1.22 | 0.17 | 6.71 | 0.47 | 7.02 | 6.90 | 2.56 |
| 32 | N1.19 | 1.12 | 63.43 | 4.66 | 66.97 | 64.33 | 1.79 | N1.17 | 0.17 | 6.71 | 0.46 | 7.01 | 6.90 | 2.53 |
| 33 | N1.09 | 1.04 | 63.35 | 4.55 | 66.87 | 64.19 | 1.66 | N3.18 | 0.15 | 6.70 | 0.45 | 7.00 | 6.87 | 2.32 |
| 34 | N6.12 | 0.99 | 63.31 | 4.45 | 66.76 | 64.10 | 1.59 | N2.22 | 0.14 | 6.68 | 0.44 | 6.99 | 6.84 | 2.07 |
| 35 | N5.21 | 0.92 | 63.23 | 4.34 | 66.66 | 63.97 | 1.47 | N1.05 | 0.12 | 6.67 | 0.43 | 6.98 | 6.81 | 1.91 |
| 36 | N5.05 | 0.86 | 63.17 | 4.25 | 66.56 | 63.87 | 1.38 | N3.03 | 0.12 | 6.66 | 0.43 | 6.97 | 6.80 | 1.79 |
| 37 | N4.21 | 0.65 | 62.96 | 4.15 | 66.47 | 63.48 | 1.04 | N5.18 | 0.11 | 6.66 | 0.42 | 6.96 | 6.78 | 1.68 |
| 38 | N3.05 | 0.57 | 62.89 | 4.06 | 66.37 | 63.35 | 0.92 | N4.15 | 0.08 | 6.62 | 0.41 | 6.95 | 6.71 | 1.18 |
| 39 | N3.19 | 0.57 | 62.88 | 3.97 | 66.28 | 63.34 | 0.91 | N5.06 | 0.06 | 6.60 | 0.40 | 6.95 | 6.67 | 0.88 |
| 40 | N4.07 | 0.15 | 62.47 | 3.87 | 66.19 | 62.59 | 0.24 | N4.13 | 0.05 | 6.60 | 0.39 | 6.94 | 6.66 | 0.81 |
| 41 | N6.08 | −0.46 | 61.85 | 3.77 | 66.08 | 61.48 | −0.74 | N3.08 | 0.02 | 6.57 | 0.38 | 6.93 | 6.59 | 0.31 |
| 42 | N3.16 | −0.53 | 61.78 | 3.66 | 65.98 | 61.36 | −0.85 | N2.02 | 0.02 | 6.56 | 0.37 | 6.92 | 6.58 | 0.27 |
| 43 | N2.13 | −0.59 | 61.73 | 3.56 | 65.88 | 61.26 | −0.94 | N4.07 | 0.02 | 6.56 | 0.36 | 6.91 | 6.58 | 0.23 |
| 44 | N4.20 | −0.74 | 61.58 | 3.47 | 65.78 | 60.98 | −1.19 | N5.09 | 0.01 | 6.55 | 0.36 | 6.90 | 6.56 | 0.08 |
| 45 | N5.03 | −0.97 | 61.34 | 3.37 | 65.68 | 60.56 | −1.56 | N3.22 | −0.02 | 6.53 | 0.35 | 6.89 | 6.51 | −0.25 |
| 46 | N2.01 | −1.03 | 61.29 | 3.27 | 65.59 | 60.46 | −1.65 | N1.16 | −0.02 | 6.53 | 0.34 | 6.89 | 6.50 | −0.30 |
| 47 | N5.18 | −1.11 | 61.21 | 3.18 | 65.49 | 60.31 | −1.78 | N6.19 | −0.04 | 6.50 | 0.33 | 6.88 | 6.46 | −0.65 |
| 48 | N1.12 | −1.16 | 61.16 | 3.09 | 65.40 | 60.23 | −1.85 | N2.16 | −0.06 | 6.49 | 0.32 | 6.87 | 6.42 | −0.88 |
| 49 | N5.16 | −1.21 | 61.11 | 3.00 | 65.32 | 60.14 | −1.93 | N6.12 | −0.06 | 6.48 | 0.32 | 6.86 | 6.41 | −0.95 |
| 50 | N5.20 | −1.26 | 61.06 | 2.92 | 65.23 | 60.04 | −2.02 | N2.11 | −0.08 | 6.47 | 0.31 | 6.85 | 6.38 | −1.16 |
| 51 | N4.13 | −1.43 | 60.89 | 2.83 | 65.15 | 59.74 | −2.29 | N4.14 | −0.09 | 6.46 | 0.30 | 6.85 | 6.35 | −1.38 |
| 52 | N2.10 | −1.46 | 60.85 | 2.75 | 65.06 | 59.68 | −2.35 | N8.03 | −0.09 | 6.45 | 0.29 | 6.84 | 6.35 | −1.40 |
| 53 | N1.17 | −1.63 | 60.69 | 2.67 | 64.98 | 59.38 | −2.61 | N2.07 | −0.11 | 6.43 | 0.28 | 6.83 | 6.31 | −1.71 |
| 54 | N4.05 | −1.72 | 60.59 | 2.58 | 64.90 | 59.20 | −2.77 | N4.08 | −0.12 | 6.43 | 0.28 | 6.82 | 6.29 | −1.84 |
| 55 | N6.05 | −2.03 | 60.29 | 2.50 | 64.82 | 58.66 | −3.25 | N2.13 | −0.13 | 6.41 | 0.27 | 6.82 | 6.26 | −2.01 |
| 56 | N1.07 | −2.14 | 60.18 | 2.42 | 64.73 | 58.45 | −3.43 | N6.05 | −0.17 | 6.37 | 0.26 | 6.81 | 6.18 | −2.63 |
| 57 | N1.16 | −2.26 | 60.06 | 2.34 | 64.65 | 58.24 | −3.63 | N2.17 | −0.17 | 6.37 | 0.25 | 6.80 | 6.17 | −2.66 |
| 58 | N4.19 | −2.35 | 59.97 | 2.25 | 64.57 | 58.07 | −3.77 | N1.10 | −0.18 | 6.36 | 0.25 | 6.79 | 6.15 | −2.80 |
| 59 | N2.22 | −2.42 | 59.89 | 2.18 | 64.49 | 57.94 | −3.89 | N4.21 | −0.24 | 6.31 | 0.24 | 6.78 | 6.04 | −3.64 |
| 60 | N5.09 | −2.65 | 59.67 | 2.10 | 64.41 | 57.54 | −4.25 | N6.18 | −0.24 | 6.31 | 0.23 | 6.78 | 6.03 | −3.66 |
| 61 | N5.19 | −2.76 | 59.56 | 2.02 | 64.33 | 57.34 | −4.43 | N7.20 | −0.26 | 6.29 | 0.22 | 6.77 | 5.99 | −3.98 |
| 62 | N6.18 | −2.83 | 59.49 | 1.94 | 64.25 | 57.21 | −4.54 | N1.12 | −0.26 | 6.29 | 0.21 | 6.76 | 5.99 | −3.98 |
| 63 | N5.10 | −2.84 | 59.48 | 1.86 | 64.18 | 57.20 | −4.55 | N2.10 | −0.31 | 6.23 | 0.21 | 6.75 | 5.88 | −4.80 |
| 64 | N7.20 | −2.89 | 59.42 | 1.79 | 64.10 | 57.10 | −4.64 | N4.10 | −0.34 | 6.21 | 0.20 | 6.74 | 5.83 | −5.13 |
| 65 | N1.14 | −2.96 | 59.36 | 1.71 | 64.03 | 56.98 | −4.74 | N6.08 | −0.35 | 6.19 | 0.19 | 6.74 | 5.79 | −5.38 |
| 66 | N5.22 | −3.15 | 59.17 | 1.64 | 63.96 | 56.63 | −5.05 | N5.22 | −0.40 | 6.15 | 0.18 | 6.73 | 5.69 | −6.11 |
| 67 | N6.19 | −3.17 | 59.15 | 1.57 | 63.88 | 56.60 | −5.09 | N1.14 | −0.45 | 6.10 | 0.17 | 6.72 | 5.59 | −6.82 |
| 68 | N1.10 | −3.47 | 58.85 | 1.49 | 63.81 | 56.06 | −5.56 | N1.07 | −0.47 | 6.07 | 0.16 | 6.71 | 5.54 | −7.23 |
| 69 | N5.11 | −3.57 | 58.75 | 1.42 | 63.74 | 55.88 | −5.73 | N4.05 | −0.47 | 6.07 | 0.15 | 6.70 | 5.53 | −7.25 |
| 70 | N4.10 | −3.82 | 58.50 | 1.35 | 63.66 | 55.43 | −6.13 | N4.19 | −0.53 | 6.01 | 0.14 | 6.69 | 5.41 | −8.14 |
| 71 | N2.17 | −3.86 | 58.46 | 1.27 | 63.59 | 55.35 | −6.19 | N5.20 | −0.57 | 5.98 | 0.13 | 6.68 | 5.34 | −8.64 |
| 72 | N5.02 | −3.98 | 58.34 | 1.20 | 63.52 | 55.14 | −6.38 | N5.02 | −0.58 | 5.97 | 0.12 | 6.67 | 5.31 | −8.82 |
| 73 | N3.07 | −4.02 | 58.29 | 1.13 | 63.44 | 55.06 | −6.46 | N6.01 | −0.60 | 5.95 | 0.11 | 6.66 | 5.27 | −9.11 |
| 74 | N4.14 | −4.83 | 57.49 | 1.05 | 63.36 | 53.61 | −7.75 | N2.18 | −0.61 | 5.94 | 0.10 | 6.65 | 5.25 | −9.28 |
| 75 | N6.01 | −5.39 | 56.93 | 0.96 | 63.28 | 52.59 | −8.65 | N5.16 | −0.62 | 5.93 | 0.09 | 6.64 | 5.22 | −9.47 |
| 76 | N3.22 | −5.40 | 56.92 | 0.88 | 63.19 | 52.58 | −8.66 | N4.12 | −0.67 | 5.87 | 0.08 | 6.63 | 5.11 | −10.30 |
| 77 | N3.06 | −5.47 | 56.84 | 0.80 | 63.11 | 52.44 | −8.78 | N3.07 | −0.68 | 5.86 | 0.07 | 6.62 | 5.08 | −10.45 |
| 78 | N5.06 | −5.78 | 56.53 | 0.71 | 63.03 | 51.88 | −9.28 | N5.05 | −0.69 | 5.86 | 0.06 | 6.61 | 5.08 | −10.49 |
| 79 | N1.15 | −6.95 | 55.36 | 0.62 | 62.93 | 49.77 | −11.16 | N7.15 | −0.72 | 5.82 | 0.05 | 6.60 | 5.00 | −11.05 |
| 80 | N1.13 | −7.78 | 54.53 | 0.51 | 62.83 | 48.27 | −12.49 | N1.15 | −0.73 | 5.82 | 0.04 | 6.59 | 4.99 | −11.10 |
| 81 | N7.15 | −8.91 | 53.41 | 0.39 | 62.71 | 46.24 | −14.30 | N1.13 | −0.73 | 5.82 | 0.03 | 6.58 | 4.99 | −11.10 |
| 82 | N4.12 | −9.95 | 52.37 | 0.27 | 62.58 | 44.37 | −15.96 | N3.06 | −0.77 | 5.77 | 0.02 | 6.57 | 4.90 | −11.78 |
| 83 | N2.18 | −10.35 | 51.96 | 0.14 | 62.46 | 43.64 | −16.61 | N5.11 | −0.84 | 5.70 | 0.01 | 6.56 | 4.74 | −12.90 |
| 84 | N4.17 | −11.60 | 50.72 | 0.00 | 62.32 | 41.39 | −18.61 | N4.17 | −1.19 | 5.36 | 0.00 | 6.55 | 4.01 | −18.12 |
| IDM (g plant−1) | TDM (g plant−1) | |||||||||||||
| Order | Ecotypes | g | u + g | Gain | µn | u + g + gem | SG | Ecotypes | g | u + g | Gain | µn | u + g + gem | SG |
| 1 | N3.10 | 0.95 | 4.82 | 0.95 | 4.82 | 5.81 | 24.70 | N3.10 | 31.75 | 107.65 | 31.75 | 107.65 | 125.44 | 41.84 |
| 2 | N3.18 | 0.90 | 4.77 | 0.93 | 4.79 | 5.71 | 23.40 | N2.06 | 15.25 | 91.14 | 23.50 | 99.40 | 99.69 | 20.09 |
| 3 | N2.16 | 0.70 | 4.57 | 0.85 | 4.72 | 5.30 | 18.21 | N3.17 | 12.64 | 88.54 | 19.88 | 95.78 | 95.62 | 16.66 |
| 4 | N2.07 | 0.69 | 4.55 | 0.81 | 4.68 | 5.27 | 17.85 | N1.05 | 10.77 | 86.66 | 17.60 | 93.50 | 92.70 | 14.19 |
| 5 | N3.17 | 0.66 | 4.53 | 0.78 | 4.65 | 5.22 | 17.19 | N1.21 | 10.30 | 86.20 | 16.14 | 92.04 | 91.97 | 13.58 |
| 6 | N3.04 | 0.60 | 4.47 | 0.75 | 4.62 | 5.09 | 15.58 | N4.09 | 9.92 | 85.81 | 15.11 | 91.00 | 91.37 | 13.07 |
| 7 | N2.11 | 0.59 | 4.46 | 0.73 | 4.59 | 5.07 | 15.29 | N3.18 | 9.64 | 85.53 | 14.33 | 90.22 | 90.93 | 12.70 |
| 8 | N4.07 | 0.59 | 4.45 | 0.71 | 4.58 | 5.06 | 15.14 | N1.06 | 8.68 | 84.57 | 13.62 | 89.51 | 89.43 | 11.43 |
| 9 | N2.02 | 0.56 | 4.43 | 0.70 | 4.56 | 5.01 | 14.58 | N5.01 | 8.35 | 84.24 | 13.03 | 88.93 | 88.92 | 11.00 |
| 10 | N1.05 | 0.49 | 4.35 | 0.67 | 4.54 | 4.86 | 12.66 | N6.22 | 8.25 | 84.14 | 12.56 | 88.45 | 88.77 | 10.87 |
| 11 | N1.18 | 0.46 | 4.32 | 0.66 | 4.52 | 4.80 | 11.84 | N3.09 | 7.81 | 83.71 | 12.12 | 88.02 | 88.08 | 10.29 |
| 12 | N3.09 | 0.41 | 4.28 | 0.63 | 4.50 | 4.71 | 10.68 | N8.04 | 7.29 | 83.19 | 11.72 | 87.62 | 87.27 | 9.61 |
| 13 | N1.21 | 0.41 | 4.27 | 0.62 | 4.48 | 4.70 | 10.54 | N2.11 | 7.28 | 83.18 | 11.38 | 87.27 | 87.26 | 9.60 |
| 14 | N5.20 | 0.37 | 4.24 | 0.60 | 4.46 | 4.63 | 9.68 | N5.12 | 7.12 | 83.02 | 11.08 | 86.97 | 87.01 | 9.38 |
| 15 | N2.06 | 0.37 | 4.24 | 0.58 | 4.45 | 4.63 | 9.66 | N2.07 | 6.94 | 82.83 | 10.80 | 86.69 | 86.72 | 9.14 |
| 16 | N5.15 | 0.36 | 4.22 | 0.57 | 4.44 | 4.59 | 9.23 | N4.06 | 6.62 | 82.51 | 10.54 | 86.43 | 86.22 | 8.72 |
| 17 | N6.08 | 0.35 | 4.22 | 0.56 | 4.42 | 4.58 | 9.08 | N1.18 | 6.00 | 81.89 | 10.27 | 86.17 | 85.26 | 7.91 |
| 18 | N4.09 | 0.34 | 4.21 | 0.55 | 4.41 | 4.56 | 8.82 | N3.04 | 5.81 | 81.70 | 10.02 | 85.92 | 84.96 | 7.66 |
| 19 | N3.03 | 0.28 | 4.15 | 0.53 | 4.40 | 4.44 | 7.33 | N3.12 | 5.28 | 81.17 | 9.77 | 85.67 | 84.12 | 6.95 |
| 20 | N4.08 | 0.26 | 4.12 | 0.52 | 4.38 | 4.39 | 6.61 | N4.08 | 4.80 | 80.69 | 9.53 | 85.42 | 83.38 | 6.32 |
| 21 | N5.01 | 0.22 | 4.09 | 0.50 | 4.37 | 4.31 | 5.70 | N3.05 | 4.54 | 80.44 | 9.29 | 85.18 | 82.98 | 5.99 |
| 22 | N5.18 | 0.21 | 4.07 | 0.49 | 4.35 | 4.29 | 5.35 | N3.03 | 4.33 | 80.22 | 9.06 | 84.96 | 82.64 | 5.70 |
| 23 | N5.09 | 0.20 | 4.06 | 0.48 | 4.34 | 4.26 | 5.07 | N2.02 | 4.08 | 79.97 | 8.85 | 84.74 | 82.25 | 5.37 |
| 24 | N8.03 | 0.16 | 4.03 | 0.46 | 4.33 | 4.20 | 4.22 | N5.15 | 3.61 | 79.51 | 8.63 | 84.52 | 81.53 | 4.76 |
| 25 | N6.22 | 0.15 | 4.02 | 0.45 | 4.32 | 4.17 | 3.93 | N3.16 | 2.56 | 78.46 | 8.39 | 84.28 | 79.89 | 3.38 |
| 26 | N1.10 | 0.14 | 4.00 | 0.44 | 4.30 | 4.14 | 3.50 | N3.19 | 2.45 | 78.34 | 8.16 | 84.05 | 79.72 | 3.23 |
| 27 | N8.04 | 0.13 | 3.99 | 0.43 | 4.29 | 4.12 | 3.30 | N2.13 | 2.38 | 78.28 | 7.94 | 83.84 | 79.61 | 3.14 |
| 28 | N3.16 | 0.12 | 3.99 | 0.42 | 4.28 | 4.12 | 3.19 | N3.15 | 2.38 | 78.27 | 7.74 | 83.64 | 79.60 | 3.13 |
| 29 | N4.05 | 0.12 | 3.99 | 0.41 | 4.27 | 4.11 | 3.13 | N1.19 | 2.28 | 78.18 | 7.56 | 83.45 | 79.45 | 3.01 |
| 30 | N4.06 | 0.10 | 3.97 | 0.40 | 4.26 | 4.07 | 2.64 | N4.07 | 2.27 | 78.16 | 7.38 | 83.27 | 79.43 | 2.99 |
| 31 | N5.19 | 0.09 | 3.95 | 0.39 | 4.25 | 4.05 | 2.30 | N3.08 | 2.24 | 78.13 | 7.21 | 83.11 | 79.38 | 2.95 |
| 32 | N5.02 | 0.08 | 3.95 | 0.38 | 4.24 | 4.03 | 2.14 | N5.21 | 1.82 | 77.71 | 7.05 | 82.94 | 78.73 | 2.40 |
| 33 | N4.13 | 0.07 | 3.93 | 0.37 | 4.23 | 4.00 | 1.73 | N5.18 | 1.75 | 77.64 | 6.88 | 82.78 | 78.62 | 2.30 |
| 34 | N2.13 | 0.06 | 3.93 | 0.36 | 4.22 | 3.99 | 1.64 | N1.22 | 1.62 | 77.51 | 6.73 | 82.62 | 78.42 | 2.13 |
| 35 | N5.12 | 0.06 | 3.92 | 0.35 | 4.22 | 3.98 | 1.51 | N2.16 | 1.50 | 77.39 | 6.58 | 82.47 | 78.23 | 1.97 |
| 36 | N1.22 | 0.05 | 3.91 | 0.34 | 4.21 | 3.97 | 1.28 | N4.21 | 1.40 | 77.30 | 6.44 | 82.33 | 78.08 | 1.85 |
| 37 | N3.19 | 0.05 | 3.91 | 0.33 | 4.20 | 3.96 | 1.27 | N5.03 | 1.19 | 77.08 | 6.29 | 82.19 | 77.74 | 1.56 |
| 38 | N1.06 | 0.01 | 3.88 | 0.33 | 4.19 | 3.89 | 0.27 | N2.01 | 0.86 | 76.75 | 6.15 | 82.05 | 77.23 | 1.13 |
| 39 | N3.02 | 0.01 | 3.87 | 0.32 | 4.18 | 3.88 | 0.19 | N5.14 | 0.83 | 76.72 | 6.02 | 81.91 | 77.19 | 1.09 |
| 40 | N1.19 | 0.01 | 3.87 | 0.31 | 4.17 | 3.88 | 0.18 | N3.02 | 0.59 | 76.49 | 5.88 | 81.77 | 76.82 | 0.78 |
| 41 | N5.21 | 0.00 | 3.86 | 0.30 | 4.17 | 3.86 | −0.04 | N1.09 | 0.28 | 76.17 | 5.74 | 81.64 | 76.33 | 0.37 |
| 42 | N5.03 | −0.01 | 3.86 | 0.29 | 4.16 | 3.85 | −0.19 | N4.15 | −0.53 | 75.37 | 5.59 | 81.49 | 75.07 | −0.70 |
| 43 | N3.12 | −0.01 | 3.85 | 0.29 | 4.15 | 3.84 | −0.27 | N5.20 | −0.63 | 75.27 | 5.45 | 81.34 | 74.92 | −0.82 |
| 44 | N3.05 | −0.01 | 3.85 | 0.28 | 4.15 | 3.84 | −0.33 | N2.10 | −0.94 | 74.95 | 5.30 | 81.20 | 74.42 | −1.24 |
| 45 | N3.08 | −0.02 | 3.84 | 0.27 | 4.14 | 3.82 | −0.59 | N6.12 | −0.99 | 74.90 | 5.16 | 81.06 | 74.34 | −1.31 |
| 46 | N2.10 | −0.04 | 3.83 | 0.27 | 4.13 | 3.79 | −1.00 | N8.03 | −1.05 | 74.85 | 5.03 | 80.92 | 74.26 | −1.38 |
| 47 | N4.14 | −0.06 | 3.81 | 0.26 | 4.13 | 3.75 | −1.50 | N1.10 | −1.36 | 74.54 | 4.89 | 80.79 | 73.77 | −1.79 |
| 48 | N3.15 | −0.07 | 3.80 | 0.25 | 4.12 | 3.72 | −1.79 | N4.20 | −1.40 | 74.50 | 4.76 | 80.66 | 73.71 | −1.84 |
| 49 | N5.06 | −0.09 | 3.78 | 0.25 | 4.11 | 3.69 | −2.22 | N1.17 | −1.45 | 74.45 | 4.64 | 80.53 | 73.64 | −1.91 |
| 50 | N6.12 | −0.10 | 3.76 | 0.24 | 4.10 | 3.66 | −2.60 | N5.09 | −1.55 | 74.35 | 4.51 | 80.41 | 73.48 | −2.04 |
