Linking Genotype to Clinical Features in SMC1A-Related Phenotypes: From Cornelia de Lange Syndrome to Developmental and Epileptic Encephalopathy, a Comprehensive Review
Abstract
1. Introduction
2. Materials and Methods
3. Genetic, Epigenetic, and Molecular Mechanisms
3.1. Molecular Mechanisms of Disease Pathogenesis
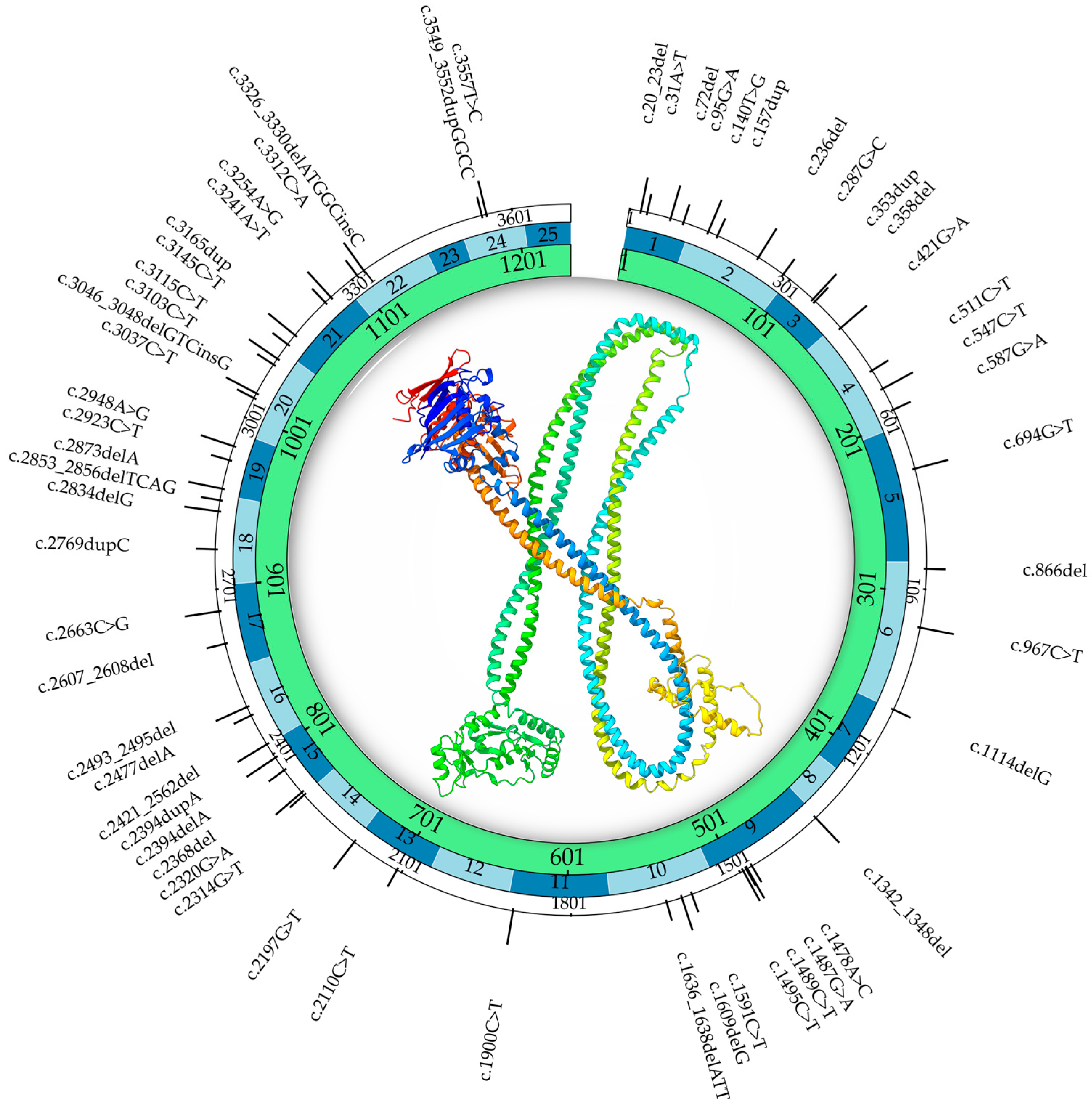
3.2. SMC1A Mutational Spectrum: Genotype–Phenotype Correlations and Clinical Implications
- Missense and in-frame deletions, typically associated with classic or mild CdLS. They preserve the reading frame and allow partial protein function. Phenotypes include minor facial dysmorphisms and some developmental delay, but absence of major limb defects [14].