| 51 | N6.05 | −0.14 | 3.72 | 0.23 | 4.10 | 3.57 | −3.72 | N2.22 | −1.66 | 74.24 | 4.39 | 80.28 | 73.31 | −2.18 |
| 52 | N1.07 | −0.15 | 3.71 | 0.22 | 4.09 | 3.56 | −3.88 | N5.19 | −2.10 | 73.80 | 4.27 | 80.16 | 72.62 | −2.76 |
| 53 | N1.17 | −0.15 | 3.71 | 0.22 | 4.08 | 3.55 | −3.96 | N6.08 | −2.11 | 73.79 | 4.15 | 80.04 | 72.61 | −2.78 |
| 54 | N2.22 | −0.16 | 3.71 | 0.21 | 4.08 | 3.54 | −4.06 | N6.19 | −2.39 | 73.51 | 4.02 | 79.92 | 72.17 | −3.14 |
| 55 | N3.22 | −0.21 | 3.66 | 0.20 | 4.07 | 3.44 | −5.35 | N4.13 | −2.82 | 73.07 | 3.90 | 79.79 | 71.49 | −3.72 |
| 56 | N4.15 | −0.21 | 3.66 | 0.20 | 4.06 | 3.44 | −5.36 | N5.05 | −2.94 | 72.95 | 3.78 | 79.67 | 71.30 | −3.88 |
| 57 | N5.22 | −0.22 | 3.64 | 0.19 | 4.05 | 3.41 | −5.73 | N4.05 | −3.09 | 72.80 | 3.66 | 79.55 | 71.07 | −4.07 |
| 58 | N4.20 | −0.23 | 3.64 | 0.18 | 4.05 | 3.40 | −5.88 | N5.22 | −3.70 | 72.19 | 3.53 | 79.42 | 70.12 | −4.87 |
| 59 | N5.16 | −0.23 | 3.63 | 0.17 | 4.04 | 3.40 | −5.96 | N6.18 | −4.00 | 71.89 | 3.40 | 79.30 | 69.65 | −5.27 |
| 60 | N3.07 | −0.24 | 3.63 | 0.17 | 4.03 | 3.38 | −6.17 | N5.16 | −4.01 | 71.89 | 3.28 | 79.17 | 69.64 | −5.28 |
| 61 | N2.01 | −0.24 | 3.62 | 0.16 | 4.03 | 3.37 | −6.28 | N5.10 | −4.34 | 71.55 | 3.15 | 79.05 | 69.12 | −5.72 |
| 62 | N6.18 | −0.25 | 3.61 | 0.15 | 4.02 | 3.36 | −6.47 | N2.17 | −4.43 | 71.46 | 3.03 | 78.93 | 68.98 | −5.84 |
| 63 | N5.14 | −0.25 | 3.61 | 0.15 | 4.01 | 3.35 | −6.56 | N6.05 | −4.70 | 71.19 | 2.91 | 78.80 | 68.56 | −6.20 |
| 64 | N2.17 | −0.28 | 3.59 | 0.14 | 4.01 | 3.30 | −7.21 | N1.14 | −4.72 | 71.17 | 2.79 | 78.68 | 68.53 | −6.22 |
| 65 | N4.21 | −0.28 | 3.59 | 0.13 | 4.00 | 3.30 | −7.22 | N7.20 | −4.76 | 71.13 | 2.67 | 78.57 | 68.46 | −6.27 |
| 66 | N1.13 | −0.32 | 3.55 | 0.13 | 3.99 | 3.22 | −8.24 | N5.02 | −5.26 | 70.64 | 2.55 | 78.45 | 67.69 | −6.92 |
| 67 | N1.14 | −0.32 | 3.54 | 0.12 | 3.99 | 3.21 | −8.27 | N3.22 | −5.48 | 70.41 | 2.43 | 78.33 | 67.34 | −7.22 |
| 68 | N1.12 | −0.34 | 3.52 | 0.11 | 3.98 | 3.17 | −8.81 | N1.16 | −5.68 | 70.22 | 2.31 | 78.21 | 67.03 | −7.48 |
| 69 | N1.16 | −0.34 | 3.52 | 0.11 | 3.97 | 3.16 | −8.92 | N1.12 | −5.75 | 70.14 | 2.20 | 78.09 | 66.92 | −7.58 |
| 70 | N7.20 | −0.35 | 3.52 | 0.10 | 3.97 | 3.16 | −8.98 | N6.01 | −5.77 | 70.13 | 2.08 | 77.98 | 66.90 | −7.60 |
| 71 | N2.18 | −0.37 | 3.50 | 0.09 | 3.96 | 3.12 | −9.47 | N3.07 | −6.09 | 69.80 | 1.97 | 77.86 | 66.39 | −8.03 |
| 72 | N1.09 | −0.37 | 3.49 | 0.09 | 3.95 | 3.10 | −9.67 | N4.10 | −6.24 | 69.65 | 1.85 | 77.75 | 66.16 | −8.22 |
| 73 | N6.19 | −0.40 | 3.46 | 0.08 | 3.95 | 3.04 | −10.47 | N4.14 | −6.59 | 69.30 | 1.74 | 77.63 | 65.61 | −8.68 |
| 74 | N4.12 | −0.41 | 3.46 | 0.07 | 3.94 | 3.04 | −10.53 | N4.19 | −6.98 | 68.92 | 1.62 | 77.51 | 65.01 | −9.19 |
| 75 | N5.10 | −0.46 | 3.41 | 0.07 | 3.93 | 2.93 | −11.81 | N1.07 | −7.16 | 68.73 | 1.50 | 77.40 | 64.72 | −9.44 |
| 76 | N6.01 | −0.46 | 3.41 | 0.06 | 3.93 | 2.93 | −11.83 | N5.06 | −7.78 | 68.11 | 1.38 | 77.28 | 63.76 | −10.25 |
| 77 | N5.11 | −0.47 | 3.40 | 0.05 | 3.92 | 2.91 | −12.14 | N5.11 | −9.61 | 66.28 | 1.24 | 77.13 | 60.90 | −12.66 |
| 78 | N7.15 | −0.47 | 3.40 | 0.05 | 3.91 | 2.91 | −12.14 | N3.06 | −9.69 | 66.20 | 1.10 | 76.99 | 60.78 | −12.77 |
| 79 | N4.19 | −0.48 | 3.38 | 0.04 | 3.90 | 2.88 | −12.54 | N1.13 | −10.11 | 65.78 | 0.96 | 76.85 | 60.12 | −13.33 |
| 80 | N4.10 | −0.53 | 3.34 | 0.03 | 3.90 | 2.79 | −13.64 | N7.15 | −12.82 | 63.07 | 0.79 | 76.68 | 55.89 | −16.90 |
| 81 | N5.05 | −0.53 | 3.33 | 0.03 | 3.89 | 2.78 | −13.77 | N1.15 | −13.59 | 62.31 | 0.61 | 76.50 | 54.70 | −17.90 |
| 82 | N4.17 | −0.64 | 3.23 | 0.02 | 3.88 | 2.57 | −16.49 | N2.18 | −14.41 | 61.49 | 0.42 | 76.32 | 53.42 | −18.98 |
| 83 | N1.15 | −0.66 | 3.20 | 0.01 | 3.87 | 2.51 | −17.17 | N4.12 | −15.22 | 60.68 | 0.24 | 76.13 | 52.15 | −20.05 |
| 84 | N3.06 | −0.83 | 3.04 | 0.00 | 3.86 | 2.18 | −21.40 | N4.17 | −19.59 | 56.30 | 0.00 | 75.89 | 45.32 | −25.82 |
| LSR | CT | |||||||||||||
| Order | Ecotypes | g | u + g | Gain | µn | u + g + gem | SG | Ecotypes | g | u + g | Gain | µn | u + g + gem | SG |
| 1 | N4.14 | 0.87 | 3.81 | 0.87 | 3.81 | 4.48 | 29.82 | N1.09 | 0.82 | 3.85 | 0.82 | 3.85 | 4.39 | 27.14 |
| 2 | N4.10 | 0.70 | 3.63 | 0.79 | 3.72 | 4.17 | 23.82 | N1.13 | 0.59 | 3.62 | 0.71 | 3.73 | 4.01 | 19.43 |
| 3 | N5.12 | 0.62 | 3.55 | 0.73 | 3.66 | 4.03 | 21.15 | N1.12 | 0.42 | 3.45 | 0.61 | 3.64 | 3.73 | 13.93 |
| 4 | N1.13 | 0.59 | 3.52 | 0.69 | 3.63 | 3.97 | 19.95 | N1.07 | 0.39 | 3.42 | 0.56 | 3.58 | 3.67 | 12.83 |
| 5 | N1.10 | 0.53 | 3.46 | 0.66 | 3.59 | 3.87 | 18.00 | N8.03 | 0.29 | 3.32 | 0.50 | 3.53 | 3.51 | 9.53 |
| 6 | N7.20 | 0.47 | 3.41 | 0.63 | 3.56 | 3.77 | 16.16 | N1.14 | 0.26 | 3.28 | 0.46 | 3.49 | 3.45 | 8.43 |
| 7 | N1.09 | 0.46 | 3.40 | 0.61 | 3.54 | 3.76 | 15.81 | N1.16 | 0.26 | 3.28 | 0.43 | 3.46 | 3.45 | 8.43 |
| 8 | N2.01 | 0.41 | 3.34 | 0.58 | 3.52 | 3.66 | 13.98 | N7.15 | 0.26 | 3.28 | 0.41 | 3.44 | 3.45 | 8.43 |
| 9 | N1.21 | 0.39 | 3.33 | 0.56 | 3.49 | 3.63 | 13.41 | N3.22 | 0.22 | 3.25 | 0.39 | 3.42 | 3.40 | 7.32 |
| 10 | N2.06 | 0.38 | 3.31 | 0.54 | 3.48 | 3.61 | 12.96 | N4.21 | 0.22 | 3.25 | 0.37 | 3.40 | 3.40 | 7.32 |
| 11 | N1.19 | 0.37 | 3.30 | 0.53 | 3.46 | 3.58 | 12.52 | N2.01 | 0.19 | 3.22 | 0.36 | 3.38 | 3.34 | 6.22 |
| 12 | N3.08 | 0.29 | 3.23 | 0.51 | 3.44 | 3.45 | 10.02 | N1.15 | 0.19 | 3.22 | 0.34 | 3.37 | 3.34 | 6.22 |
| 13 | N6.08 | 0.25 | 3.18 | 0.49 | 3.42 | 3.37 | 8.41 | N1.06 | 0.19 | 3.22 | 0.33 | 3.36 | 3.34 | 6.22 |
| 14 | N1.14 | 0.24 | 3.18 | 0.47 | 3.40 | 3.36 | 8.26 | N8.04 | 0.16 | 3.18 | 0.32 | 3.35 | 3.29 | 5.13 |
| 15 | N5.22 | 0.23 | 3.16 | 0.45 | 3.39 | 3.34 | 7.84 | N3.19 | 0.16 | 3.18 | 0.31 | 3.34 | 3.29 | 5.13 |
| 16 | N2.18 | 0.22 | 3.15 | 0.44 | 3.37 | 3.32 | 7.51 | N5.05 | 0.16 | 3.18 | 0.30 | 3.33 | 3.29 | 5.13 |
| 17 | N4.12 | 0.21 | 3.15 | 0.43 | 3.36 | 3.31 | 7.25 | N6.19 | 0.16 | 3.18 | 0.29 | 3.32 | 3.29 | 5.13 |
| 18 | N1.15 | 0.21 | 3.14 | 0.41 | 3.35 | 3.31 | 7.15 | N1.10 | 0.16 | 3.18 | 0.28 | 3.31 | 3.29 | 5.13 |
| 19 | N2.07 | 0.20 | 3.14 | 0.40 | 3.34 | 3.29 | 6.92 | N1.19 | 0.12 | 3.15 | 0.27 | 3.30 | 3.23 | 4.02 |
| 20 | N3.02 | 0.19 | 3.13 | 0.39 | 3.33 | 3.27 | 6.56 | N4.06 | 0.12 | 3.15 | 0.27 | 3.29 | 3.23 | 4.02 |
| 21 | N5.14 | 0.19 | 3.12 | 0.38 | 3.32 | 3.27 | 6.46 | N1.17 | 0.09 | 3.12 | 0.26 | 3.29 | 3.18 | 2.92 |
| 22 | N5.06 | 0.19 | 3.12 | 0.37 | 3.31 | 3.26 | 6.32 | N5.11 | 0.09 | 3.12 | 0.25 | 3.28 | 3.18 | 2.92 |
| 23 | N4.21 | 0.16 | 3.09 | 0.36 | 3.30 | 3.21 | 5.34 | N3.18 | 0.09 | 3.12 | 0.24 | 3.27 | 3.18 | 2.92 |
| 24 | N1.12 | 0.15 | 3.08 | 0.36 | 3.29 | 3.20 | 5.08 | N3.12 | 0.06 | 3.08 | 0.23 | 3.26 | 3.12 | 1.82 |
| 25 | N6.22 | 0.13 | 3.07 | 0.35 | 3.28 | 3.17 | 4.58 | N3.07 | 0.06 | 3.08 | 0.23 | 3.26 | 3.12 | 1.82 |
| 26 | N3.04 | 0.13 | 3.06 | 0.34 | 3.27 | 3.16 | 4.42 | N2.17 | 0.06 | 3.08 | 0.22 | 3.25 | 3.12 | 1.82 |
| 27 | N3.19 | 0.12 | 3.06 | 0.33 | 3.26 | 3.15 | 4.16 | N6.05 | 0.02 | 3.05 | 0.21 | 3.24 | 3.07 | 0.72 |
| 28 | N6.19 | 0.12 | 3.05 | 0.32 | 3.26 | 3.14 | 4.07 | N3.03 | 0.02 | 3.05 | 0.21 | 3.24 | 3.07 | 0.72 |
| 29 | N7.15 | 0.11 | 3.04 | 0.32 | 3.25 | 3.12 | 3.63 | N2.18 | 0.02 | 3.05 | 0.20 | 3.23 | 3.07 | 0.72 |
| 30 | N5.19 | 0.08 | 3.01 | 0.31 | 3.24 | 3.07 | 2.60 | N5.16 | 0.02 | 3.05 | 0.19 | 3.22 | 3.07 | 0.72 |
| 31 | N4.13 | 0.08 | 3.01 | 0.30 | 3.23 | 3.07 | 2.59 | N3.04 | −0.01 | 3.02 | 0.19 | 3.22 | 3.01 | −0.38 |
| 32 | N3.16 | 0.08 | 3.01 | 0.29 | 3.23 | 3.07 | 2.57 | N5.14 | −0.01 | 3.02 | 0.18 | 3.21 | 3.01 | −0.38 |
| 33 | N5.10 | 0.07 | 3.01 | 0.29 | 3.22 | 3.06 | 2.50 | N5.01 | −0.01 | 3.02 | 0.18 | 3.20 | 3.01 | −0.38 |
| 34 | N4.08 | 0.07 | 3.00 | 0.28 | 3.21 | 3.06 | 2.35 | N4.20 | −0.01 | 3.02 | 0.17 | 3.20 | 3.01 | −0.38 |
| 35 | N5.21 | 0.06 | 3.00 | 0.27 | 3.21 | 3.05 | 2.19 | N4.14 | −0.01 | 3.02 | 0.16 | 3.19 | 3.01 | −0.38 |
| 36 | N3.22 | 0.06 | 3.00 | 0.27 | 3.20 | 3.05 | 2.17 | N3.08 | −0.01 | 3.02 | 0.16 | 3.19 | 3.01 | −0.38 |
| 37 | N3.15 | 0.05 | 2.99 | 0.26 | 3.20 | 3.03 | 1.80 | N1.18 | −0.01 | 3.02 | 0.16 | 3.18 | 3.01 | −0.38 |
| 38 | N8.04 | 0.04 | 2.97 | 0.26 | 3.19 | 3.00 | 1.34 | N3.09 | −0.01 | 3.02 | 0.15 | 3.18 | 3.01 | −0.38 |
| 39 | N5.02 | 0.03 | 2.97 | 0.25 | 3.18 | 2.99 | 1.11 | N3.15 | −0.01 | 3.02 | 0.15 | 3.18 | 3.01 | −0.38 |
| 40 | N6.12 | 0.03 | 2.96 | 0.24 | 3.18 | 2.98 | 0.92 | N4.05 | −0.01 | 3.02 | 0.14 | 3.17 | 3.01 | −0.38 |
| 41 | N5.20 | 0.03 | 2.96 | 0.24 | 3.17 | 2.98 | 0.86 | N4.13 | −0.04 | 2.98 | 0.14 | 3.17 | 2.95 | −1.48 |
| 42 | N3.18 | 0.02 | 2.95 | 0.23 | 3.17 | 2.96 | 0.53 | N4.15 | −0.04 | 2.98 | 0.13 | 3.16 | 2.95 | −1.48 |
| 43 | N6.01 | −0.03 | 2.90 | 0.23 | 3.16 | 2.88 | −0.99 | N3.05 | −0.04 | 2.98 | 0.13 | 3.16 | 2.95 | −1.48 |
| 44 | N5.09 | −0.03 | 2.90 | 0.22 | 3.16 | 2.88 | −1.08 | N4.19 | −0.04 | 2.98 | 0.13 | 3.15 | 2.95 | −1.48 |
| 45 | N3.06 | −0.04 | 2.89 | 0.22 | 3.15 | 2.87 | −1.30 | N2.10 | −0.04 | 2.98 | 0.12 | 3.15 | 2.95 | −1.48 |
| 46 | N3.17 | −0.04 | 2.89 | 0.21 | 3.14 | 2.86 | −1.34 | N2.16 | −0.04 | 2.98 | 0.12 | 3.15 | 2.95 | −1.48 |
| 47 | N1.22 | −0.04 | 2.89 | 0.21 | 3.14 | 2.86 | −1.43 | N6.01 | −0.04 | 2.98 | 0.11 | 3.14 | 2.95 | −1.48 |
| 48 | N4.05 | −0.05 | 2.88 | 0.20 | 3.13 | 2.85 | −1.65 | N4.12 | −0.04 | 2.98 | 0.11 | 3.14 | 2.95 | −1.48 |
| 49 | N2.11 | −0.05 | 2.88 | 0.19 | 3.13 | 2.84 | −1.77 | N5.15 | −0.04 | 2.98 | 0.11 | 3.14 | 2.95 | −1.48 |
| 50 | N4.07 | −0.06 | 2.88 | 0.19 | 3.12 | 2.83 | −1.98 | N5.18 | −0.04 | 2.98 | 0.11 | 3.13 | 2.95 | −1.48 |
| 51 | N1.06 | −0.08 | 2.85 | 0.18 | 3.12 | 2.79 | −2.68 | N2.13 | −0.04 | 2.98 | 0.10 | 3.13 | 2.95 | −1.48 |
| 52 | N6.05 | −0.08 | 2.85 | 0.18 | 3.11 | 2.79 | −2.84 | N3.06 | −0.04 | 2.98 | 0.10 | 3.13 | 2.95 | −1.48 |
| 53 | N5.18 | −0.14 | 2.80 | 0.17 | 3.11 | 2.69 | −4.62 | N6.22 | −0.04 | 2.98 | 0.10 | 3.13 | 2.95 | −1.48 |
| 54 | N5.03 | −0.14 | 2.79 | 0.17 | 3.10 | 2.68 | −4.87 | N5.06 | −0.08 | 2.95 | 0.09 | 3.12 | 2.90 | −2.58 |
| 55 | N3.07 | −0.14 | 2.79 | 0.16 | 3.10 | 2.68 | −4.89 | N3.17 | −0.08 | 2.95 | 0.09 | 3.12 | 2.90 | −2.58 |
| 56 | N4.20 | −0.17 | 2.77 | 0.16 | 3.09 | 2.64 | −5.67 | N6.18 | −0.08 | 2.95 | 0.09 | 3.12 | 2.90 | −2.58 |
| 57 | N5.11 | −0.19 | 2.75 | 0.15 | 3.08 | 2.60 | −6.37 | N5.09 | −0.08 | 2.95 | 0.08 | 3.11 | 2.90 | −2.58 |
| 58 | N2.17 | −0.19 | 2.75 | 0.14 | 3.08 | 2.60 | −6.38 | N6.12 | −0.08 | 2.95 | 0.08 | 3.11 | 2.90 | −2.58 |
| 59 | N3.10 | −0.19 | 2.74 | 0.14 | 3.07 | 2.60 | −6.44 | N1.22 | −0.08 | 2.95 | 0.08 | 3.11 | 2.90 | −2.58 |
| 60 | N3.03 | −0.19 | 2.74 | 0.13 | 3.07 | 2.60 | −6.49 | N5.22 | −0.08 | 2.95 | 0.08 | 3.11 | 2.90 | −2.58 |
| 61 | N2.02 | −0.19 | 2.74 | 0.13 | 3.06 | 2.59 | −6.59 | N4.09 | −0.11 | 2.92 | 0.07 | 3.10 | 2.84 | −3.68 |
| 62 | N3.12 | −0.20 | 2.73 | 0.12 | 3.06 | 2.58 | −6.76 | N3.16 | −0.11 | 2.92 | 0.07 | 3.10 | 2.84 | −3.68 |
| 63 | N4.06 | −0.21 | 2.72 | 0.12 | 3.05 | 2.56 | −7.25 | N5.02 | −0.14 | 2.88 | 0.07 | 3.10 | 2.79 | −4.78 |
| 64 | N1.17 | −0.23 | 2.70 | 0.11 | 3.04 | 2.53 | −7.81 | N5.19 | −0.14 | 2.88 | 0.06 | 3.09 | 2.79 | −4.78 |
| 65 | N4.09 | −0.24 | 2.70 | 0.11 | 3.04 | 2.51 | −8.10 | N5.03 | −0.14 | 2.88 | 0.06 | 3.09 | 2.79 | −4.78 |
| 66 | N5.05 | −0.24 | 2.69 | 0.10 | 3.03 | 2.51 | −8.16 | N2.11 | −0.14 | 2.88 | 0.06 | 3.09 | 2.79 | −4.78 |
| 67 | N5.16 | −0.25 | 2.69 | 0.10 | 3.03 | 2.50 | −8.40 | N5.20 | −0.14 | 2.88 | 0.05 | 3.08 | 2.79 | −4.78 |
| 68 | N1.05 | −0.25 | 2.68 | 0.09 | 3.02 | 2.49 | −8.51 | N5.21 | −0.14 | 2.88 | 0.05 | 3.08 | 2.79 | −4.78 |
| 69 | N1.07 | −0.25 | 2.68 | 0.09 | 3.02 | 2.48 | −8.68 | N4.17 | −0.14 | 2.88 | 0.05 | 3.08 | 2.79 | −4.78 |
| 70 | N6.18 | −0.26 | 2.67 | 0.08 | 3.01 | 2.47 | −8.82 | N3.02 | −0.14 | 2.88 | 0.05 | 3.07 | 2.79 | −4.78 |
| 71 | N4.15 | −0.27 | 2.66 | 0.08 | 3.01 | 2.45 | −9.36 | N5.10 | −0.18 | 2.85 | 0.04 | 3.07 | 2.73 | −5.88 |
| 72 | N4.17 | −0.28 | 2.66 | 0.07 | 3.00 | 2.44 | −9.41 | N7.20 | −0.18 | 2.85 | 0.04 | 3.07 | 2.73 | −5.88 |
| 73 | N4.19 | −0.29 | 2.65 | 0.07 | 3.00 | 2.43 | −9.71 | N5.12 | −0.18 | 2.85 | 0.04 | 3.07 | 2.73 | −5.88 |
| 74 | N3.05 | −0.29 | 2.65 | 0.06 | 2.99 | 2.43 | −9.73 | N1.21 | −0.18 | 2.85 | 0.03 | 3.06 | 2.73 | −5.88 |
| 75 | N8.03 | −0.32 | 2.61 | 0.06 | 2.99 | 2.36 | −11.04 | N6.08 | −0.21 | 2.82 | 0.03 | 3.06 | 2.68 | −6.99 |
| 76 | N2.16 | −0.34 | 2.59 | 0.05 | 2.98 | 2.33 | −11.59 | N4.08 | −0.21 | 2.82 | 0.03 | 3.06 | 2.68 | −6.99 |
| 77 | N5.01 | −0.39 | 2.54 | 0.05 | 2.98 | 2.23 | −13.43 | N1.05 | −0.21 | 2.82 | 0.02 | 3.05 | 2.68 | −6.99 |
| 78 | N1.16 | −0.40 | 2.53 | 0.04 | 2.97 | 2.22 | −13.73 | N2.22 | −0.21 | 2.82 | 0.02 | 3.05 | 2.68 | −6.99 |
| 79 | N1.18 | −0.41 | 2.52 | 0.03 | 2.97 | 2.20 | −14.14 | N2.07 | −0.21 | 2.82 | 0.02 | 3.05 | 2.68 | −6.99 |
| 80 | N3.09 | −0.45 | 2.48 | 0.03 | 2.96 | 2.13 | −15.39 | N4.07 | −0.24 | 2.78 | 0.01 | 3.04 | 2.62 | −8.08 |
| 81 | N5.15 | −0.50 | 2.44 | 0.02 | 2.95 | 2.05 | −16.92 | N4.10 | −0.24 | 2.78 | 0.01 | 3.04 | 2.62 | −8.08 |
| 82 | N2.22 | −0.52 | 2.41 | 0.01 | 2.95 | 2.01 | −17.68 | N2.06 | −0.28 | 2.75 | 0.01 | 3.04 | 2.57 | −9.19 |
| 83 | N2.10 | −0.56 | 2.37 | 0.01 | 2.94 | 1.94 | −19.18 | N2.02 | −0.31 | 2.72 | 0.00 | 3.03 | 2.51 | −10.29 |
| 84 | N2.13 | −0.64 | 2.29 | 0.00 | 2.93 | 1.79 | −21.89 | N3.10 | −0.34 | 2.68 | 0.00 | 3.03 | 2.46 | −11.39 |
| GH | PH (cm) | |||||||||||||
| Order | Ecotypes | g | u + g | Gain | µn | u + g + gem | SG | Ecotypes | g | u + g | Gain | µn | u + g + gem | SG |
| 1 | N5.14 | 0.73 | 3.60 | 0.73 | 3.60 | 3.65 | 25.33 | N2.02 | 2.97 | 59.99 | 2.97 | 59.99 | 65.46 | 5.21 |
| 2 | N5.15 | 0.73 | 3.60 | 0.73 | 3.60 | 3.65 | 25.33 | N1.18 | 2.09 | 59.11 | 2.53 | 59.55 | 62.96 | 3.66 |
| 3 | N5.16 | 0.73 | 3.60 | 0.73 | 3.60 | 3.65 | 25.33 | N3.09 | 1.71 | 58.73 | 2.26 | 59.28 | 61.89 | 3.00 |
| 4 | N4.21 | 0.73 | 3.60 | 0.73 | 3.60 | 3.65 | 25.33 | N2.06 | 1.71 | 58.73 | 2.12 | 59.14 | 61.88 | 3.00 |
| 5 | N7.20 | 0.73 | 3.60 | 0.73 | 3.60 | 3.65 | 25.33 | N2.10 | 1.32 | 58.34 | 1.96 | 58.98 | 60.77 | 2.31 |
| 6 | N6.01 | 0.66 | 3.53 | 0.72 | 3.59 | 3.58 | 22.93 | N5.09 | 1.18 | 58.20 | 1.83 | 58.85 | 60.38 | 2.07 |
| 7 | N6.12 | 0.66 | 3.53 | 0.71 | 3.58 | 3.58 | 22.93 | N1.21 | 1.16 | 58.18 | 1.73 | 58.76 | 60.32 | 2.03 |
| 8 | N6.19 | 0.66 | 3.53 | 0.70 | 3.57 | 3.58 | 22.93 | N3.02 | 1.12 | 58.14 | 1.66 | 58.68 | 60.20 | 1.96 |
| 9 | N1.21 | 0.66 | 3.53 | 0.70 | 3.57 | 3.58 | 22.93 | N4.07 | 1.03 | 58.05 | 1.59 | 58.61 | 59.94 | 1.80 |
| 10 | N2.17 | 0.66 | 3.53 | 0.69 | 3.56 | 3.58 | 22.93 | N3.10 | 0.90 | 57.92 | 1.52 | 58.54 | 59.58 | 1.58 |
| 11 | N2.13 | 0.66 | 3.53 | 0.69 | 3.56 | 3.58 | 22.93 | N5.15 | 0.86 | 57.88 | 1.46 | 58.48 | 59.47 | 1.51 |
| 12 | N4.17 | 0.66 | 3.53 | 0.69 | 3.56 | 3.58 | 22.93 | N5.22 | 0.80 | 57.82 | 1.40 | 58.43 | 59.29 | 1.40 |
| 13 | N5.01 | 0.66 | 3.53 | 0.68 | 3.56 | 3.58 | 22.93 | N2.16 | 0.78 | 57.80 | 1.36 | 58.38 | 59.24 | 1.37 |
| 14 | N1.15 | 0.66 | 3.53 | 0.68 | 3.55 | 3.58 | 22.93 | N4.08 | 0.75 | 57.77 | 1.31 | 58.33 | 59.15 | 1.31 |
| 15 | N3.12 | 0.66 | 3.53 | 0.68 | 3.55 | 3.58 | 22.93 | N8.04 | 0.75 | 57.77 | 1.28 | 58.30 | 59.15 | 1.31 |
| 16 | N1.14 | 0.66 | 3.53 | 0.68 | 3.55 | 3.58 | 22.93 | N2.07 | 0.74 | 57.76 | 1.24 | 58.26 | 59.13 | 1.30 |
| 17 | N4.19 | 0.52 | 3.39 | 0.67 | 3.54 | 3.43 | 18.13 | N5.01 | 0.72 | 57.74 | 1.21 | 58.23 | 59.06 | 1.26 |
| 18 | N1.12 | 0.31 | 3.18 | 0.65 | 3.52 | 3.21 | 10.92 | N1.06 | 0.64 | 57.66 | 1.18 | 58.20 | 58.83 | 1.12 |
| 19 | N5.05 | 0.24 | 3.12 | 0.63 | 3.50 | 3.13 | 8.52 | N1.10 | 0.60 | 57.62 | 1.15 | 58.17 | 58.72 | 1.05 |
| 20 | N4.14 | 0.24 | 3.12 | 0.61 | 3.48 | 3.13 | 8.52 | N3.08 | 0.56 | 57.59 | 1.12 | 58.14 | 58.62 | 0.99 |
| 21 | N3.09 | 0.24 | 3.12 | 0.59 | 3.46 | 3.13 | 8.52 | N1.13 | 0.55 | 57.57 | 1.09 | 58.11 | 58.59 | 0.97 |
| 22 | N3.05 | 0.24 | 3.12 | 0.58 | 3.45 | 3.13 | 8.52 | N6.22 | 0.55 | 57.57 | 1.07 | 58.09 | 58.59 | 0.97 |
| 23 | N7.15 | 0.24 | 3.12 | 0.56 | 3.43 | 3.13 | 8.52 | N2.01 | 0.55 | 57.57 | 1.04 | 58.07 | 58.58 | 0.96 |
| 24 | N3.15 | 0.24 | 3.12 | 0.55 | 3.42 | 3.13 | 8.52 | N3.12 | 0.55 | 57.57 | 1.02 | 58.05 | 58.58 | 0.96 |
| 25 | N5.06 | 0.24 | 3.12 | 0.54 | 3.41 | 3.13 | 8.52 | N3.17 | 0.55 | 57.57 | 1.01 | 58.03 | 58.57 | 0.96 |
| 26 | N4.06 | 0.24 | 3.12 | 0.53 | 3.40 | 3.13 | 8.52 | N1.05 | 0.44 | 57.46 | 0.98 | 58.01 | 58.27 | 0.77 |
| 27 | N5.21 | 0.18 | 3.05 | 0.51 | 3.38 | 3.06 | 6.12 | N3.04 | 0.40 | 57.42 | 0.96 | 57.98 | 58.15 | 0.70 |
| 28 | N5.22 | 0.18 | 3.05 | 0.50 | 3.37 | 3.06 | 6.12 | N3.07 | 0.38 | 57.40 | 0.94 | 57.96 | 58.10 | 0.67 |
| 29 | N5.18 | 0.18 | 3.05 | 0.49 | 3.36 | 3.06 | 6.12 | N5.06 | 0.33 | 57.35 | 0.92 | 57.94 | 57.96 | 0.58 |
| 30 | N6.22 | 0.18 | 3.05 | 0.48 | 3.35 | 3.06 | 6.12 | N4.13 | 0.31 | 57.34 | 0.90 | 57.92 | 57.91 | 0.55 |
| 31 | N6.18 | 0.18 | 3.05 | 0.47 | 3.34 | 3.06 | 6.12 | N4.10 | 0.30 | 57.32 | 0.88 | 57.90 | 57.86 | 0.52 |
| 32 | N4.12 | 0.18 | 3.05 | 0.46 | 3.33 | 3.06 | 6.12 | N1.07 | 0.25 | 57.27 | 0.86 | 57.88 | 57.72 | 0.43 |
| 33 | N3.02 | 0.18 | 3.05 | 0.45 | 3.32 | 3.06 | 6.12 | N2.22 | 0.22 | 57.24 | 0.84 | 57.86 | 57.65 | 0.39 |
| 34 | N3.16 | 0.18 | 3.05 | 0.44 | 3.31 | 3.06 | 6.12 | N5.05 | 0.22 | 57.24 | 0.82 | 57.84 | 57.64 | 0.38 |
| 35 | N4.10 | 0.18 | 3.05 | 0.44 | 3.31 | 3.06 | 6.12 | N2.17 | 0.22 | 57.24 | 0.81 | 57.83 | 57.64 | 0.38 |
| 36 | N1.05 | 0.18 | 3.05 | 0.43 | 3.30 | 3.06 | 6.12 | N4.12 | 0.21 | 57.24 | 0.79 | 57.81 | 57.63 | 0.37 |
| 37 | N5.10 | 0.18 | 3.05 | 0.42 | 3.29 | 3.06 | 6.12 | N5.19 | 0.21 | 57.24 | 0.77 | 57.80 | 57.63 | 0.37 |
| 38 | N4.05 | 0.18 | 3.05 | 0.42 | 3.29 | 3.06 | 6.12 | N4.05 | 0.18 | 57.20 | 0.76 | 57.78 | 57.54 | 0.32 |
| 39 | N1.07 | 0.18 | 3.05 | 0.41 | 3.28 | 3.06 | 6.12 | N4.09 | 0.09 | 57.11 | 0.74 | 57.76 | 57.28 | 0.16 |
| 40 | N3.07 | 0.18 | 3.05 | 0.40 | 3.27 | 3.06 | 6.12 | N4.20 | 0.09 | 57.11 | 0.72 | 57.75 | 57.27 | 0.15 |
| 41 | N1.22 | 0.11 | 2.98 | 0.40 | 3.27 | 2.99 | 3.72 | N4.17 | 0.06 | 57.08 | 0.71 | 57.73 | 57.18 | 0.10 |
| 42 | N1.13 | 0.11 | 2.98 | 0.39 | 3.26 | 2.99 | 3.72 | N7.20 | 0.02 | 57.04 | 0.69 | 57.71 | 57.08 | 0.04 |
| 43 | N2.11 | 0.11 | 2.98 | 0.38 | 3.25 | 2.99 | 3.72 | N3.06 | 0.02 | 57.04 | 0.68 | 57.70 | 57.07 | 0.03 |
| 44 | N4.13 | 0.11 | 2.98 | 0.38 | 3.25 | 2.99 | 3.72 | N3.03 | 0.01 | 57.03 | 0.66 | 57.68 | 57.04 | 0.01 |
| 45 | N4.07 | 0.11 | 2.98 | 0.37 | 3.24 | 2.99 | 3.72 | N4.14 | −0.08 | 56.95 | 0.64 | 57.67 | 56.81 | −0.13 |
| 46 | N6.08 | 0.11 | 2.98 | 0.36 | 3.24 | 2.99 | 3.72 | N4.06 | −0.14 | 56.88 | 0.63 | 57.65 | 56.63 | −0.24 |
| 47 | N5.02 | 0.11 | 2.98 | 0.36 | 3.23 | 2.99 | 3.72 | N1.22 | −0.16 | 56.87 | 0.61 | 57.63 | 56.58 | −0.27 |
| 48 | N5.09 | 0.11 | 2.98 | 0.35 | 3.22 | 2.99 | 3.72 | N5.12 | −0.18 | 56.84 | 0.59 | 57.62 | 56.51 | −0.32 |
| 49 | N1.19 | 0.11 | 2.98 | 0.35 | 3.22 | 2.99 | 3.72 | N6.08 | −0.19 | 56.83 | 0.58 | 57.60 | 56.49 | −0.33 |
| 50 | N3.04 | 0.11 | 2.98 | 0.34 | 3.22 | 2.99 | 3.72 | N5.20 | −0.23 | 56.79 | 0.56 | 57.58 | 56.36 | −0.41 |
| 51 | N2.07 | 0.11 | 2.98 | 0.34 | 3.21 | 2.99 | 3.72 | N1.19 | −0.28 | 56.75 | 0.55 | 57.57 | 56.24 | −0.48 |
| 52 | N2.16 | 0.11 | 2.98 | 0.33 | 3.21 | 2.99 | 3.72 | N5.02 | −0.29 | 56.73 | 0.53 | 57.55 | 56.20 | −0.51 |
| 53 | N2.18 | 0.11 | 2.98 | 0.33 | 3.20 | 2.99 | 3.72 | N1.12 | −0.35 | 56.67 | 0.51 | 57.53 | 56.03 | −0.61 |
| 54 | N2.10 | −0.17 | 2.70 | 0.32 | 3.19 | 2.69 | −5.89 | N5.14 | −0.37 | 56.66 | 0.50 | 57.52 | 55.98 | −0.64 |
| 55 | N5.03 | −0.24 | 2.63 | 0.31 | 3.18 | 2.62 | −8.29 | N3.18 | −0.37 | 56.65 | 0.48 | 57.50 | 55.98 | −0.65 |
| 56 | N3.06 | −0.24 | 2.63 | 0.30 | 3.17 | 2.62 | −8.29 | N5.16 | −0.39 | 56.63 | 0.47 | 57.49 | 55.91 | −0.69 |
| 57 | N3.03 | −0.31 | 2.56 | 0.29 | 3.16 | 2.54 | −10.69 | N3.05 | −0.42 | 56.60 | 0.45 | 57.47 | 55.82 | −0.74 |
| 58 | N5.19 | −0.31 | 2.56 | 0.28 | 3.15 | 2.54 | −10.69 | N4.19 | −0.47 | 56.55 | 0.43 | 57.46 | 55.68 | −0.83 |
| 59 | N4.20 | −0.31 | 2.56 | 0.27 | 3.14 | 2.54 | −10.69 | N1.09 | −0.49 | 56.53 | 0.42 | 57.44 | 55.64 | −0.86 |
| 60 | N3.18 | −0.31 | 2.56 | 0.26 | 3.13 | 2.54 | −10.69 | N5.18 | −0.51 | 56.51 | 0.40 | 57.42 | 55.57 | −0.90 |
| 61 | N3.17 | −0.31 | 2.56 | 0.25 | 3.12 | 2.54 | −10.69 | N2.11 | −0.58 | 56.44 | 0.39 | 57.41 | 55.38 | −1.02 |
| 62 | N5.20 | −0.31 | 2.56 | 0.24 | 3.11 | 2.54 | −10.69 | N5.03 | −0.63 | 56.40 | 0.37 | 57.39 | 55.24 | −1.10 |
| 63 | N4.08 | −0.38 | 2.50 | 0.23 | 3.10 | 2.47 | −13.09 | N3.22 | −0.68 | 56.35 | 0.35 | 57.38 | 55.10 | −1.19 |
| 64 | N3.22 | −0.38 | 2.50 | 0.22 | 3.09 | 2.47 | −13.09 | N3.16 | −0.69 | 56.33 | 0.34 | 57.36 | 55.05 | −1.22 |
| 65 | N4.15 | −0.38 | 2.50 | 0.21 | 3.08 | 2.47 | −13.09 | N3.15 | −0.71 | 56.31 | 0.32 | 57.34 | 55.01 | −1.24 |
| 66 | N1.09 | −0.38 | 2.50 | 0.20 | 3.08 | 2.47 | −13.09 | N3.19 | −0.72 | 56.30 | 0.31 | 57.33 | 54.96 | −1.27 |
| 67 | N4.09 | −0.44 | 2.43 | 0.20 | 3.07 | 2.39 | −15.50 | N1.17 | −0.77 | 56.26 | 0.29 | 57.31 | 54.84 | −1.34 |
| 68 | N1.18 | −0.44 | 2.43 | 0.19 | 3.06 | 2.39 | −15.50 | N8.03 | −0.80 | 56.23 | 0.27 | 57.30 | 54.76 | −1.39 |
| 69 | N1.17 | −0.44 | 2.43 | 0.18 | 3.05 | 2.39 | −15.50 | N2.18 | −0.80 | 56.23 | 0.26 | 57.28 | 54.76 | −1.39 |
| 70 | N5.12 | −0.44 | 2.43 | 0.17 | 3.04 | 2.39 | −15.50 | N7.15 | −0.80 | 56.22 | 0.24 | 57.26 | 54.75 | −1.40 |
| 71 | N2.01 | −0.44 | 2.43 | 0.16 | 3.03 | 2.39 | −15.50 | N6.19 | −0.81 | 56.22 | 0.23 | 57.25 | 54.73 | −1.41 |
| 72 | N5.11 | −0.44 | 2.43 | 0.15 | 3.02 | 2.39 | −15.50 | N6.05 | −0.82 | 56.20 | 0.21 | 57.24 | 54.69 | −1.44 |
| 73 | N2.06 | −0.72 | 2.15 | 0.14 | 3.01 | 2.10 | −25.10 | N5.21 | −0.82 | 56.20 | 0.20 | 57.22 | 54.68 | −1.44 |
| 74 | N2.22 | −0.72 | 2.15 | 0.13 | 3.00 | 2.10 | −25.10 | N1.15 | −0.86 | 56.16 | 0.19 | 57.21 | 54.58 | −1.51 |
| 75 | N8.03 | −0.79 | 2.08 | 0.12 | 2.99 | 2.03 | −27.51 | N4.15 | −0.89 | 56.13 | 0.17 | 57.19 | 54.49 | −1.56 |
| 76 | N3.19 | −0.86 | 2.01 | 0.10 | 2.97 | 1.95 | −29.91 | N2.13 | −1.01 | 56.01 | 0.16 | 57.18 | 54.15 | −1.77 |
| 77 | N2.02 | −0.93 | 1.94 | 0.09 | 2.96 | 1.88 | −32.31 | N6.12 | −1.05 | 55.97 | 0.14 | 57.16 | 54.03 | −1.85 |
| 78 | N1.10 | −0.93 | 1.94 | 0.08 | 2.95 | 1.88 | −32.31 | N5.10 | −1.11 | 55.91 | 0.12 | 57.15 | 53.88 | −1.94 |
| 79 | N3.08 | −0.93 | 1.94 | 0.06 | 2.93 | 1.88 | −32.31 | N5.11 | −1.29 | 55.73 | 0.11 | 57.13 | 53.34 | −2.27 |
| 80 | N1.06 | −1.00 | 1.87 | 0.05 | 2.92 | 1.80 | −34.71 | N1.16 | −1.45 | 55.57 | 0.09 | 57.11 | 52.90 | −2.54 |
| 81 | N3.10 | −1.00 | 1.87 | 0.04 | 2.91 | 1.80 | −34.71 | N6.01 | −1.56 | 55.46 | 0.07 | 57.09 | 52.59 | −2.74 |
| 82 | N1.16 | −1.00 | 1.87 | 0.02 | 2.90 | 1.80 | −34.71 | N6.18 | −1.69 | 55.33 | 0.04 | 57.07 | 52.21 | −2.97 |
| 83 | N8.04 | −1.00 | 1.87 | 0.01 | 2.88 | 1.80 | −34.71 | N4.21 | −1.79 | 55.23 | 0.02 | 57.04 | 51.94 | −3.14 |
| 84 | N6.05 | −1.00 | 1.87 | 0.00 | 2.87 | 1.80 | −34.71 | N1.14 | −1.84 | 55.18 | 0.00 | 57.02 | 51.79 | −3.23 |
Appendix B
| NT (Tillers Plant−1) | FM (g Plant−1) | ||||||||||
| Ecotype | MHVG | PRVG | PRVG*µ | MHPRVG | MHPRVG*µ | Ecotype | MHVG | PRVG | PRVG*µ | MHPRVG | MHPRVG*µ |
| N3.10 | 159.48 | 2.40 | 499.68 | 1.53 | 319.02 | N3.10 | 67.13 | 1.79 | 278.14 | 1.42 | 220.60 |
| N3.17 | 124.32 | 1.43 | 297.21 | 1.32 | 273.85 | N2.06 | 77.58 | 1.38 | 214.81 | 1.31 | 203.89 |
| N1.21 | 140.72 | 1.42 | 295.25 | 1.22 | 252.71 | N4.09 | 60.76 | 1.30 | 201.99 | 1.27 | 197.44 |
| N5.01 | 115.90 | 1.34 | 277.76 | 1.21 | 252.52 | N1.21 | 78.37 | 1.44 | 223.82 | 1.27 | 196.44 |
| N5.09 | 113.62 | 1.24 | 257.41 | 1.19 | 248.19 | N1.05 | 72.52 | 1.32 | 205.49 | 1.24 | 192.90 |
| N8.04 | 114.36 | 1.27 | 265.02 | 1.19 | 248.16 | N5.01 | 66.08 | 1.32 | 204.50 | 1.24 | 192.64 |