- Nonsense and splice-site mutations. These often result in more severe neurological presentations like early-onset epileptic encephalopathy, with seizure onset before age 1 and features resembling the RTT (e.g., stereotypies, regression, profound intellectual disability) [19,49]. Notably, nonsense mutations cluster in regions crucial for cohesin function and have been shown to cause mRNA instability or altered splicing, resulting in loss of function [50].
3.3. X-Chromosome Inactivation Skewing and Other Epigenetic Modifiers
4. Clinical Phenotypes of SMC1A-Related Disorders
4.1. X-Chromosome Inactivation and Dosage Effects
4.2. Cdls vs. SMC1A-DEE: Clinical and Molecular Distinctions
4.3. Case Series
5. Diagnostic Approaches in Cornelia de Lange Syndrome and SMC1A-Related Developmental and Epileptic Encephalopathy
5.1. Current Molecular Strategies
5.2. Limits
6. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| ATM | Ataxia telangiectasia-mutated |
| ATR | Ataxia telangiectasia- and rad3-related |
| CdLS | Cornelia de Lange syndrome |
| CTCF | CCCTC-binding factor |
| DEE | Developmental and epileptic encephalopathy |
| iPSCs | Induced pluripotent stem cells |
| LoF | loss of function |
| NGS | Next-generation sequencing |
| NIPBL | Nipped-B like protein |
| RC-1 | Recombination protein complex |
| RT-PCR | Reverse transcription polymerase chain reaction |
| RTT | Rett syndrome |
| SMCs | Structural maintenance of chromosomes |
| SMC1A | Structural maintenance of chromosomes 1A |
| WES | Whole-exome sequencing |
| WGS | Whole-genome sequencing |
| Xa | X active |
| XCI | X-chromosome inactivation |
| Xi | X inactive |
References
- Yatskevich, S.; Rhodes, J.; Nasmyth, K. Organization of Chromosomal DNA by SMC Complexes. Annu. Rev. Genet. 2019, 53, 445–482. [Google Scholar] [CrossRef]
- Parenti, I.; Rovina, D.; Masciadri, M.; Cereda, A.; Azzollini, J.; Picinelli, C.; Limongelli, G.; Finelli, P.; Selicorni, A.; Russo, S.; et al. Overall and allele-specific expression of the SMC1A gene in female Cornelia de Lange syndrome patients and healthy controls. Epigenetics 2014, 9, 973–979. [Google Scholar] [CrossRef]
- Yuen, K.C.; Gerton, J.L. Taking cohesin and condensin in context. PLoS Genet. 2018, 14, e1007118. [Google Scholar] [CrossRef] [PubMed]
- Mannini, L.; Cucco, F.; Quarantotti, V.; Amato, C.; Tinti, M.; Tana, L.; Frattini, A.; Delia, D.; Krantz, I.D.; Jessberger, R.; et al. SMC1B is present in mammalian somatic cells and interacts with mitotic cohesin proteins. Sci. Rep. 2015, 5, 18472. [Google Scholar] [CrossRef] [PubMed]
- Gligoris, T.G.; Scheinost, J.C.; Burmann, F.; Petela, N.; Chan, K.L.; Uluocak, P.; Beckouet, F.; Gruber, S.; Nasmyth, K.; Lowe, J. Closing the cohesin ring: Structure and function of its Smc3-kleisin interface. Science 2014, 346, 963–967. [Google Scholar] [CrossRef]