| N3.04 | 131.70 | 1.19 | 248.36 | 1.17 | 244.24 | N3.09 | 55.73 | 1.25 | 194.02 | 1.17 | 180.88 |
| N4.09 | 111.85 | 1.21 | 251.39 | 1.17 | 242.38 | N1.06 | 65.84 | 1.31 | 203.36 | 1.13 | 175.70 |
| N3.09 | 96.85 | 1.35 | 280.26 | 1.17 | 242.36 | N4.08 | 59.24 | 1.17 | 182.31 | 1.13 | 175.31 |
| N1.05 | 131.93 | 1.21 | 251.27 | 1.14 | 237.30 | N3.16 | 58.03 | 1.13 | 174.69 | 1.11 | 172.48 |
| N3.03 | 115.64 | 1.20 | 249.94 | 1.14 | 237.24 | N2.07 | 61.42 | 1.17 | 181.21 | 1.09 | 168.45 |
| N3.05 | 128.67 | 1.44 | 300.37 | 1.12 | 233.30 | N2.02 | 57.95 | 1.13 | 174.97 | 1.08 | 168.00 |
| N4.20 | 127.99 | 1.26 | 262.22 | 1.09 | 226.82 | N3.05 | 59.99 | 1.22 | 189.83 | 1.08 | 167.43 |
| N3.16 | 108.41 | 1.10 | 229.33 | 1.08 | 223.81 | N1.22 | 52.48 | 1.09 | 169.65 | 1.07 | 166.78 |
| N1.17 | 130.37 | 1.19 | 246.62 | 1.07 | 223.00 | N2.16 | 52.36 | 1.12 | 173.25 | 1.06 | 164.70 |
| N2.16 | 110.36 | 1.11 | 230.49 | 1.07 | 222.01 | N3.04 | 53.08 | 1.17 | 181.30 | 1.05 | 162.56 |
| N4.08 | 106.39 | 1.09 | 227.41 | 1.05 | 218.18 | N3.12 | 50.26 | 1.10 | 170.37 | 1.05 | 162.34 |
| N6.12 | 111.96 | 1.08 | 224.72 | 1.05 | 217.85 | N2.11 | 55.34 | 1.14 | 176.53 | 1.03 | 160.49 |
| N1.06 | 119.02 | 1.15 | 240.03 | 1.04 | 216.78 | N2.10 | 55.25 | 1.05 | 163.42 | 1.03 | 159.94 |
| N5.20 | 105.60 | 1.06 | 221.14 | 1.04 | 215.86 | N3.02 | 62.88 | 1.14 | 176.34 | 1.03 | 159.45 |
| N3.12 | 103.03 | 1.19 | 247.09 | 1.04 | 215.70 | N1.19 | 73.63 | 1.36 | 211.24 | 1.02 | 158.79 |
| N3.02 | 115.60 | 1.10 | 228.71 | 1.03 | 213.55 | N3.19 | 49.33 | 1.04 | 161.96 | 1.02 | 157.72 |
| N1.10 | 104.72 | 1.05 | 217.84 | 1.01 | 210.99 | N8.04 | 51.22 | 1.13 | 176.03 | 1.02 | 157.59 |
| N5.19 | 104.02 | 1.02 | 212.43 | 1.00 | 207.09 | N4.06 | 51.01 | 1.06 | 164.78 | 1.00 | 155.86 |
| N2.06 | 101.65 | 1.08 | 223.87 | 0.99 | 206.22 | N1.17 | 52.87 | 1.07 | 165.40 | 1.00 | 155.80 |
| N2.01 | 122.21 | 1.07 | 222.77 | 0.99 | 205.71 | N3.03 | 45.89 | 1.03 | 159.30 | 1.00 | 155.70 |
| N4.15 | 101.61 | 1.09 | 227.47 | 0.98 | 203.82 | N3.17 | 54.60 | 1.17 | 181.73 | 1.00 | 155.57 |
| N2.10 | 104.18 | 1.00 | 208.91 | 0.96 | 199.12 | N1.18 | 53.69 | 1.05 | 163.56 | 1.00 | 154.73 |
| N2.07 | 95.51 | 1.07 | 222.32 | 0.95 | 198.17 | N5.12 | 47.56 | 1.16 | 179.76 | 0.98 | 152.77 |
| N5.12 | 98.23 | 1.10 | 228.45 | 0.95 | 197.68 | N6.08 | 56.56 | 1.03 | 159.44 | 0.97 | 150.48 |
| N5.03 | 92.12 | 1.05 | 217.97 | 0.95 | 196.57 | N2.01 | 52.69 | 1.01 | 155.96 | 0.97 | 150.11 |
| N2.02 | 100.46 | 0.99 | 205.09 | 0.94 | 196.46 | N5.19 | 57.53 | 1.03 | 159.92 | 0.96 | 149.21 |
| N4.06 | 89.82 | 0.96 | 200.32 | 0.94 | 194.89 | N5.22 | 55.98 | 0.99 | 152.94 | 0.96 | 148.97 |
| N2.11 | 92.13 | 1.05 | 218.80 | 0.93 | 193.04 | N4.21 | 54.56 | 1.01 | 157.36 | 0.96 | 148.49 |
| N5.21 | 103.26 | 0.94 | 195.09 | 0.92 | 190.79 | N5.15 | 48.76 | 1.02 | 157.75 | 0.95 | 148.14 |
| N4.21 | 106.70 | 0.98 | 203.50 | 0.91 | 188.42 | N6.19 | 51.94 | 0.97 | 150.97 | 0.95 | 148.02 |
| N3.15 | 98.48 | 0.96 | 198.62 | 0.90 | 188.09 | N6.22 | 63.59 | 1.09 | 168.80 | 0.94 | 146.36 |
| N5.15 | 96.11 | 0.92 | 192.18 | 0.90 | 186.55 | N6.12 | 44.46 | 0.99 | 153.51 | 0.93 | 145.04 |
| N1.19 | 115.12 | 1.06 | 220.68 | 0.89 | 185.51 | N2.13 | 47.32 | 0.98 | 151.29 | 0.92 | 143.32 |
| N4.13 | 90.30 | 0.96 | 199.82 | 0.89 | 184.31 | N3.15 | 48.86 | 1.01 | 156.49 | 0.92 | 142.44 |
| N5.05 | 91.74 | 0.91 | 189.37 | 0.88 | 183.59 | N5.03 | 45.22 | 1.00 | 155.64 | 0.91 | 141.92 |
| N4.07 | 88.21 | 0.98 | 203.69 | 0.88 | 183.52 | N5.20 | 44.42 | 0.97 | 150.56 | 0.90 | 140.20 |
| N2.13 | 88.52 | 0.92 | 191.09 | 0.86 | 179.55 | N5.21 | 39.96 | 0.91 | 140.98 | 0.89 | 138.22 |
| N1.18 | 89.11 | 0.97 | 201.91 | 0.86 | 178.54 | N3.18 | 47.37 | 1.06 | 164.22 | 0.89 | 137.96 |
| N5.22 | 89.97 | 0.93 | 193.15 | 0.86 | 178.43 | N4.15 | 48.50 | 0.93 | 144.77 | 0.88 | 137.21 |
| N4.10 | 94.85 | 0.89 | 185.05 | 0.86 | 178.17 | N4.13 | 41.20 | 0.95 | 147.19 | 0.88 | 136.77 |
| N1.13 | 102.18 | 0.91 | 189.86 | 0.86 | 177.91 | N6.18 | 42.82 | 0.89 | 138.45 | 0.87 | 134.33 |
| N2.17 | 91.68 | 0.91 | 188.70 | 0.85 | 177.56 | N4.20 | 40.85 | 0.92 | 143.33 | 0.86 | 133.93 |
| N1.22 | 93.89 | 0.92 | 191.24 | 0.85 | 176.19 | N5.18 | 40.09 | 0.99 | 153.39 | 0.84 | 130.52 |
| N1.07 | 94.71 | 0.88 | 182.80 | 0.84 | 175.53 | N5.09 | 36.81 | 0.96 | 149.45 | 0.83 | 128.73 |
| N6.05 | 99.16 | 0.90 | 186.35 | 0.84 | 174.61 | N6.05 | 56.18 | 1.19 | 184.89 | 0.82 | 127.49 |
| N3.07 | 86.48 | 0.89 | 185.20 | 0.84 | 174.59 | N3.08 | 47.21 | 0.93 | 144.98 | 0.82 | 126.98 |
| N5.10 | 89.63 | 0.85 | 177.74 | 0.84 | 174.09 | N5.14 | 35.78 | 0.86 | 134.00 | 0.82 | 126.56 |
| N3.18 | 78.63 | 0.98 | 204.61 | 0.83 | 171.94 | N1.16 | 47.53 | 0.87 | 134.27 | 0.81 | 126.42 |
| N6.01 | 90.21 | 0.90 | 187.81 | 0.82 | 171.52 | N5.06 | 40.23 | 0.83 | 128.32 | 0.81 | 125.86 |
| N5.16 | 91.12 | 0.98 | 203.40 | 0.82 | 169.51 | N5.02 | 42.23 | 0.85 | 131.93 | 0.81 | 125.48 |
| N1.16 | 95.41 | 0.84 | 175.54 | 0.81 | 168.79 | N5.10 | 37.52 | 0.88 | 136.66 | 0.81 | 125.14 |
| N3.19 | 86.72 | 0.84 | 175.15 | 0.81 | 167.81 | N4.07 | 43.00 | 0.97 | 150.02 | 0.80 | 124.48 |
| N5.14 | 89.81 | 0.83 | 172.09 | 0.81 | 167.48 | N7.20 | 40.43 | 0.84 | 129.66 | 0.79 | 122.86 |
| N5.11 | 92.09 | 0.84 | 174.47 | 0.79 | 164.23 | N1.10 | 38.05 | 0.88 | 135.91 | 0.78 | 121.48 |
| N5.02 | 84.15 | 0.81 | 168.68 | 0.78 | 162.38 | N4.05 | 39.99 | 0.90 | 139.48 | 0.78 | 121.41 |
| N5.18 | 78.01 | 0.97 | 201.03 | 0.77 | 160.16 | N4.10 | 39.40 | 0.83 | 129.28 | 0.78 | 120.99 |
| N6.19 | 83.10 | 0.86 | 179.35 | 0.76 | 157.44 | N5.16 | 42.27 | 0.94 | 146.32 | 0.78 | 120.86 |
| N4.12 | 80.81 | 0.81 | 168.46 | 0.75 | 155.05 | N5.05 | 39.54 | 0.85 | 132.43 | 0.77 | 120.01 |
| N5.06 | 80.74 | 0.77 | 160.74 | 0.74 | 154.58 | N4.19 | 42.79 | 0.80 | 123.63 | 0.77 | 119.58 |
| N3.22 | 77.17 | 0.82 | 171.55 | 0.74 | 154.15 | N8.03 | 31.25 | 0.83 | 128.37 | 0.76 | 118.63 |
| N8.03 | 80.49 | 0.79 | 164.06 | 0.74 | 154.10 | N1.14 | 53.62 | 0.90 | 139.01 | 0.76 | 117.32 |
| N1.12 | 98.59 | 0.90 | 187.83 | 0.74 | 153.30 | N6.01 | 45.77 | 0.88 | 137.10 | 0.76 | 117.27 |
| N1.15 | 86.77 | 0.80 | 165.48 | 0.73 | 151.43 | N2.17 | 40.61 | 0.83 | 128.26 | 0.74 | 114.90 |
| N7.20 | 75.00 | 0.83 | 172.45 | 0.72 | 150.34 | N3.07 | 36.46 | 0.85 | 132.57 | 0.73 | 113.83 |
| N1.14 | 100.90 | 0.92 | 191.92 | 0.70 | 145.34 | N1.07 | 47.25 | 0.84 | 130.02 | 0.72 | 112.00 |
| N1.09 | 80.93 | 1.06 | 220.59 | 0.68 | 142.09 | N4.14 | 38.77 | 0.82 | 127.78 | 0.72 | 111.66 |
| N4.14 | 79.63 | 0.87 | 180.42 | 0.68 | 140.62 | N3.22 | 39.17 | 0.84 | 130.08 | 0.71 | 109.96 |
| N4.05 | 69.16 | 0.90 | 187.87 | 0.67 | 139.14 | N1.13 | 44.02 | 0.78 | 121.80 | 0.70 | 108.73 |
| N6.18 | 77.00 | 0.72 | 148.81 | 0.66 | 137.83 | N5.11 | 39.10 | 0.75 | 116.80 | 0.70 | 108.37 |
| N6.08 | 68.61 | 0.81 | 168.41 | 0.65 | 134.62 | N2.18 | 34.23 | 0.79 | 123.09 | 0.67 | 104.62 |
| N4.19 | 68.48 | 0.75 | 155.01 | 0.63 | 130.52 | N1.12 | 33.54 | 0.88 | 135.98 | 0.66 | 103.04 |
| N6.22 | 65.19 | 0.87 | 179.93 | 0.60 | 123.78 | N2.22 | 37.06 | 0.83 | 129.18 | 0.63 | 98.08 |
| N3.08 | 66.15 | 0.72 | 150.09 | 0.59 | 122.55 | N7.15 | 38.86 | 0.72 | 111.97 | 0.62 | 96.52 |
| N2.22 | 56.93 | 0.91 | 189.43 | 0.57 | 118.69 | N1.15 | 39.47 | 0.70 | 107.97 | 0.60 | 93.73 |
| N2.18 | 55.99 | 0.71 | 147.71 | 0.55 | 114.86 | N4.12 | 25.46 | 0.63 | 97.73 | 0.54 | 83.18 |
| N3.06 | 55.74 | 0.71 | 148.38 | 0.53 | 111.22 | N1.09 | 38.26 | 0.91 | 140.97 | 0.52 | 80.50 |
| N7.15 | 70.49 | 0.64 | 134.00 | 0.49 | 101.25 | N4.17 | 32.88 | 0.59 | 90.95 | 0.48 | 74.45 |
| N4.17 | 58.07 | 0.57 | 119.34 | 0.46 | 95.50 | N3.06 | 24.74 | 0.69 | 107.30 | 0.47 | 72.35 |
| LDM (g plant−1) | SDM (g plant−1) | ||||||||||
| Ecotype | MHVG | PRVG | PRVG*µ | MHPRVG | MHPRVG*µ | Ecotype | MHVG | PRVG | PRVG*µ | MHPRVG | MHPRVG*µ |
| N1.05 | 34.42 | 1.36 | 84.56 | 1.28 | 79.46 | N2.06 | 4.86 | 1.49 | 9.73 | 1.40 | 9.17 |
| N2.06 | 31.72 | 1.33 | 82.91 | 1.27 | 79.03 | N3.10 | 3.83 | 1.84 | 12.02 | 1.19 | 7.76 |
| N3.10 | 20.88 | 1.75 | 108.95 | 1.26 | 78.36 | N5.01 | 3.50 | 1.21 | 7.93 | 1.13 | 7.38 |
| N5.01 | 26.80 | 1.20 | 74.89 | 1.16 | 72.55 | N3.17 | 3.71 | 1.50 | 9.79 | 1.12 | 7.31 |
| N4.09 | 22.86 | 1.15 | 71.66 | 1.12 | 69.93 | N3.05 | 2.82 | 1.21 | 7.93 | 1.11 | 7.25 |
| N5.15 | 27.61 | 1.14 | 70.99 | 1.12 | 69.61 | N3.09 | 3.14 | 1.23 | 8.06 | 1.10 | 7.20 |
| N2.11 | 27.73 | 1.20 | 74.74 | 1.11 | 69.13 | N1.21 | 3.11 | 1.21 | 7.91 | 1.08 | 7.09 |
| N1.21 | 30.59 | 1.20 | 74.73 | 1.10 | 68.47 | N3.04 | 2.51 | 1.21 | 7.93 | 1.08 | 7.07 |
| N3.12 | 25.22 | 1.15 | 71.81 | 1.09 | 68.23 | N6.22 | 3.71 | 1.20 | 7.84 | 1.08 | 7.05 |
| N4.08 | 24.85 | 1.09 | 68.18 | 1.07 | 66.51 | N2.01 | 3.44 | 1.13 | 7.37 | 1.07 | 7.04 |
| N3.09 | 22.55 | 1.16 | 72.27 | 1.07 | 66.41 | N4.09 | 3.20 | 1.18 | 7.72 | 1.06 | 6.94 |
| N3.03 | 24.72 | 1.13 | 70.19 | 1.06 | 66.19 | N5.21 | 2.54 | 1.12 | 7.30 | 1.04 | 6.78 |
| N3.04 | 22.78 | 1.09 | 68.17 | 1.06 | 66.11 | N5.19 | 3.96 | 1.14 | 7.46 | 1.02 | 6.65 |
| N2.07 | 23.74 | 1.09 | 67.92 | 1.06 | 65.99 | N5.15 | 2.63 | 1.10 | 7.21 | 1.01 | 6.63 |
| N2.02 | 22.73 | 1.11 | 69.15 | 1.04 | 65.04 | N1.22 | 2.85 | 1.05 | 6.85 | 1.01 | 6.62 |
| N1.06 | 23.74 | 1.21 | 75.31 | 1.04 | 64.71 | N1.18 | 3.33 | 1.08 | 7.05 | 1.01 | 6.60 |
| N2.16 | 21.83 | 1.06 | 66.16 | 1.03 | 64.33 | N3.19 | 2.87 | 1.07 | 6.97 | 1.01 | 6.58 |
| N1.22 | 23.34 | 1.03 | 64.39 | 1.02 | 63.87 | N8.04 | 2.42 | 1.14 | 7.44 | 1.00 | 6.53 |
| N3.17 | 23.86 | 1.19 | 73.97 | 1.01 | 63.23 | N1.17 | 3.50 | 1.06 | 6.93 | 0.99 | 6.49 |
| N1.18 | 23.03 | 1.06 | 65.77 | 1.01 | 63.19 | N5.14 | 2.60 | 1.04 | 6.83 | 0.99 | 6.48 |
| N6.22 | 28.01 | 1.11 | 68.91 | 1.01 | 62.89 | N4.20 | 2.78 | 1.02 | 6.67 | 0.98 | 6.40 |
| N3.02 | 27.90 | 1.08 | 67.41 | 1.00 | 62.32 | N1.06 | 3.54 | 1.11 | 7.29 | 0.96 | 6.28 |
| N5.14 | 22.94 | 1.08 | 67.41 | 0.99 | 61.75 | N2.16 | 2.40 | 1.00 | 6.58 | 0.95 | 6.24 |
| N8.04 | 20.15 | 1.18 | 73.49 | 0.98 | 61.29 | N4.13 | 2.86 | 1.11 | 7.27 | 0.95 | 6.22 |
| N3.19 | 21.58 | 1.01 | 63.12 | 0.98 | 61.24 | N3.02 | 3.55 | 1.12 | 7.36 | 0.95 | 6.20 |
| N3.08 | 26.90 | 1.11 | 69.11 | 0.98 | 61.17 | N2.07 | 3.38 | 1.01 | 6.62 | 0.94 | 6.18 |
| N4.15 | 26.96 | 1.03 | 64.27 | 0.98 | 61.11 | N2.11 | 2.87 | 1.01 | 6.62 | 0.94 | 6.15 |
| N8.03 | 21.31 | 1.00 | 62.11 | 0.97 | 60.48 | N1.05 | 3.84 | 1.20 | 7.83 | 0.94 | 6.14 |
| N5.21 | 18.48 | 1.01 | 63.14 | 0.97 | 60.35 | N3.16 | 2.32 | 1.02 | 6.68 | 0.94 | 6.12 |
| N4.06 | 22.86 | 1.02 | 63.65 | 0.96 | 59.67 | N4.15 | 3.02 | 0.99 | 6.47 | 0.92 | 6.05 |
| N3.16 | 24.14 | 0.97 | 60.62 | 0.96 | 59.52 | N3.18 | 3.03 | 1.06 | 6.94 | 0.92 | 6.04 |
| N2.10 | 22.99 | 0.97 | 60.43 | 0.95 | 59.49 | N4.08 | 2.95 | 0.98 | 6.43 | 0.91 | 5.96 |
| N3.15 | 22.26 | 1.01 | 63.20 | 0.95 | 59.40 | N5.12 | 3.21 | 1.10 | 7.21 | 0.91 | 5.94 |
| N6.08 | 24.84 | 0.99 | 61.76 | 0.95 | 59.02 | N3.08 | 2.73 | 1.00 | 6.54 | 0.90 | 5.92 |
| N3.05 | 19.79 | 1.05 | 65.68 | 0.94 | 58.71 | N4.06 | 2.88 | 1.14 | 7.46 | 0.90 | 5.92 |
| N2.01 | 23.96 | 0.97 | 60.74 | 0.94 | 58.45 | N1.19 | 3.97 | 1.35 | 8.84 | 0.89 | 5.82 |
| N5.22 | 24.85 | 0.95 | 59.02 | 0.93 | 58.19 | N3.12 | 2.55 | 1.06 | 6.92 | 0.88 | 5.77 |
| N4.21 | 22.07 | 0.99 | 61.86 | 0.93 | 58.01 | N3.03 | 2.41 | 0.98 | 6.44 | 0.87 | 5.70 |
| N6.12 | 24.42 | 0.97 | 60.48 | 0.92 | 57.46 | N3.15 | 2.71 | 1.03 | 6.76 | 0.87 | 5.69 |
| N4.13 | 20.41 | 0.98 | 60.83 | 0.92 | 57.18 | N5.03 | 2.23 | 1.00 | 6.56 | 0.87 | 5.68 |
| N1.17 | 21.45 | 0.96 | 59.66 | 0.92 | 57.10 | N5.10 | 2.26 | 0.96 | 6.26 | 0.86 | 5.65 |
| N3.18 | 18.52 | 1.12 | 69.78 | 0.91 | 56.58 | N2.02 | 2.10 | 0.99 | 6.48 | 0.86 | 5.65 |
| N2.13 | 22.04 | 0.95 | 59.06 | 0.91 | 56.57 | N4.21 | 3.27 | 0.96 | 6.26 | 0.86 | 5.61 |
| N5.12 | 20.18 | 0.98 | 61.20 | 0.91 | 56.44 | N4.07 | 2.36 | 0.94 | 6.15 | 0.85 | 5.56 |
| N4.19 | 25.48 | 0.95 | 58.93 | 0.89 | 55.71 | N8.03 | 2.49 | 0.91 | 5.93 | 0.83 | 5.44 |
| N5.05 | 22.10 | 0.97 | 60.32 | 0.88 | 55.12 | N2.17 | 2.57 | 1.00 | 6.53 | 0.83 | 5.41 |
| N4.05 | 19.47 | 0.93 | 58.26 | 0.88 | 54.96 | N6.08 | 2.79 | 0.89 | 5.83 | 0.82 | 5.38 |
| N5.19 | 24.74 | 0.91 | 56.83 | 0.88 | 54.72 | N6.19 | 2.94 | 0.94 | 6.18 | 0.82 | 5.35 |
| N5.02 | 19.76 | 0.92 | 57.09 | 0.88 | 54.61 | N5.22 | 2.98 | 0.89 | 5.80 | 0.82 | 5.35 |
| N1.07 | 27.69 | 0.98 | 61.00 | 0.87 | 54.38 | N5.06 | 2.32 | 0.93 | 6.09 | 0.82 | 5.34 |
| N1.19 | 26.80 | 1.12 | 69.99 | 0.87 | 54.29 | N4.10 | 2.60 | 0.87 | 5.73 | 0.81 | 5.30 |
| N1.14 | 21.85 | 0.99 | 61.69 | 0.87 | 54.18 | N1.14 | 2.16 | 0.88 | 5.74 | 0.81 | 5.30 |
| N4.20 | 18.49 | 0.94 | 58.80 | 0.87 | 54.13 | N2.10 | 2.66 | 0.96 | 6.30 | 0.81 | 5.30 |
| N5.03 | 19.42 | 0.92 | 57.14 | 0.87 | 54.12 | N5.18 | 1.97 | 0.98 | 6.41 | 0.80 | 5.27 |
| N1.16 | 23.24 | 0.92 | 57.41 | 0.87 | 54.11 | N1.10 | 1.81 | 0.87 | 5.69 | 0.80 | 5.26 |
| N6.05 | 25.96 | 1.08 | 67.51 | 0.87 | 54.10 | N2.13 | 2.02 | 0.91 | 5.96 | 0.80 | 5.23 |
| N5.20 | 16.63 | 0.95 | 59.23 | 0.86 | 53.85 | N4.19 | 3.11 | 0.87 | 5.71 | 0.80 | 5.22 |
| N2.17 | 18.95 | 0.91 | 56.47 | 0.86 | 53.61 | N4.14 | 2.45 | 0.91 | 5.98 | 0.79 | 5.18 |
| N5.18 | 18.30 | 0.92 | 57.63 | 0.86 | 53.48 | N7.20 | 2.49 | 0.87 | 5.70 | 0.79 | 5.17 |
| N4.07 | 18.15 | 0.98 | 60.91 | 0.85 | 53.05 | N5.09 | 2.31 | 0.93 | 6.10 | 0.78 | 5.12 |
| N7.20 | 18.87 | 0.93 | 57.73 | 0.84 | 52.60 | N1.09 | 2.12 | 0.96 | 6.25 | 0.78 | 5.08 |
| N6.18 | 18.77 | 0.90 | 56.27 | 0.84 | 52.41 | N3.22 | 2.19 | 0.92 | 5.99 | 0.77 | 5.07 |
| N5.16 | 21.73 | 0.93 | 58.03 | 0.83 | 51.68 | N6.05 | 3.22 | 1.15 | 7.54 | 0.77 | 5.03 |
| N6.19 | 23.95 | 0.88 | 54.86 | 0.83 | 51.54 | N6.12 | 2.93 | 0.99 | 6.48 | 0.76 | 5.00 |
| N4.10 | 18.59 | 0.90 | 56.26 | 0.82 | 51.38 | N1.16 | 2.00 | 0.87 | 5.71 | 0.76 | 5.00 |
| N5.11 | 25.21 | 0.89 | 55.65 | 0.82 | 50.84 | N5.02 | 2.06 | 0.84 | 5.49 | 0.75 | 4.93 |
| N5.06 | 19.41 | 0.82 | 50.90 | 0.80 | 49.91 | N5.05 | 2.30 | 0.77 | 5.07 | 0.74 | 4.86 |
| N1.12 | 21.62 | 0.96 | 59.63 | 0.80 | 49.72 | N6.18 | 2.14 | 0.81 | 5.29 | 0.74 | 4.83 |
| N1.10 | 19.07 | 0.84 | 52.39 | 0.80 | 49.64 | N6.01 | 2.91 | 0.88 | 5.75 | 0.73 | 4.81 |
| N3.07 | 17.96 | 0.87 | 54.49 | 0.79 | 49.50 | N5.20 | 1.72 | 0.82 | 5.40 | 0.73 | 4.79 |
| N5.10 | 17.24 | 0.90 | 56.37 | 0.79 | 49.45 | N4.05 | 1.65 | 0.81 | 5.32 | 0.72 | 4.68 |
| N1.09 | 21.46 | 0.95 | 59.47 | 0.78 | 48.89 | N1.13 | 2.98 | 0.80 | 5.26 | 0.71 | 4.66 |
| N5.09 | 17.37 | 0.87 | 54.49 | 0.76 | 47.64 | N1.07 | 1.84 | 0.86 | 5.63 | 0.71 | 4.66 |
| N6.01 | 22.36 | 0.84 | 52.26 | 0.76 | 47.56 | N3.07 | 2.19 | 0.82 | 5.36 | 0.69 | 4.50 |
| N4.14 | 18.89 | 0.83 | 51.95 | 0.76 | 47.45 | N7.15 | 2.30 | 0.76 | 4.96 | 0.67 | 4.40 |
| N3.22 | 20.28 | 0.83 | 51.57 | 0.76 | 47.27 | N4.12 | 1.64 | 0.76 | 4.99 | 0.67 | 4.39 |
| N1.13 | 23.67 | 0.80 | 49.74 | 0.72 | 45.07 | N1.12 | 1.57 | 0.84 | 5.51 | 0.67 | 4.37 |
| N1.15 | 23.65 | 0.81 | 50.72 | 0.71 | 44.46 | N2.22 | 2.48 | 1.02 | 6.65 | 0.66 | 4.33 |
| N3.06 | 17.29 | 0.82 | 51.37 | 0.71 | 44.45 | N5.16 | 2.23 | 0.76 | 4.98 | 0.66 | 4.33 |
| N4.12 | 17.50 | 0.73 | 45.73 | 0.69 | 43.27 | N5.11 | 2.47 | 0.72 | 4.73 | 0.66 | 4.30 |
| N7.15 | 20.50 | 0.74 | 46.31 | 0.68 | 42.61 | N2.18 | 1.32 | 0.79 | 5.19 | 0.66 | 4.29 |
| N2.22 | 18.22 | 0.89 | 55.52 | 0.68 | 42.15 | N3.06 | 2.11 | 0.78 | 5.09 | 0.60 | 3.94 |
| N2.18 | 18.47 | 0.72 | 45.12 | 0.67 | 42.05 | N1.15 | 1.96 | 0.72 | 4.69 | 0.58 | 3.80 |
| N4.17 | 20.24 | 0.72 | 44.80 | 0.61 | 38.31 | N4.17 | 1.24 | 0.61 | 3.96 | 0.47 | 3.08 |
| IDM (g plant−1) | TDM (g plant−1) | ||||||||||
| Ecotype | MHVG | PRVG | PRVG*µ | MHPRVG | MHPRVG*µ | Ecotype | MHVG | PRVG | PRVG*µ | MHPRVG | MHPRVG*µ |
| N3.10 | 2.47 | 1.84 | 7.12 | 1.36 | 5.24 | N3.10 | 41.40 | 1.93 | 146.27 | 1.43 | 108.77 |
| N1.05 | 2.15 | 1.34 | 5.19 | 1.23 | 4.76 | N2.06 | 43.26 | 1.36 | 103.47 | 1.29 | 97.80 |
| N1.21 | 2.81 | 1.48 | 5.70 | 1.21 | 4.68 | N1.05 | 42.32 | 1.32 | 100.19 | 1.26 | 95.67 |
| N5.15 | 2.00 | 1.21 | 4.70 | 1.18 | 4.54 | N1.21 | 42.23 | 1.34 | 101.43 | 1.24 | 93.82 |
| N3.04 | 2.47 | 1.38 | 5.32 | 1.18 | 4.54 | N4.09 | 33.09 | 1.23 | 93.24 | 1.21 | 91.48 |
| N4.09 | 1.81 | 1.18 | 4.54 | 1.12 | 4.32 | N5.01 | 35.03 | 1.21 | 91.51 | 1.16 | 88.39 |
| N2.16 | 1.92 | 1.25 | 4.84 | 1.11 | 4.30 | N3.09 | 31.34 | 1.23 | 93.14 | 1.12 | 85.32 |
| N2.02 | 2.35 | 1.29 | 4.97 | 1.10 | 4.26 | N3.04 | 31.14 | 1.16 | 87.67 | 1.12 | 84.73 |
| N2.11 | 1.88 | 1.23 | 4.75 | 1.10 | 4.24 | N2.07 | 32.33 | 1.14 | 86.29 | 1.10 | 83.44 |
| N2.07 | 2.18 | 1.32 | 5.11 | 1.08 | 4.18 | N2.11 | 33.03 | 1.16 | 87.71 | 1.09 | 82.91 |
| N2.06 | 1.67 | 1.15 | 4.46 | 1.08 | 4.16 | N1.06 | 35.59 | 1.25 | 94.78 | 1.09 | 82.68 |
| N1.18 | 1.76 | 1.16 | 4.50 | 1.07 | 4.14 | N2.02 | 32.43 | 1.13 | 86.09 | 1.09 | 82.49 |
| N3.09 | 1.84 | 1.25 | 4.82 | 1.06 | 4.10 | N5.15 | 31.44 | 1.10 | 83.76 | 1.08 | 82.25 |
| N3.03 | 2.04 | 1.19 | 4.59 | 1.06 | 4.09 | N4.08 | 31.20 | 1.08 | 82.20 | 1.06 | 80.11 |
| N3.17 | 2.16 | 1.27 | 4.92 | 1.06 | 4.09 | N3.12 | 29.35 | 1.10 | 83.28 | 1.06 | 80.07 |
| N4.08 | 1.85 | 1.13 | 4.36 | 1.02 | 3.96 | N4.06 | 30.42 | 1.11 | 84.57 | 1.05 | 79.94 |
| N5.01 | 2.04 | 1.23 | 4.74 | 1.01 | 3.89 | N1.18 | 31.31 | 1.09 | 82.91 | 1.05 | 79.65 |
| N6.08 | 1.83 | 1.14 | 4.41 | 1.00 | 3.86 | N6.22 | 37.58 | 1.16 | 88.01 | 1.05 | 79.46 |
| N6.22 | 1.57 | 1.09 | 4.21 | 0.98 | 3.78 | N3.03 | 28.91 | 1.08 | 81.72 | 1.04 | 79.08 |
| N3.02 | 1.70 | 1.08 | 4.16 | 0.97 | 3.76 | N8.04 | 29.72 | 1.17 | 88.80 | 1.03 | 78.44 |
| N5.19 | 1.65 | 0.99 | 3.84 | 0.95 | 3.69 | N3.05 | 26.10 | 1.15 | 87.01 | 1.03 | 78.14 |
| N3.18 | 1.49 | 1.23 | 4.74 | 0.95 | 3.69 | N3.17 | 32.56 | 1.21 | 92.15 | 1.03 | 78.09 |
| N8.03 | 1.61 | 1.04 | 4.00 | 0.95 | 3.66 | N2.16 | 26.14 | 1.06 | 80.11 | 1.03 | 77.87 |
| N8.04 | 1.80 | 1.15 | 4.45 | 0.95 | 3.66 | N3.16 | 30.47 | 1.04 | 78.72 | 1.03 | 77.87 |
| N1.22 | 1.41 | 1.03 | 3.97 | 0.94 | 3.62 | N1.22 | 28.25 | 1.02 | 77.41 | 1.01 | 76.64 |
| N4.07 | 1.60 | 1.13 | 4.38 | 0.92 | 3.57 | N3.19 | 27.86 | 1.04 | 79.16 | 1.01 | 76.58 |
| N1.06 | 1.83 | 1.18 | 4.58 | 0.92 | 3.56 | N5.21 | 25.13 | 1.01 | 76.79 | 1.00 | 76.13 |
| N3.16 | 1.44 | 0.99 | 3.82 | 0.92 | 3.56 | N2.01 | 30.02 | 1.03 | 78.32 | 1.00 | 75.64 |
| N1.07 | 2.07 | 1.04 | 4.00 | 0.92 | 3.56 | N5.12 | 27.14 | 1.08 | 81.97 | 0.98 | 74.28 |
| N5.21 | 1.39 | 0.97 | 3.76 | 0.91 | 3.52 | N3.02 | 33.84 | 1.07 | 81.07 | 0.98 | 74.18 |
| N5.20 | 1.47 | 1.05 | 4.04 | 0.91 | 3.51 | N2.10 | 29.01 | 0.99 | 75.36 | 0.98 | 74.14 |
| N2.10 | 1.37 | 0.97 | 3.75 | 0.91 | 3.51 | N2.13 | 27.32 | 1.01 | 76.78 | 0.97 | 73.65 |
| N1.17 | 1.83 | 1.01 | 3.89 | 0.91 | 3.50 | N3.18 | 27.77 | 1.14 | 86.24 | 0.97 | 73.45 |
| N3.08 | 1.80 | 1.06 | 4.08 | 0.90 | 3.49 | N4.21 | 29.68 | 0.99 | 75.38 | 0.96 | 72.86 |
| N2.13 | 1.30 | 0.99 | 3.83 | 0.89 | 3.45 | N5.22 | 30.67 | 0.96 | 72.77 | 0.94 | 71.41 |
| N3.12 | 1.40 | 0.97 | 3.75 | 0.89 | 3.43 | N5.14 | 25.06 | 1.00 | 76.14 | 0.93 | 70.71 |
| N5.02 | 1.28 | 0.98 | 3.77 | 0.88 | 3.40 | N4.15 | 29.73 | 0.98 | 74.18 | 0.93 | 70.65 |
| N4.06 | 1.17 | 1.04 | 4.02 | 0.87 | 3.36 | N3.08 | 30.18 | 1.04 | 79.27 | 0.93 | 70.64 |
| N6.05 | 2.02 | 1.21 | 4.67 | 0.86 | 3.32 | N3.15 | 27.04 | 1.00 | 75.81 | 0.93 | 70.60 |
| N4.05 | 1.61 | 1.03 | 3.97 | 0.86 | 3.31 | N1.17 | 26.47 | 0.98 | 74.03 | 0.92 | 70.13 |
| N3.05 | 1.18 | 1.11 | 4.28 | 0.85 | 3.29 | N6.08 | 29.00 | 0.96 | 72.87 | 0.92 | 69.64 |
| N5.18 | 1.34 | 1.02 | 3.95 | 0.85 | 3.29 | N8.03 | 22.88 | 0.94 | 70.97 | 0.91 | 69.22 |
| N4.13 | 1.31 | 0.98 | 3.78 | 0.85 | 3.28 | N5.19 | 30.12 | 0.95 | 72.24 | 0.91 | 68.95 |
| N3.19 | 1.27 | 0.93 | 3.61 | 0.85 | 3.27 | N5.03 | 23.54 | 0.97 | 73.46 | 0.91 | 68.92 |
| N5.03 | 1.33 | 0.91 | 3.52 | 0.84 | 3.26 | N5.18 | 22.12 | 0.99 | 75.32 | 0.90 | 68.02 |
| N1.10 | 1.36 | 0.99 | 3.84 | 0.84 | 3.24 | N4.13 | 22.83 | 0.95 | 72.24 | 0.88 | 67.11 |
| N5.22 | 1.51 | 0.89 | 3.44 | 0.83 | 3.21 | N6.12 | 26.98 | 0.93 | 70.83 | 0.88 | 66.67 |
| N3.15 | 1.67 | 1.01 | 3.92 | 0.83 | 3.19 | N4.20 | 23.57 | 0.93 | 70.23 | 0.88 | 66.42 |
| N1.19 | 2.01 | 1.22 | 4.73 | 0.82 | 3.18 | N5.02 | 24.03 | 0.91 | 69.02 | 0.87 | 66.27 |
| N5.12 | 1.12 | 1.04 | 4.03 | 0.82 | 3.17 | N6.19 | 28.66 | 0.92 | 69.67 | 0.87 | 65.84 |
| N2.17 | 1.35 | 0.85 | 3.28 | 0.81 | 3.14 | N5.20 | 20.81 | 0.94 | 71.39 | 0.86 | 65.25 |
| N5.09 | 1.30 | 0.99 | 3.81 | 0.81 | 3.14 | N4.07 | 24.28 | 0.99 | 75.09 | 0.86 | 65.20 |
| N6.12 | 1.38 | 0.91 | 3.51 | 0.81 | 3.13 | N1.14 | 27.56 | 0.97 | 73.76 | 0.86 | 65.14 |
| N6.18 | 1.24 | 0.84 | 3.25 | 0.79 | 3.05 | N1.19 | 34.74 | 1.16 | 87.83 | 0.86 | 64.92 |
| N1.14 | 1.74 | 0.99 | 3.82 | 0.79 | 3.04 | N2.17 | 23.29 | 0.91 | 68.81 | 0.85 | 64.71 |
| N4.15 | 1.54 | 0.90 | 3.50 | 0.78 | 3.03 | N6.18 | 23.55 | 0.88 | 66.69 | 0.85 | 64.50 |
| N4.20 | 1.05 | 0.85 | 3.27 | 0.78 | 3.01 | N1.10 | 22.77 | 0.91 | 68.80 | 0.85 | 64.45 |
| N2.01 | 0.84 | 0.86 | 3.32 | 0.78 | 3.00 | N4.05 | 21.37 | 0.91 | 69.17 | 0.84 | 63.69 |
| N5.06 | 1.48 | 0.91 | 3.54 | 0.78 | 3.00 | N4.19 | 26.58 | 0.87 | 65.98 | 0.84 | 63.67 |
| N5.14 | 1.04 | 0.91 | 3.53 | 0.76 | 2.94 | N7.20 | 23.07 | 0.89 | 67.49 | 0.84 | 63.51 |
| N3.22 | 1.32 | 0.85 | 3.30 | 0.76 | 2.93 | N5.05 | 23.07 | 0.89 | 67.73 | 0.83 | 63.12 |
| N1.16 | 1.06 | 0.79 | 3.06 | 0.74 | 2.85 | N4.10 | 22.34 | 0.86 | 65.45 | 0.82 | 62.01 |
| N1.12 | 1.47 | 0.87 | 3.37 | 0.74 | 2.84 | N6.05 | 29.78 | 1.05 | 80.00 | 0.82 | 61.91 |
| N7.20 | 1.25 | 0.80 | 3.10 | 0.73 | 2.81 | N1.16 | 25.21 | 0.87 | 65.81 | 0.81 | 61.49 |
| N1.13 | 1.58 | 0.89 | 3.45 | 0.73 | 2.80 | N1.07 | 28.76 | 0.90 | 67.99 | 0.81 | 61.44 |
| N3.07 | 1.23 | 0.85 | 3.28 | 0.72 | 2.76 | N5.06 | 22.00 | 0.81 | 61.37 | 0.79 | 60.27 |
| N6.19 | 1.09 | 0.79 | 3.06 | 0.71 | 2.75 | N5.09 | 19.99 | 0.91 | 69.26 | 0.79 | 59.98 |
| N5.11 | 1.32 | 0.78 | 3.00 | 0.71 | 2.72 | N6.01 | 26.05 | 0.87 | 66.19 | 0.79 | 59.82 |
| N5.16 | 0.94 | 0.86 | 3.33 | 0.68 | 2.62 | N3.07 | 21.11 | 0.84 | 63.91 | 0.78 | 59.07 |
| N4.19 | 1.06 | 0.77 | 2.97 | 0.67 | 2.58 | N5.16 | 24.23 | 0.89 | 67.29 | 0.78 | 58.98 |
| N4.14 | 0.78 | 0.88 | 3.40 | 0.66 | 2.55 | N3.22 | 22.58 | 0.86 | 65.08 | 0.77 | 58.60 |
| N2.18 | 0.77 | 0.77 | 2.96 | 0.65 | 2.53 | N5.10 | 20.55 | 0.87 | 66.08 | 0.77 | 58.45 |
| N1.09 | 0.76 | 0.82 | 3.19 | 0.65 | 2.50 | N5.11 | 25.52 | 0.80 | 60.50 | 0.75 | 56.73 |
| N5.10 | 0.86 | 0.77 | 2.97 | 0.64 | 2.48 | N4.14 | 21.33 | 0.82 | 62.40 | 0.74 | 55.95 |
| N7.15 | 1.06 | 0.75 | 2.88 | 0.64 | 2.47 | N1.09 | 25.43 | 0.94 | 71.23 | 0.73 | 55.55 |
| N4.12 | 1.25 | 0.75 | 2.89 | 0.64 | 2.47 | N1.13 | 27.03 | 0.81 | 61.42 | 0.73 | 55.41 |
| N3.06 | −1.38 | 0.54 | 2.07 | 0.61 | 2.35 | N1.12 | 21.33 | 0.88 | 66.45 | 0.72 | 54.73 |
| N2.22 | 0.83 | 0.78 | 3.03 | 0.60 | 2.33 | N7.15 | 22.03 | 0.72 | 54.36 | 0.65 | 49.39 |
| N4.10 | 0.72 | 0.76 | 2.94 | 0.60 | 2.30 | N2.18 | 17.47 | 0.71 | 54.09 | 0.65 | 49.30 |
| N1.15 | 0.96 | 0.62 | 2.40 | 0.57 | 2.22 | N4.12 | 15.47 | 0.69 | 52.03 | 0.63 | 47.67 |
| N5.05 | 0.63 | 0.69 | 2.66 | 0.57 | 2.19 | N3.06 | 15.90 | 0.76 | 58.02 | 0.62 | 47.39 |
| N4.21 | 0.38 | 0.86 | 3.32 | 0.56 | 2.18 | N2.22 | 21.42 | 0.91 | 68.86 | 0.62 | 46.94 |
| N4.17 | 0.79 | 0.67 | 2.59 | 0.52 | 1.99 | N1.15 | 22.37 | 0.72 | 54.62 | 0.62 | 46.94 |
| N6.01 | 0.09 | 0.72 | 2.76 | 0.25 | 0.97 | N4.17 | 17.96 | 0.62 | 47.32 | 0.51 | 38.63 |
| LSR | CT | ||||||||||
| Ecotype | MHVG | PRVG | PRVG*µ | MHPRVG | MHPRVG*µ | Ecotype | MHVG | PRVG | PRVG*µ | MHPRVG | MHPRVG*µ |
| N1.13 | 3.13 | 1.35 | 3.96 | 1.28 | 3.74 | N1.09 | 4.35 | 1.51 | 4.59 | 1.45 | 4.39 |
| N5.12 | 2.92 | 1.33 | 3.91 | 1.23 | 3.61 | N1.13 | 3.95 | 1.36 | 4.12 | 1.34 | 4.05 |
| N1.21 | 2.83 | 1.23 | 3.61 | 1.15 | 3.38 | N1.12 | 3.68 | 1.27 | 3.85 | 1.24 | 3.76 |
| N4.10 | 2.61 | 1.34 | 3.94 | 1.15 | 3.37 | N1.07 | 3.61 | 1.24 | 3.76 | 1.23 | 3.71 |
| N2.01 | 2.67 | 1.21 | 3.56 | 1.14 | 3.34 | N8.03 | 3.40 | 1.17 | 3.55 | 1.17 | 3.54 |
| N7.20 | 2.63 | 1.23 | 3.62 | 1.10 | 3.23 | N1.16 | 3.36 | 1.16 | 3.51 | 1.15 | 3.49 |
| N2.07 | 2.77 | 1.15 | 3.36 | 1.08 | 3.17 | N7.15 | 3.23 | 1.13 | 3.43 | 1.13 | 3.42 |
| N4.21 | 2.65 | 1.10 | 3.22 | 1.07 | 3.14 | N4.21 | 3.34 | 1.15 | 3.50 | 1.13 | 3.42 |
| N5.14 | 2.56 | 1.10 | 3.23 | 1.06 | 3.12 | N1.14 | 3.39 | 1.19 | 3.62 | 1.12 | 3.39 |
| N3.02 | 2.54 | 1.11 | 3.24 | 1.05 | 3.09 | N3.22 | 3.20 | 1.12 | 3.39 | 1.12 | 3.38 |
| N5.22 | 2.55 | 1.13 | 3.31 | 1.05 | 3.07 | N1.06 | 3.27 | 1.13 | 3.41 | 1.11 | 3.37 |
| N4.14 | 2.42 | 1.42 | 4.15 | 1.04 | 3.04 | N1.15 | 3.27 | 1.13 | 3.41 | 1.11 | 3.37 |
| N2.06 | 2.52 | 1.20 | 3.53 | 1.04 | 3.04 | N2.01 | 3.27 | 1.13 | 3.41 | 1.11 | 3.37 |
| N5.06 | 2.56 | 1.10 | 3.24 | 1.04 | 3.04 | N8.04 | 3.19 | 1.10 | 3.33 | 1.10 | 3.32 |
| N6.19 | 2.48 | 1.06 | 3.11 | 1.03 | 3.02 | N1.10 | 3.19 | 1.10 | 3.33 | 1.10 | 3.32 |
| N1.14 | 2.45 | 1.11 | 3.26 | 1.03 | 3.01 | N6.19 | 3.13 | 1.09 | 3.29 | 1.09 | 3.29 |
| N1.09 | 2.53 | 1.24 | 3.63 | 1.02 | 3.00 | N5.05 | 3.13 | 1.09 | 3.29 | 1.09 | 3.29 |
| N2.18 | 2.56 | 1.11 | 3.27 | 1.02 | 2.99 | N3.19 | 3.13 | 1.09 | 3.29 | 1.09 | 3.29 |
| N3.19 | 2.52 | 1.07 | 3.13 | 1.01 | 2.98 | N4.06 | 3.15 | 1.08 | 3.29 | 1.08 | 3.26 |
| N1.12 | 2.40 | 1.06 | 3.11 | 1.01 | 2.95 | N1.19 | 3.17 | 1.10 | 3.32 | 1.07 | 3.25 |
| N8.04 | 2.51 | 1.03 | 3.04 | 1.01 | 2.95 | N1.17 | 2.99 | 1.04 | 3.16 | 1.04 | 3.16 |
| N3.16 | 2.43 | 1.04 | 3.05 | 1.00 | 2.94 | N5.11 | 2.99 | 1.04 | 3.16 | 1.04 | 3.16 |
| N1.10 | 2.42 | 1.26 | 3.69 | 1.00 | 2.94 | N3.18 | 2.99 | 1.04 | 3.16 | 1.04 | 3.16 |
| N3.04 | 2.55 | 1.10 | 3.22 | 1.00 | 2.93 | N2.17 | 2.95 | 1.03 | 3.12 | 1.03 | 3.12 |
| N5.10 | 2.38 | 1.02 | 3.00 | 1.00 | 2.93 | N3.12 | 2.95 | 1.03 | 3.12 | 1.03 | 3.12 |
| N3.18 | 2.52 | 1.02 | 3.00 | 1.00 | 2.92 | N3.07 | 2.95 | 1.03 | 3.12 | 1.03 | 3.12 |
| N3.15 | 2.36 | 1.01 | 2.96 | 0.99 | 2.92 | N3.03 | 2.92 | 1.01 | 3.07 | 1.01 | 3.07 |
| N1.15 | 2.44 | 1.11 | 3.26 | 0.99 | 2.91 | N2.18 | 2.84 | 1.00 | 3.04 | 1.00 | 3.03 |
| N6.08 | 2.38 | 1.11 | 3.25 | 0.99 | 2.89 | N6.05 | 2.84 | 1.00 | 3.04 | 1.00 | 3.03 |
| N3.08 | 2.38 | 1.14 | 3.33 | 0.99 | 2.89 | N5.01 | 2.88 | 1.00 | 3.03 | 1.00 | 3.02 |
| N5.21 | 2.36 | 1.03 | 3.01 | 0.97 | 2.86 | N4.20 | 2.88 | 1.00 | 3.03 | 1.00 | 3.02 |
| N3.22 | 2.38 | 1.03 | 3.02 | 0.97 | 2.84 | N3.04 | 2.95 | 1.02 | 3.10 | 1.00 | 3.02 |
| N5.09 | 2.40 | 0.99 | 2.89 | 0.97 | 2.84 | N1.18 | 2.81 | 0.99 | 2.99 | 0.99 | 2.99 |
| N1.19 | 2.37 | 1.18 | 3.46 | 0.97 | 2.83 | N3.08 | 2.81 | 0.99 | 2.99 | 0.99 | 2.99 |
| N5.19 | 2.39 | 1.04 | 3.06 | 0.96 | 2.82 | N3.09 | 2.81 | 0.99 | 2.99 | 0.99 | 2.99 |
| N5.20 | 2.37 | 1.01 | 2.97 | 0.96 | 2.82 | N5.14 | 2.81 | 0.99 | 2.99 | 0.99 | 2.99 |
| N2.11 | 2.40 | 0.98 | 2.87 | 0.96 | 2.81 | N3.15 | 2.81 | 0.99 | 2.99 | 0.99 | 2.99 |
| N7.15 | 2.41 | 1.07 | 3.15 | 0.96 | 2.81 | N4.05 | 2.81 | 0.99 | 2.99 | 0.99 | 2.99 |
| N4.13 | 2.36 | 1.04 | 3.04 | 0.96 | 2.80 | N4.14 | 2.81 | 0.99 | 2.99 | 0.99 | 2.99 |
| N3.06 | 2.37 | 0.98 | 2.88 | 0.95 | 2.79 | N3.05 | 2.88 | 1.00 | 3.02 | 0.98 | 2.98 |
| N5.02 | 2.35 | 1.02 | 2.99 | 0.95 | 2.78 | N6.22 | 2.84 | 0.98 | 2.98 | 0.98 | 2.97 |
| N4.12 | 2.39 | 1.11 | 3.25 | 0.94 | 2.76 | N2.16 | 2.84 | 0.98 | 2.98 | 0.98 | 2.97 |
| N1.22 | 2.39 | 0.99 | 2.90 | 0.94 | 2.75 | N4.19 | 2.84 | 0.98 | 2.98 | 0.98 | 2.97 |
| N1.06 | 2.31 | 0.96 | 2.81 | 0.93 | 2.73 | N5.16 | 2.74 | 0.99 | 3.00 | 0.98 | 2.96 |
| N4.05 | 2.33 | 0.99 | 2.90 | 0.92 | 2.70 | N2.13 | 2.78 | 0.97 | 2.94 | 0.97 | 2.94 |
| N6.12 | 2.28 | 1.01 | 2.95 | 0.92 | 2.69 | N4.13 | 2.78 | 0.97 | 2.94 | 0.97 | 2.94 |
| N4.08 | 2.29 | 1.03 | 3.02 | 0.91 | 2.68 | N3.06 | 2.78 | 0.97 | 2.94 | 0.97 | 2.94 |
| N6.01 | 2.25 | 0.97 | 2.86 | 0.90 | 2.65 | N4.15 | 2.78 | 0.97 | 2.94 | 0.97 | 2.94 |
| N5.18 | 2.22 | 0.92 | 2.70 | 0.90 | 2.63 | N2.10 | 2.78 | 0.97 | 2.94 | 0.97 | 2.94 |
| N5.03 | 2.28 | 0.94 | 2.75 | 0.89 | 2.61 | N1.22 | 2.74 | 0.96 | 2.90 | 0.96 | 2.90 |
| N3.07 | 2.19 | 0.91 | 2.67 | 0.89 | 2.61 | N5.22 | 2.74 | 0.96 | 2.90 | 0.96 | 2.90 |
| N4.07 | 2.24 | 0.97 | 2.85 | 0.89 | 2.61 | N6.01 | 2.69 | 0.96 | 2.91 | 0.95 | 2.89 |
| N6.05 | 2.25 | 0.96 | 2.80 | 0.89 | 2.60 | N4.12 | 2.69 | 0.96 | 2.91 | 0.95 | 2.89 |
| N3.12 | 2.25 | 0.91 | 2.67 | 0.88 | 2.58 | N5.15 | 2.69 | 0.96 | 2.91 | 0.95 | 2.89 |
| N6.22 | 2.21 | 1.07 | 3.13 | 0.88 | 2.57 | N5.18 | 2.69 | 0.96 | 2.91 | 0.95 | 2.89 |
| N3.10 | 2.15 | 0.89 | 2.61 | 0.87 | 2.56 | N4.09 | 2.76 | 0.95 | 2.89 | 0.95 | 2.86 |
| N3.03 | 2.23 | 0.91 | 2.66 | 0.87 | 2.54 | N6.12 | 2.66 | 0.94 | 2.86 | 0.94 | 2.85 |
| N2.17 | 2.20 | 0.90 | 2.65 | 0.86 | 2.53 | N6.18 | 2.66 | 0.94 | 2.86 | 0.94 | 2.85 |
| N4.06 | 2.24 | 0.90 | 2.65 | 0.86 | 2.52 | N5.06 | 2.66 | 0.94 | 2.86 | 0.94 | 2.85 |
| N1.07 | 2.21 | 0.88 | 2.59 | 0.85 | 2.51 | N3.16 | 2.63 | 0.93 | 2.81 | 0.93 | 2.81 |
| N5.11 | 2.18 | 0.91 | 2.66 | 0.85 | 2.48 | N3.02 | 2.73 | 0.95 | 2.88 | 0.92 | 2.78 |
| N4.20 | 2.09 | 0.90 | 2.64 | 0.83 | 2.43 | N5.09 | 2.56 | 0.93 | 2.82 | 0.92 | 2.77 |
| N1.17 | 2.12 | 0.88 | 2.59 | 0.83 | 2.43 | N3.17 | 2.56 | 0.93 | 2.82 | 0.92 | 2.77 |
| N3.05 | 2.20 | 0.87 | 2.57 | 0.83 | 2.42 | N5.21 | 2.60 | 0.91 | 2.77 | 0.91 | 2.77 |
| N6.18 | 2.12 | 0.87 | 2.54 | 0.82 | 2.40 | N1.21 | 2.63 | 0.91 | 2.76 | 0.91 | 2.75 |
| N2.02 | 2.07 | 0.89 | 2.61 | 0.82 | 2.40 | N5.12 | 2.57 | 0.90 | 2.72 | 0.90 | 2.72 |
| N4.15 | 2.04 | 0.84 | 2.47 | 0.81 | 2.37 | N2.11 | 2.51 | 0.90 | 2.73 | 0.89 | 2.70 |
| N1.05 | 2.08 | 0.87 | 2.55 | 0.81 | 2.36 | N5.02 | 2.51 | 0.90 | 2.73 | 0.89 | 2.70 |
| N4.19 | 2.04 | 0.85 | 2.48 | 0.80 | 2.33 | N5.19 | 2.51 | 0.90 | 2.73 | 0.89 | 2.70 |
| N2.16 | 2.01 | 0.81 | 2.38 | 0.79 | 2.32 | N5.03 | 2.51 | 0.90 | 2.73 | 0.89 | 2.70 |
| N4.17 | 2.12 | 0.88 | 2.59 | 0.79 | 2.31 | N5.20 | 2.51 | 0.90 | 2.73 | 0.89 | 2.70 |
| N8.03 | 2.00 | 0.82 | 2.40 | 0.79 | 2.30 | N1.05 | 2.59 | 0.90 | 2.71 | 0.89 | 2.69 |
| N3.17 | 1.97 | 0.96 | 2.82 | 0.77 | 2.26 | N7.20 | 2.48 | 0.89 | 2.68 | 0.88 | 2.67 |
| N1.16 | 2.05 | 0.81 | 2.37 | 0.76 | 2.23 | N5.10 | 2.48 | 0.89 | 2.68 | 0.88 | 2.67 |
| N4.09 | 1.95 | 0.86 | 2.51 | 0.76 | 2.23 | N2.07 | 2.45 | 0.87 | 2.64 | 0.87 | 2.63 |
| N5.16 | 1.94 | 0.87 | 2.55 | 0.74 | 2.18 | N4.08 | 2.45 | 0.87 | 2.64 | 0.87 | 2.63 |
| N3.09 | 1.91 | 0.76 | 2.22 | 0.74 | 2.16 | N4.17 | 2.40 | 0.89 | 2.69 | 0.86 | 2.62 |
| N5.01 | 1.89 | 0.78 | 2.29 | 0.74 | 2.16 | N6.08 | 2.35 | 0.86 | 2.60 | 0.84 | 2.55 |
| N5.05 | 1.92 | 0.88 | 2.59 | 0.71 | 2.07 | N2.06 | 2.39 | 0.84 | 2.55 | 0.84 | 2.54 |
| N1.18 | 1.85 | 0.78 | 2.28 | 0.70 | 2.06 | N4.10 | 2.32 | 0.84 | 2.56 | 0.83 | 2.52 |
| N5.15 | 1.81 | 0.73 | 2.14 | 0.69 | 2.04 | N2.02 | 2.35 | 0.83 | 2.50 | 0.82 | 2.50 |
| N2.22 | 1.77 | 0.72 | 2.12 | 0.67 | 1.97 | N3.10 | 2.37 | 0.82 | 2.49 | 0.81 | 2.46 |
| N2.10 | 1.68 | 0.71 | 2.09 | 0.61 | 1.79 | N2.22 | 2.22 | 0.85 | 2.56 | 0.81 | 2.45 |
| N2.13 | 1.59 | 0.67 | 1.96 | 0.57 | 1.68 | N4.07 | 2.20 | 0.83 | 2.52 | 0.80 | 2.42 |
| GH | PH (cm) | ||||||||||
| Ecotype | MHVG | PRVG | PRVG*µ | MHPRVG | MHPRVG*µ | Ecotype | MHVG | PRVG | PRVG*µ | MHPRVG | MHPRVG*µ |
| N7.20 | 3.65 | 1.27 | 3.65 | 1.27 | 3.65 | N2.02 | 64.21 | 1.15 | 65.46 | 1.15 | 65.37 |
| N5.15 | 3.65 | 1.27 | 3.66 | 1.27 | 3.65 | N1.18 | 61.11 | 1.10 | 62.65 | 1.10 | 62.56 |
| N5.16 | 3.65 | 1.27 | 3.66 | 1.27 | 3.65 | N3.09 | 61.53 | 1.09 | 62.39 | 1.09 | 62.11 |
| N5.14 | 3.65 | 1.27 | 3.66 | 1.27 | 3.65 | N2.06 | 60.73 | 1.09 | 61.87 | 1.08 | 61.81 |
| N4.21 | 3.65 | 1.27 | 3.66 | 1.27 | 3.65 | N2.10 | 60.12 | 1.07 | 61.02 | 1.07 | 60.93 |
| N6.01 | 3.57 | 1.25 | 3.59 | 1.24 | 3.57 | N5.09 | 59.55 | 1.06 | 60.54 | 1.06 | 60.44 |
| N6.12 | 3.57 | 1.25 | 3.59 | 1.24 | 3.57 | N3.02 | 58.99 | 1.06 | 60.16 | 1.05 | 60.11 |
| N6.19 | 3.57 | 1.25 | 3.59 | 1.24 | 3.57 | N1.21 | 58.83 | 1.05 | 60.13 | 1.05 | 60.09 |
| N1.21 | 3.57 | 1.25 | 3.59 | 1.24 | 3.57 | N4.07 | 59.13 | 1.05 | 60.08 | 1.05 | 60.05 |
| N2.17 | 3.57 | 1.25 | 3.59 | 1.24 | 3.57 | N5.15 | 58.83 | 1.05 | 59.71 | 1.05 | 59.64 |
| N2.13 | 3.57 | 1.25 | 3.59 | 1.24 | 3.57 | N3.10 | 58.48 | 1.05 | 59.59 | 1.04 | 59.50 |
| N4.17 | 3.57 | 1.25 | 3.59 | 1.24 | 3.57 | N2.16 | 58.36 | 1.04 | 59.41 | 1.04 | 59.20 |
| N5.01 | 3.57 | 1.25 | 3.59 | 1.24 | 3.57 | N2.07 | 58.23 | 1.04 | 59.25 | 1.04 | 59.13 |
| N1.15 | 3.57 | 1.25 | 3.59 | 1.24 | 3.57 | N5.22 | 57.95 | 1.04 | 59.17 | 1.04 | 59.11 |
| N3.12 | 3.57 | 1.25 | 3.59 | 1.24 | 3.57 | N4.08 | 57.91 | 1.04 | 59.13 | 1.03 | 58.93 |
| N1.14 | 3.57 | 1.25 | 3.59 | 1.24 | 3.57 | N8.04 | 57.69 | 1.03 | 58.99 | 1.03 | 58.93 |
| N4.19 | 3.42 | 1.20 | 3.44 | 1.19 | 3.42 | N1.06 | 58.01 | 1.03 | 59.00 | 1.03 | 58.85 |
| N1.12 | 3.21 | 1.12 | 3.21 | 1.12 | 3.21 | N5.01 | 57.87 | 1.04 | 59.08 | 1.03 | 58.84 |
| N5.05 | 3.13 | 1.09 | 3.13 | 1.09 | 3.13 | N3.12 | 57.82 | 1.03 | 58.73 | 1.03 | 58.71 |
| N4.06 | 3.13 | 1.09 | 3.13 | 1.09 | 3.13 | N1.13 | 57.84 | 1.03 | 58.75 | 1.03 | 58.70 |
| N3.05 | 3.13 | 1.09 | 3.13 | 1.09 | 3.13 | N3.08 | 57.72 | 1.03 | 58.71 | 1.03 | 58.66 |
| N7.15 | 3.13 | 1.09 | 3.14 | 1.09 | 3.13 | N3.17 | 57.68 | 1.03 | 58.65 | 1.03 | 58.64 |
| N3.09 | 3.12 | 1.09 | 3.13 | 1.09 | 3.13 | N2.01 | 57.65 | 1.03 | 58.63 | 1.03 | 58.62 |
| N5.06 | 3.12 | 1.09 | 3.13 | 1.09 | 3.13 | N5.06 | 57.49 | 1.02 | 58.31 | 1.02 | 58.17 |
| N3.15 | 3.12 | 1.09 | 3.13 | 1.09 | 3.13 | N3.07 | 57.21 | 1.02 | 58.18 | 1.02 | 58.17 |
| N4.14 | 3.12 | 1.09 | 3.13 | 1.09 | 3.13 | N3.04 | 57.15 | 1.02 | 58.19 | 1.02 | 58.15 |
| N6.22 | 3.06 | 1.07 | 3.06 | 1.07 | 3.06 | N1.05 | 56.72 | 1.02 | 58.05 | 1.02 | 57.96 |
| N6.18 | 3.06 | 1.07 | 3.06 | 1.07 | 3.06 | N4.10 | 57.00 | 1.02 | 57.95 | 1.02 | 57.93 |
| N1.07 | 3.06 | 1.07 | 3.06 | 1.07 | 3.06 | N1.07 | 57.25 | 1.02 | 58.07 | 1.02 | 57.92 |
| N1.05 | 3.06 | 1.07 | 3.06 | 1.07 | 3.06 | N2.22 | 57.21 | 1.02 | 58.00 | 1.02 | 57.87 |
| N5.22 | 3.06 | 1.07 | 3.06 | 1.07 | 3.06 | N4.13 | 56.65 | 1.01 | 57.80 | 1.01 | 57.79 |
| N5.10 | 3.06 | 1.07 | 3.06 | 1.07 | 3.06 | N5.05 | 56.82 | 1.01 | 57.75 | 1.01 | 57.73 |
| N4.12 | 3.06 | 1.07 | 3.06 | 1.07 | 3.06 | N5.19 | 56.66 | 1.01 | 57.65 | 1.01 | 57.65 |
| N5.21 | 3.06 | 1.07 | 3.06 | 1.07 | 3.06 | N4.12 | 56.48 | 1.01 | 57.58 | 1.01 | 57.53 |
| N5.18 | 3.06 | 1.07 | 3.06 | 1.07 | 3.06 | N1.10 | 55.80 | 1.02 | 58.18 | 1.01 | 57.46 |
| N4.05 | 3.06 | 1.07 | 3.06 | 1.07 | 3.06 | N2.17 | 56.32 | 1.01 | 57.51 | 1.01 | 57.45 |
| N4.10 | 3.06 | 1.07 | 3.06 | 1.07 | 3.06 | N4.05 | 56.13 | 1.01 | 57.38 | 1.01 | 57.33 |
| N3.02 | 3.06 | 1.07 | 3.06 | 1.07 | 3.06 | N4.17 | 56.46 | 1.01 | 57.34 | 1.01 | 57.31 |
| N3.07 | 3.06 | 1.07 | 3.06 | 1.07 | 3.06 | N3.06 | 56.14 | 1.00 | 57.12 | 1.00 | 57.11 |
| N3.16 | 3.06 | 1.07 | 3.06 | 1.07 | 3.06 | N4.20 | 56.12 | 1.00 | 57.27 | 1.00 | 57.10 |
| N6.08 | 2.98 | 1.04 | 2.99 | 1.04 | 2.99 | N3.03 | 56.13 | 1.00 | 57.11 | 1.00 | 57.06 |
| N1.22 | 2.98 | 1.04 | 2.99 | 1.04 | 2.99 | N4.09 | 56.02 | 1.00 | 57.23 | 1.00 | 57.04 |
| N1.13 | 2.98 | 1.04 | 2.99 | 1.04 | 2.99 | N4.06 | 56.39 | 1.00 | 57.15 | 1.00 | 56.87 |
| N2.07 | 2.98 | 1.04 | 2.99 | 1.04 | 2.99 | N4.14 | 56.07 | 1.00 | 57.02 | 1.00 | 56.86 |
| N2.11 | 2.98 | 1.04 | 2.99 | 1.04 | 2.99 | N7.20 | 55.66 | 1.00 | 56.90 | 1.00 | 56.86 |
| N2.16 | 2.98 | 1.04 | 2.99 | 1.04 | 2.99 | N6.22 | 54.58 | 1.01 | 57.69 | 0.99 | 56.62 |
| N5.02 | 2.98 | 1.04 | 2.99 | 1.04 | 2.99 | N1.22 | 55.50 | 0.99 | 56.55 | 0.99 | 56.54 |
| N2.18 | 2.98 | 1.04 | 2.99 | 1.04 | 2.99 | N5.12 | 55.14 | 0.99 | 56.35 | 0.99 | 56.32 |
| N4.13 | 2.98 | 1.04 | 2.99 | 1.04 | 2.99 | N1.19 | 55.22 | 0.99 | 56.23 | 0.99 | 56.23 |
| N4.07 | 2.98 | 1.04 | 2.99 | 1.04 | 2.99 | N5.20 | 55.07 | 0.99 | 56.26 | 0.99 | 56.18 |
| N5.09 | 2.98 | 1.04 | 2.99 | 1.04 | 2.99 | N1.12 | 54.98 | 0.98 | 56.01 | 0.98 | 56.00 |
| N1.19 | 2.98 | 1.04 | 2.99 | 1.04 | 2.99 | N5.02 | 54.59 | 0.98 | 55.94 | 0.98 | 55.84 |
| N3.04 | 2.98 | 1.04 | 2.99 | 1.04 | 2.99 | N4.19 | 55.12 | 0.98 | 55.96 | 0.98 | 55.82 |
| N2.10 | 2.69 | 0.94 | 2.70 | 0.93 | 2.68 | N6.08 | 54.33 | 0.98 | 56.03 | 0.98 | 55.79 |
| N5.03 | 2.58 | 0.91 | 2.61 | 0.90 | 2.59 | N3.05 | 54.29 | 0.98 | 55.63 | 0.97 | 55.49 |
| N3.06 | 2.57 | 0.91 | 2.61 | 0.90 | 2.59 | N5.14 | 54.09 | 0.98 | 55.68 | 0.97 | 55.42 |
| N3.03 | 2.51 | 0.88 | 2.54 | 0.88 | 2.52 | N5.18 | 54.29 | 0.97 | 55.45 | 0.97 | 55.41 |
| N3.18 | 2.51 | 0.88 | 2.54 | 0.88 | 2.52 | N1.09 | 54.92 | 0.98 | 56.13 | 0.97 | 55.37 |
| N5.20 | 2.51 | 0.88 | 2.54 | 0.88 | 2.52 | N3.22 | 54.91 | 0.98 | 55.66 | 0.97 | 55.32 |
| N5.19 | 2.51 | 0.88 | 2.54 | 0.88 | 2.52 | N3.16 | 54.58 | 0.97 | 55.40 | 0.97 | 55.20 |
| N4.20 | 2.51 | 0.88 | 2.54 | 0.88 | 2.52 | N3.19 | 54.43 | 0.97 | 55.22 | 0.97 | 55.14 |
| N3.17 | 2.51 | 0.88 | 2.54 | 0.88 | 2.52 | N1.17 | 54.19 | 0.97 | 55.04 | 0.96 | 54.97 |
| N4.08 | 2.45 | 0.86 | 2.46 | 0.86 | 2.46 | N5.16 | 53.36 | 0.97 | 55.37 | 0.96 | 54.95 |
| N3.22 | 2.45 | 0.86 | 2.46 | 0.86 | 2.46 | N5.03 | 54.36 | 0.98 | 55.60 | 0.96 | 54.94 |
| N4.15 | 2.45 | 0.86 | 2.46 | 0.86 | 2.46 | N8.03 | 54.13 | 0.96 | 54.95 | 0.96 | 54.91 |
| N1.09 | 2.45 | 0.86 | 2.46 | 0.86 | 2.46 | N7.15 | 54.13 | 0.96 | 54.98 | 0.96 | 54.86 |
| N1.18 | 2.38 | 0.83 | 2.39 | 0.83 | 2.39 | N3.18 | 53.14 | 0.97 | 55.31 | 0.96 | 54.80 |
| N1.17 | 2.38 | 0.83 | 2.39 | 0.83 | 2.39 | N6.05 | 53.93 | 0.96 | 54.80 | 0.96 | 54.79 |
| N5.11 | 2.38 | 0.83 | 2.39 | 0.83 | 2.39 | N6.19 | 53.92 | 0.96 | 54.84 | 0.96 | 54.77 |
| N5.12 | 2.38 | 0.83 | 2.39 | 0.83 | 2.39 | N1.15 | 53.88 | 0.96 | 54.74 | 0.96 | 54.70 |
| N4.09 | 2.38 | 0.83 | 2.39 | 0.83 | 2.39 | N4.15 | 53.85 | 0.96 | 54.80 | 0.96 | 54.49 |
| N2.01 | 2.38 | 0.83 | 2.39 | 0.83 | 2.39 | N3.15 | 53.07 | 0.96 | 54.64 | 0.96 | 54.45 |
| N2.22 | 1.98 | 0.73 | 2.08 | 0.70 | 2.00 | N2.11 | 52.77 | 0.96 | 54.79 | 0.95 | 54.38 |
| N2.06 | 1.97 | 0.73 | 2.08 | 0.69 | 1.99 | N5.21 | 52.53 | 0.95 | 54.31 | 0.95 | 53.92 |
| N8.03 | 1.92 | 0.70 | 2.01 | 0.67 | 1.93 | N2.13 | 52.91 | 0.95 | 54.14 | 0.95 | 53.90 |
| N3.19 | 1.86 | 0.68 | 1.94 | 0.66 | 1.88 | N5.10 | 53.09 | 0.95 | 54.01 | 0.95 | 53.89 |
| N2.02 | 1.80 | 0.65 | 1.87 | 0.63 | 1.82 | N2.18 | 52.19 | 0.95 | 54.18 | 0.94 | 53.76 |
| N3.08 | 1.80 | 0.65 | 1.87 | 0.63 | 1.82 | N5.11 | 52.61 | 0.94 | 53.47 | 0.94 | 53.42 |
| N1.10 | 1.80 | 0.65 | 1.87 | 0.63 | 1.82 | N6.12 | 51.25 | 0.94 | 53.39 | 0.93 | 52.89 |
| N1.06 | 1.74 | 0.62 | 1.79 | 0.61 | 1.75 | N1.16 | 51.89 | 0.93 | 52.87 | 0.93 | 52.87 |
| N1.16 | 1.74 | 0.62 | 1.79 | 0.61 | 1.75 | N6.01 | 51.83 | 0.92 | 52.70 | 0.92 | 52.63 |
| N3.10 | 1.74 | 0.62 | 1.79 | 0.61 | 1.75 | N4.21 | 51.17 | 0.91 | 52.03 | 0.91 | 51.99 |
| N8.04 | 1.74 | 0.62 | 1.79 | 0.61 | 1.75 | N1.14 | 51.23 | 0.91 | 52.00 | 0.91 | 51.93 |
| N6.05 | 1.74 | 0.62 | 1.79 | 0.61 | 1.75 | N6.18 | 50.40 | 0.91 | 51.85 | 0.91 | 51.69 |
References
- Bengtsson, J.; Bullock, J.M.; Egoh, B.; Everson, C.; Everson, T.; O’Connor, T.; O’Farrell, P.J.; Smith, H.G.; Lindborg, R. Grasslands—More Important for Ecosystem Services than You Might Think. Ecosphere 2019, 10, e02582. [Google Scholar] [CrossRef]
- Matta, F.D.P.; Fávero, A.P.; Vigna, B.B.Z.; Cavallari, M.M.; Alves, F.; Oliveira, F.A.D.; Souza, A.P.D.; Pozzobon, M.T.; Azevedo, A.L.S.; Gusmão, M.R. Characterization of Paspalum Genotypes for Turfgrass Cultivars Development. Crop Sci. 2024, 64, 1928–1941. [Google Scholar] [CrossRef]
- Zuloaga, F.O.; Morrone, O. Revisión de las Especies de Paspalum para América del Sur Austral (Argentina, Bolivia, Sur del Brasil, Chile, Paraguay y Uruguay); Monographs in Systematic Botany from the Missouri Botanical Garden; Missouri Botanical Garden Press: St. Louis, MO, USA, 2005; Volume 102, pp. 1–297. Available online: https://floradobrasil.jbrj.gov.br/consulta/ficha.html?idDadosListaBrasil=13432 (accessed on 22 August 2025).
- Valls, J.F.M.; Maciel, J.R.; Sousa, M.W.D.S.; Oliveira, R.C.; Pimenta, K.M.; Rua, G.H. Paspalum. In Flora e Funga do Brasil; Jardim Botânico do Rio de Janeiro: Rio de Janeiro, Brazil, 2023. Available online: https://floradobrasil.jbrj.gov.br/reflora/listaBrasil/PrincipalUC/PrincipalUC.do;jsessionid=B1CE6B5E8E3876997C2B9DAA1616F15D#CondicaoTaxonCP (accessed on 22 August 2025).
- Bonasora, M.G.; Pozzobon, M.T.; Honfi, A.I.; Rua, G.H. Paspalum schesslii (Poaceae, Paspaleae), a New Species from Mato Grosso (Brazil) with an Unusual Base Chromosome Number. Plant Syst. Evol. 2015, 301, 2325–2339. [Google Scholar] [CrossRef]
- Chase, A. The North American Species of Paspalum; Contributions from the United States National Herbarium; United States Government Printing Office: Washington, DC, USA, 1929; Volume 28, pp. 1–310. Available online: https://repository.si.edu/handle/10088/27051 (accessed on 22 August 2025).
- Judziewicz, E.J. Family 187. Poaceae (Gramineae). In Flora of the Guianas. Series A: Phanerogams; Görts-van Rijn, A.R.A., Ed.; Koeltz Scientific Books: Königstein, Germany, 1990; Volume 8, pp. 1–727. [Google Scholar]
- Maciel, J.R.; Oliveira, R.C.; Alves, M. Paspalum L. (Poaceae: Panicoideae: Paniceae) no Estado de Pernambuco, Brasil. Acta Bot. Bras. 2009, 23, 1145–1161. [Google Scholar] [CrossRef]
- Denham, S.S.; Zuloaga, F.O.; Morrone, O. Systematic Revision and Phylogeny of Paspalum subgenus Ceresia (Poaceae: Panicoideae: Paniceae). Ann. Mo. Bot. Gard. 2002, 89, 337–399. [Google Scholar] [CrossRef]
- Rua, G.H.; Valls, J.F.M.; Graciano-Ribeiro, D.; Oliveira, R.C. Four New Species of Paspalum (Poaceae, Paniceae) from Central Brazil, and Resurrection of an Old One. Syst. Bot. 2008, 33, 267–276. Available online: https://www.alice.cnptia.embrapa.br/alice/bitstream/doc/190393/1/SP19577ID30672.pdf (accessed on 22 August 2025). [CrossRef]
- Pereira, E.A.; Dall’Agnol, M.; Nabinger, C.; Huber, K.G.C.; Montardo, D.P.; Genro, T.C.M. Produção Agronômica de uma Coleção de Acessos de Paspalum nicorae Parodi. Rev. Bras. Zootec. 2011, 40, 498–508. [Google Scholar] [CrossRef]
- Oliveira, R.C.; Valls, J.F.M. Paspalum. In Lista de Espécies da Flora do Brasil; Jardim Botânico do Rio de Janeiro: Rio de Janeiro, Brazil, 2015. Available online: http://floradobrasil2015.jbrj.gov.br/FB13489 (accessed on 22 August 2025).
- Filho, C.C.F.; Barrios, S.C.L.; Santos, M.F.; Nunes, J.A.R.; do Valle, C.B.; Jank, L.; Rios, E.F. Assessing Genotype Adaptability and Stability in Perennial Forage Breeding Trials Using Random Regression Models for Longitudinal Dry Matter Yield Data. G3 Genes Genomes Genet. 2025, 15, jkae306. [Google Scholar] [CrossRef]
- Cook, B.G.; Lloyd, D.L.; Scattini, W.J.; Robertson, A.D. Paspalum lepton—A Valuable Adjunct to the Suite of Grasses Used in Grazing Systems in the Subtropics or a Potential Weed. Trop. Grassl.-Forrajes Trop. 2024, 12, 124–141. [Google Scholar] [CrossRef]
- Machado, J.M.; Dall’Agnol, M.; da Motta, E.A.M.; Pereira, E.A.; Simioni, C.; Weiler, R.L.; Zuñeda, M.P.; Ferreira, P.B. Agronomic Evaluation of Paspalum notatum Flügge under the Influence of Photoperiod. Rev. Bras. Zootec. 2017, 46, 8–12. [Google Scholar] [CrossRef]
- Motta, E.A.M.d.; Dall’Agnol, M.; Pereira, E.A.; Machado, J.M.; Simioni, C. Valor Forrageiro de Híbridos Interespecíficos Superiores de Paspalum. Rev. Cienc. Agron. 2017, 48, 191–198. [Google Scholar] [CrossRef][Green Version]
- Weiler, R.L.; Dall’Agnol, M.; Simioni, C.; Krycki, K.C.; Pereira, E.A.; Machado, J.M.; Motta, E.A.M.d. Intraspecific Tetraploid Hybrids of Paspalum notatum: Agronomic Evaluation of Segregating Progeny. Sci. Agric. 2018, 75, 36–42. [Google Scholar] [CrossRef]
- Saraiva, K.M.; Dall’Agnol, M.; da Motta, E.A.; Pereira, E.A.; de Souza, C.H.; Simioni, C.; Weiler, R.L.; Kopp, M.M.; Schneider-Canny, R.; Barbosa, M.R. Hybrids of Paspalum plicatulum × P. guenoarum: Selection for Forage Yield and Cold Tolerance in a Subtropical Environment. Trop. Grassl.-Forrajes Trop. 2021, 9, 138–143. [Google Scholar] [CrossRef]
- Krycki, K.C.; da Motta, E.A.M.; Brunes, A.P.; Mills, A.; Weiler, R.L.; Simioni, C. Hybrid Progenies of Bahiagrass: Agronomic Evaluation. Sci. Agrar. Parana. 2022, 21, 99–105. [Google Scholar] [CrossRef]
- Eeuwijk, F.A.; Bustos-Korts, D.V.; Malosetti, M. What Should Students in Plant Breeding Know about the Statistical Aspects of Genotype × Environment Interactions? Crop Sci. 2016, 56, 2119–2140. [Google Scholar] [CrossRef]
- Marcón, F.; Brugnoli, E.A.; Nunes, J.A.R.; Gutierrez, V.A.; Martínez, E.J.; Acuña, C.A. Evaluating General Combining Ability for Agro-Morphological Traits in Tetraploid Bahiagrass. Euphytica 2021, 217, 208. [Google Scholar] [CrossRef]
- Silveira, D.C.; Machado, J.M.; da Motta, E.A.M.; Barbosa, M.R.; Simioni, C.; Weiler, R.L.; Mills, A.; Sampaio, R.; Brunes, A.P.; Dall’Agnol, M. Genetic Parameters, Prediction of Gains and Intraspecific Hybrid Selection of Paspalum notatum Flügge for Forage Using REML/BLUP. Agronomy 2022, 12, 1654. [Google Scholar] [CrossRef]
- Silveira, D.C.; Weiler, R.L.; Brunes, A.P.; Simioni, C.; Mills, A.; Longhi, J.; Corrêa, M.V.S.; Nauderer, C.; Valentini, A.; dos Santos, W.M.; et al. REML/BLUP Methodology for Selection of Intraspecific Hybrids of Paspalum notatum Flügge by Multivariate Analysis. An. Acad. Bras. Cienc. 2023, 95 (Suppl. S2), e20230137. [Google Scholar] [CrossRef]
- Olivoto, T.; Lúcio, A.D.; Silva, J.A.d.; Marchioro, V.S.; Souza, V.Q.d.; Jost, E. Mean Performance and Stability in Multi-Environment Trials I: Combining Features of AMMI and BLUP Techniques. Agron. J. 2019, 111, 2949–2960. [Google Scholar] [CrossRef]
- Viana, A.P.; Resende, M.D.V. Genética Quantitativa no Melhoramento de Fruteiras; Interciência: Rio de Janeiro, Brazil, 2014. [Google Scholar]
- Olivoto, T.; Lúcio, A.D.C. metan: An R Package for Multi-Environment Trial Analysis. Methods Ecol. Evol. 2020, 11, 783–789. [Google Scholar] [CrossRef]
- dos Santos, H.G.; Jacomine, P.K.T.; dos Anjos, L.H.C.; de Oliveira, V.A.; Lumbreras, J.F.; Coelho, M.R.; de Almeida, J.A.; de Araujo Filho, J.C.; de Oliveira, J.B.; Cunha, T.J.F. Sistema Brasileiro de Classificação de Solos, 5th ed.; Embrapa: Brasília, Brazil, 2018; p. 356. [Google Scholar]
- Wrege, M.S.; Steinmetz, S.; Reisser Júnior, C.; Almeida, I.R. Atlas Climático da Região Sul do Brasil: Estados do Paraná, Santa Catarina e Rio Grande do Sul; Embrapa Clima Temperado, Embrapa Florestas: Pelotas and Colombo, Brazil, 2011; p. 336. [Google Scholar]
- Bergamaschi, H.; Melo, R.D.; Guadagnin, M.R.; Cardoso, L.S.; Silva, M.D.; Comiran, F.; Brauner, P.C. Boletins Agrometeorológicos da Estação Experimental Agronômica da UFRGS: Série Histórica 1970–2012. In Boletim Agrometeorológico: Estação Experimental Agronômica/UFRGS: Eldorado do Sul; Faculdade de Agronomia da UFRGS: Porto Alegre, Brazil, 1970; Universidade Federal do Rio Grande do Sul: Porto Alegre, Brazil, 2013; 1 CD-ROM. [Google Scholar]
- Silveira, D.C.; Mills, A.; Sampaio, R.; Longhi, J.; Amaral, E.C.; Ávila, V.S.; Weiler, R.L.; Brunes, A.P.; Simioni, C.; Dall’Agnol, M. Correlation and path analysis based on multi-trait BLUP as selection criteria for forage in Paspalum nicorae Parodi. Rev. Bras. De Zootec. Braz. 2025, 54, e20240151. [Google Scholar] [CrossRef]
- Comissão de Química e Fertilidade do Solo. Manual de Calagem e Adubação para os Estados do Rio Grande do Sul e de Santa Catarina; Sociedade Brasileira de Ciência do Solo–Núcleo Regional Sul, Comissão de Química e Fertilidade do Solo–RS/SC: Rio de Janeiro, Brazil, 2016; p. 376. [Google Scholar]
- Brasil. Ministério da Agricultura, Pecuária e Abastecimento. Regras para Análise de Sementes; Secretaria de Defesa Agropecuária, MAPA: Brasília, Brazil, 2009.
- Rao, C.R. Linear Statistical Inference and Its Applications, 2nd ed.; John Wiley & Sons: New York, NY, USA, 1973. [Google Scholar]
- Dempster, A.P.; Laird, N.M.; Rubin, D.B. Maximum Likelihood from Incomplete Data via the EM Algorithm. J. R. Stat. Soc. Ser. B Methodol. 1977, 39, 1–22. [Google Scholar] [CrossRef]
- Pimentel-Gomes, F. Índice de Variação: Um Substituto Vantajoso do Coeficiente de Variação. Circ. Téc. IPEF 1991, 178, 1–4. [Google Scholar]
- Resende, M.D.V. Matemática e Estatística na Análise de Experimentos e no Melhoramento Genético; Embrapa Florestas: Colombo, Brazil, 2007; p. 561. Available online: https://revistas.ufg.br/pat/article/view/1867 (accessed on 22 August 2025).
- Olivoto, T.; Nardino, M. MGIDI: Toward an Effective Multivariate Selection in Biological Experiments. Bioinformatics 2021, 37, 1383–1389. [Google Scholar] [CrossRef]
- R Core Team. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2025; Available online: https://www.R-project.org/ (accessed on 22 August 2025).
- Resende, M.D.V. Software Selegen-REML/BLUP: A Useful Tool for Plant Breeding. Crop Breed. Appl. Biotechnol. 2016, 16, 330–339. [Google Scholar] [CrossRef]
- Cruz, C.D. Genes Software—Extended and Integrated with the R, Matlab and Selegen. Acta Sci. Agron. 2016, 38, 547–552. [Google Scholar] [CrossRef]
- Sebbenn, A.M.; Siqueira, A.C.M.d.F.; Kageyama, P.Y.; Machado, J.A.R. Parâmetros Genéticos na Conservação da Cabreúva—Myroxylon peruiferum L.F. Allemão. Sci. For. 1998, 53, 31–38. Available online: https://www.ipef.br/publicacoes/scientia/nr53/cap3.pdf (accessed on 12 June 2025).
- Vencovsky, R. Melhoramento e Produção de Milho no Brasil; Paterniani, E., Ed.; Fundação Cargill: Campinas, Brazil, 1987; Volume 2, pp. 137–214. [Google Scholar]
- Massaro, R.A.M.; Bonine, C.A.V.; Scarpinati, E.A.; Paula, R.C. Viabilidade de Aplicação da Seleção Precoce em Testes Clonais de Eucalyptus spp. Cienc. Florest. 2010, 20, 597–609. [Google Scholar] [CrossRef]
- Valadares, N.R.; Fernandes, A.C.G.; Rodrigues, C.H.O.; Guedes, L.L.M.; Magalhães, J.R.; Alves, R.A.; de Andrade, V.C.; Azevedo, A.M. Estimation of Genetic Parameters and Selection Gains for Sweet Potato Using Bayesian Inference with A Priori Information. Acta Sci. Agron. 2022, 45, e56160. [Google Scholar] [CrossRef]
- Resende, M.D.V.; Duarte, J.B. Precisão e Controle de Qualidade em Experimentos de Avaliação de Cultivares. Pesqui. Agropecu. Trop. 2007, 37, 182–194. Available online: https://revistas.ufg.br/pat/article/view/1867 (accessed on 22 August 2025).
- Oliveira, E.J.; Santana, F.A.; Oliveira, L.A.; Santos, V.S. Genetic Parameters and Prediction of Genotypic Values for Root Quality Traits in Cassava Using REML/BLUP. Genet. Mol. Res. 2014, 13, 6683–6700. [Google Scholar] [CrossRef] [PubMed]
- Azevedo, A.M.; Andrade Júnior, V.C.; Fernandes, J.S.C.; Pedrosa, C.E.; Oliveira, C.M. Desempenho Agronômico e Parâmetros Genéticos em Genótipos de Batata-Doce. Hortic. Bras. 2015, 33, 84–90. [Google Scholar] [CrossRef]
- Falconer, D.S.; Mackay, T.F.C. Introduction to Quantitative Genetics, 4th ed.; Addison Wesley Longman: Harlow, UK, 1996. [Google Scholar]
- Miranda, D.P.; Ramos, H.C.C.; Santa-Catarina, R.; Vettorazzi, J.C.F.; Santana, J.G.S.; Poltronieri, T.P.S.; Pirovani, A.A.V.; Azevedo, A.O.N.; Duarte, R.P.; Rodrigues, A.S.; et al. Topcross Hybrids in Papaya (Carica papaya L.): Evaluation of the Potential for Increasing Fruit Quality in New Cultivars. Arch. Agron. Soil Sci. 2022, 68, 1473–1486. [Google Scholar] [CrossRef]
- Fischer, S.; Möhring, J.; Schön, C.C.; Piepho, H.P.; Klein, D.; Schipprack, W.; Utz, H.F.; Melchinger, A.E.; Reif, J.C. Trends in Genetic Variance Components during 30 Years of Hybrid Maize Breeding at the University of Hohenheim. Plant Breed. 2008, 127, 446–451. [Google Scholar] [CrossRef]
- Gonçalves, Z.S.; Lima, L.K.S.; Soares, T.L.; Souza, E.H.; Jesus, O.N. Leaf Anatomical Aspects of CABMV Infection in Passiflora spp. by Light and Fluorescence Microscopy. Australas. Plant Pathol. 2021, 50, 203–215. [Google Scholar] [CrossRef]
- Rosado, R.D.; Rosado, L.D.; Borges, L.L.; Bruckner, C.H.; Cruz, C.D.; Santos, C.E. Genetic Diversity of Sour Passion Fruit Revealed by Predicted Genetic Values. Agron. J. 2019, 111, 165–174. [Google Scholar] [CrossRef]
- Latheef, A.; Pugalendhi, L.; Rani, A.; Jeyakumar, P.; Kumar, M.; Devi, M. Genetic Analysis of Mango (Mangifera indica L.) Genotypes for Year-Round Flowering and Yield Characters. Madras Agric. J. 2022, 109, 23–27. [Google Scholar] [CrossRef]
- Baker, R.J. Selection Indices in Plant Breeding, 1st ed.; CRC Press: Boca Raton, FL, USA, 2020. [Google Scholar]
- Donald, C.M. The Breeding of Crop Ideotypes. Euphytica 1968, 17, 385–403. [Google Scholar] [CrossRef]
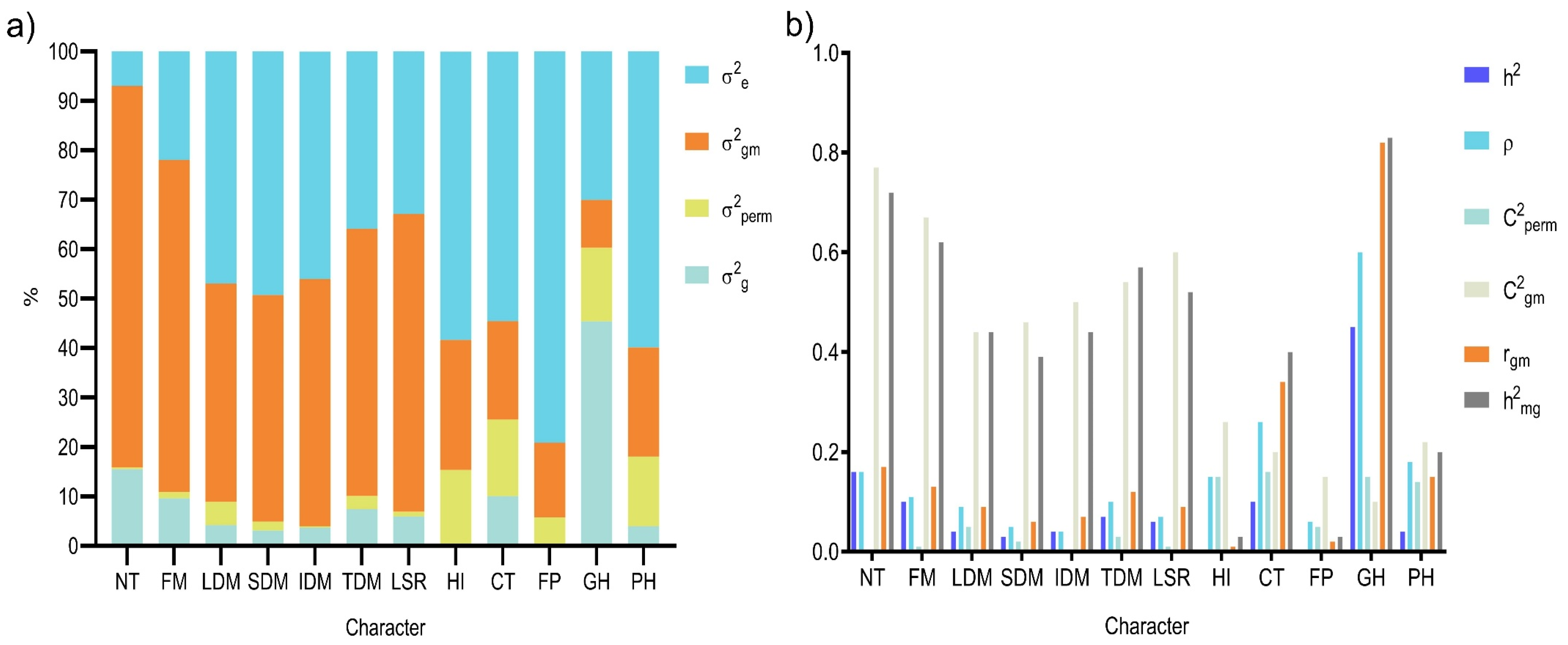
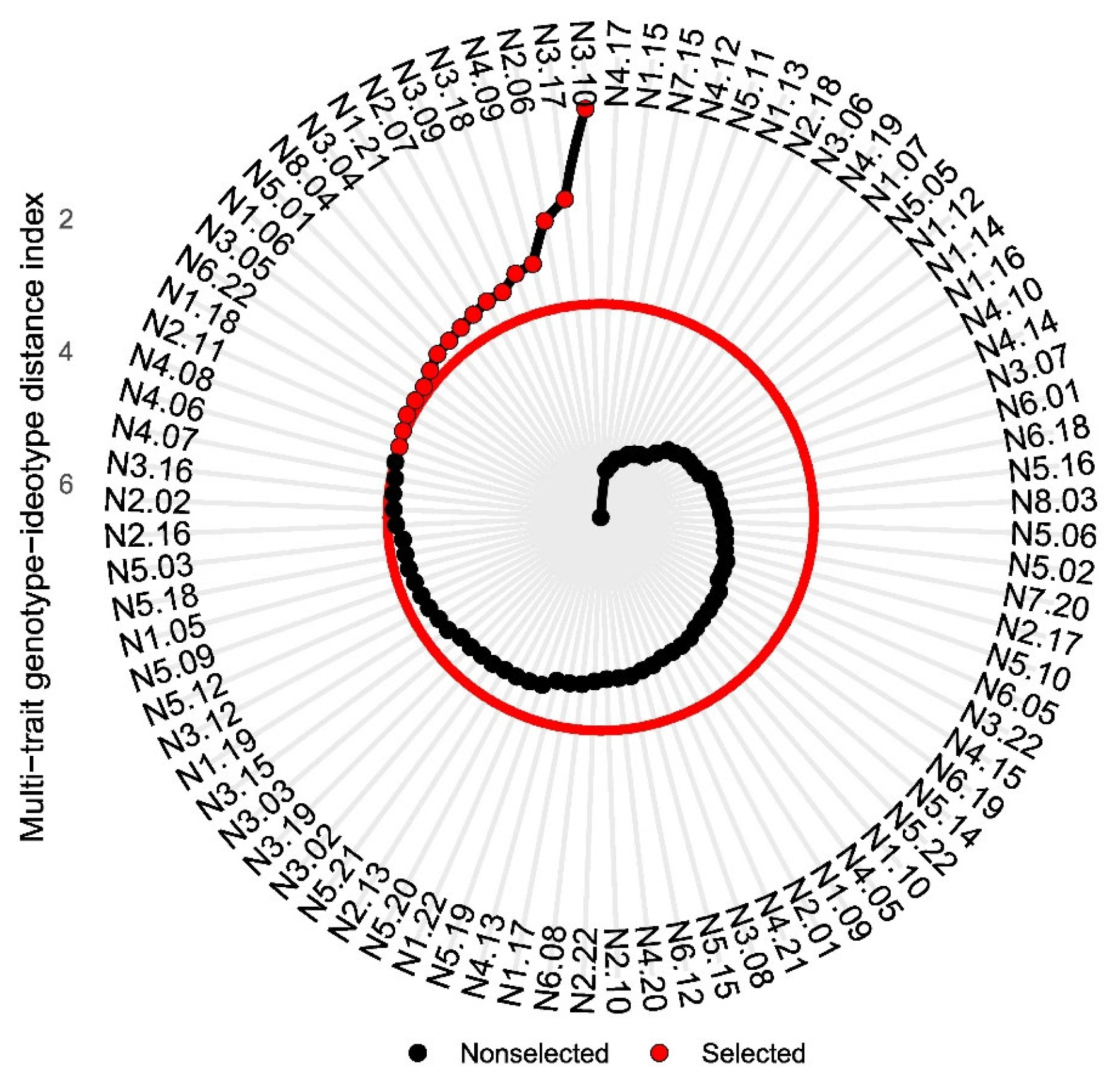
| Months | Temperature (°C, Min–Max) | Rainfall (mm) | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| 2020 | 2021 | 2022 | 2023 | 54 yr avg. | 2020 | 2021 | 2022 | 2023 | 54 yr avg. | |
| Jan | 20.3–30.0 | 20.2–33.3 | 25.8–33.0 | 19.3–30.2 | 67.8 | 110.1 | 31.2 | 109 | ||
| Feb | 18.8–29.7 | 19.0–30.7 | 24.0–30.6 | 19.0–29.4 | 31.2 | 77.2 | 67.8 | 100 | ||
| Mar | 18.9–28.7 | 17.6–27.7 | 25.0–31.5 | 18.0–28.2 | 86.4 | 98.1 | 5.8 | 83 | ||
| Apr | 15.4–27.4 | 14.1–24.3 | 14.5–25.2 | 5.8 | 118.2 | 74 | ||||
| May | 8.4–20.8 | 10.9–19.7 | 11.2–21.8 | 147.6 | 94.4 | 85 | ||||
| Jun | 9.3–18.6 | 7.9–17.3 | 8.7–18.6 | 120.8 | 97.6 | 109 | ||||
| Jul | 6.7–19.2 | 11.1–21.1 | 8.5–18.7 | 52.6 | 131.4 | 145 | ||||
| Aug | 11.0–21.5 | 9.2–20.1 | 9.7–20.2 | 121.4 | 92.2 | 115 | ||||
| Sep | 13.7–22.4 | 10.7–21.6 | 11.2–21.4 | 176.4 | 58.6 | 159 | ||||
| Oct | 15.5–25.0 | 13.5–25.4 | 13.4–24.1 | 13.8–24.0 | 28.8 | 44.4 | 128 | |||
| Nov | 16.7–27.3 | 15.9–27.6 | 14.7–27.1 | 15.4–26.6 | 109.6 | 56.4 | 49.8 | 107 | ||
| Dec | 17.2–32.0 | 17.9–29.1 | 17.1–31.9 | 17.5–29.0 | 44.2 | 42.8 | 53.6 | 94 | ||
| Variable | Units | 2020 | 2021 | 2022 |
|---|---|---|---|---|
| Clay | (%) | 17 | 22 | 29 |
| pH | (H2O) | 4.4 | 4.6 | 4.2 |
| SMP index | - | 6 | 5.7 | 5.5 |
| O.M. | (%) | 1.2 | 1.6 | 1.1 |
| P | mg/dm3 | 6.6 | 9.5 | 11 |
| K | mg/dm3 | 51 | 64 | 87 |
| Al | cmolc/dm3 | 1.1 | 0.8 | 1.1 |
| Ca | cmolc/dm3 | 0.6 | 1.1 | 0.3 |
| Mg | cmolc/dm3 | 0.2 | 0.3 | 0.1 |
| H + Al | cmolc/dm3 | 4.4 | 6.2 | 7.7 |
| CTCeffective | cmolc/dm3 | 5.36 | 7.78 | 8.4 |
| CTCpH7 | cmolc/dm3 | 17 | 19 | 7 |
| V | % | 17 | 19 | 7 |
| M | % | 53 | 33 | 66 |
| S | mg/dm3 | 5.6 | 11 | 18 |
| Cu | mg/dm3 | 1 | 1.1 | 1.2 |
| Zn | mg/dm3 | 1 | 1.3 | 1 |
| Mn | mg/dm3 | 18 | 67 | 43 |
| B | mg/dm3 | 0.3 | 0.2 | 0.4 |
| Effects | NT (Tillers Plant−1) | FM (g Plant−1) | LDM (g Plant−1) | SDM (g Plant−1) | IDM (g Plant−1) | TDM (g Plant−1) | LSR | HI | CT | FP | GH | PH (cm) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Full model | 36,404.77 | 37,909.20 | 34,132.21 | 16,407.65 | 12,348.90 | 33,497.38 | 6635.83 | −11,662.29 | 801.24 | −1197.18 | 90.26 | 5486.26 |
| Genotype | 36,489.74 | 37,956.31 | 34,146.62 | 16,418.08 | 12,364.09 | 33,532.62 | 6661.55 | −11,662.22 | 809.63 | −1197.25 | 211.52 | 5497.80 |
| Environment | 36,432.95 | 37,952.24 | 34,234.11 | 16,426.91 | 12,349.53 | 33,561.22 | 6648.39 | −11,264.48 | 834.62 | −1193.77 | 164.35 | 5511.13 |
| Interaction GxA | 42,801.97 | 40,979.57 | 35,216.43 | 17,486.52 | 13,648.72 | 35,223.72 | 8718.61 | −11,204.75 | 858.19 | −1171.75 | 138.45 | 5544.04 |
| LRTg (X2) | 84.97 ** | 47.11 ** | 14.41 ** | 10.43 ** | 15.19 ** | 35.24 ** | 25.72 ** | 0.07 ns | 8.39 ** | −0.07 ns | 121.26 ** | 11.54 ** |
| LRTe(X2) | 28.18 ** | 43.04 ** | 101.90 ** | 19.26 ** | 0.63 ns | 63.84 ** | 12.56 ** | 397.81 ** | 33.38 ** | 3.41 ns | 74.09 ** | 24.87 ** |
| LRTGxE(X2) | 6397.20 ** | 3070.37 ** | 1084.22 ** | 1078.87 ** | 1299.82 ** | 1726.34 ** | 2082.78 ** | 457.54 ** | 56.95 ** | 25.43 ** | 48.19 ** | 57.78 ** |
| µ | 207.96 | 155.18 | 62.32 | 6.55 | 3.86 | 75.89 | 2.93 | 0.82 | 3.03 | 0.86 | 2.87 | 57.02 |
| CVe (%) | 11.73 | 21.71 | 39.43 | 49.13 | 51.45 | 28.73 | 32.80 | 15.65 | 23.32 | 34.02 | 15.76 | 13.32 |
| CVg (%) | 17.51 | 14.39 | 11.81 | 12.33 | 14.58 | 13.07 | 13.99 | 0.99 | 10.00 | 2.34 | 19.37 | 3.44 |
| VIe (%) | 5.87 | 10.85 | 19.71 | 24.57 | 25.73 | 14.36 | 16.40 | 7.82 | 11.66 | 17.01 | 7.88 | 6.66 |
| VIg (%) | 8.76 | 7.19 | 5.91 | 6.17 | 7.29 | 6.54 | 7.00 | 0.50 | 5.00 | 1.17 | 9.69 | 1.72 |
| CVr | 1.49 | 0.66 | 0.30 | 0.25 | 0.28 | 0.45 | 0.43 | 0.06 | 0.43 | 0.07 | 1.23 | 0.26 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Silveira, D.C.; Mills, A.; Antoniolli, J.; Schneider de Ávila, V.; Pagani Sangineto, M.E.; Machado, J.M.; Weiler, R.L.; Brunes, A.P.; Simioni, C.; Dall’Agnol, M. Genetic Parameters, Prediction of Genotypic Values, and Forage Stability in Paspalum nicorae Parodi Ecotypes via REML/BLUP. Genes 2025, 16, 1164. https://doi.org/10.3390/genes16101164
Silveira DC, Mills A, Antoniolli J, Schneider de Ávila V, Pagani Sangineto ME, Machado JM, Weiler RL, Brunes AP, Simioni C, Dall’Agnol M. Genetic Parameters, Prediction of Genotypic Values, and Forage Stability in Paspalum nicorae Parodi Ecotypes via REML/BLUP. Genes. 2025; 16(10):1164. https://doi.org/10.3390/genes16101164
Chicago/Turabian StyleSilveira, Diógenes Cecchin, Annamaria Mills, Júlio Antoniolli, Victor Schneider de Ávila, Maria Eduarda Pagani Sangineto, Juliana Medianeira Machado, Roberto Luis Weiler, André Pich Brunes, Carine Simioni, and Miguel Dall’Agnol. 2025. "Genetic Parameters, Prediction of Genotypic Values, and Forage Stability in Paspalum nicorae Parodi Ecotypes via REML/BLUP" Genes 16, no. 10: 1164. https://doi.org/10.3390/genes16101164
APA StyleSilveira, D. C., Mills, A., Antoniolli, J., Schneider de Ávila, V., Pagani Sangineto, M. E., Machado, J. M., Weiler, R. L., Brunes, A. P., Simioni, C., & Dall’Agnol, M. (2025). Genetic Parameters, Prediction of Genotypic Values, and Forage Stability in Paspalum nicorae Parodi Ecotypes via REML/BLUP. Genes, 16(10), 1164. https://doi.org/10.3390/genes16101164