- Schwarzer, W.; Abdennur, N.; Goloborodko, A.; Pekowska, A.; Fudenberg, G.; Loe-Mie, Y.; Fonseca, N.A.; Huber, W.; Haering, C.H.; Mirny, L.; et al. Two independent modes of chromatin organization revealed by cohesin removal. Nature 2017, 551, 51–56. [Google Scholar] [CrossRef]
- Cukrov, D.; Newman, T.A.C.; Leask, M.; Leeke, B.; Sarogni, P.; Patimo, A.; Kline, A.D.; Krantz, I.D.; Horsfield, J.A.; Musio, A. Antioxidant treatment ameliorates phenotypic features of SMC1A-mutated Cornelia de Lange syndrome in vitro and in vivo. Hum. Mol. Genet. 2018, 27, 3002–3011. [Google Scholar] [CrossRef]
- Musio, A. The multiple facets of the SMC1A gene. Gene 2020, 743, 144612. [Google Scholar] [CrossRef] [PubMed]
- Stursberg, S.; Riwar, B.; Jessberger, R. Cloning and characterization of mammalian SMC1 and SMC3 genes and proteins, components of the DNA recombination complexes RC-1. Gene 1999, 228, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Baranano, K.W.; Kimball, A.; Fong, S.L.; Egense, A.S.; Hudon, C.; Kline, A.D. Further Characterization of SMC1A Loss of Function Epilepsy Distinct From Cornelia de Lange Syndrome. J. Child. Neurol. 2022, 37, 390–396. [Google Scholar] [CrossRef]
- Cooper, D.N.; Ball, E.V.; Stenson, P.D.; Phillips, A.D.; Evans, K.; Heywood, S.; Chapman, M.; Hayden, M.J.; Shiel, J.; Beynon, S.; et al. The Human Gene Mutation Database. Available online: http://www.hgmd.cf.ac.uk/ac/index.php (accessed on 10 August 2025).
- Luppino, G.; Wasniewska, M.; Pepe, G.; Morabito, L.A.; Briuglia, S.; Moschella, A.; Franchina, F.; Lugarà, C.; Aversa, T.; Corica, D. Two Years of Growth Hormone Therapy in a Child with Severe Short Stature Due to Overlap Syndrome with a Novel SETD5 Gene Mutation: Case Report and Review of the Literature. Genes 2025, 16, 859. [Google Scholar] [CrossRef] [PubMed]
- Mannini, L.; Cucco, F.; Quarantotti, V.; Krantz, I.D.; Musio, A. Mutation spectrum and genotype-phenotype correlation in Cornelia de Lange syndrome. Hum. Mutat. 2013, 34, 1589–1596. [Google Scholar] [CrossRef] [PubMed]
- Huisman, S.; Mulder, P.A.; Redeker, E.; Bader, I.; Bisgaard, A.M.; Brooks, A.; Cereda, A.; Cinca, C.; Clark, D.; Cormier-Daire, V.; et al. Phenotypes and genotypes in individuals with SMC1A variants. Am. J. Med. Genet. A 2017, 173, 2108–2125. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.; Chen, L.; Wang, Z.; Ding, Y.; Liu, Y. A De Novo Frameshift Variant in SMC1A Causes Non-Classic Cornelia de Lange Syndrome with Epilepsy: A Case Report and Literature Review. Mol. Genet. Genomic Med. 2025, 13, e70058. [Google Scholar] [CrossRef]
- Pepe, G.; Coco, R.; Corica, D.; Luppino, G.; Morabito, L.A.; Lugara, C.; Abbate, T.; Zirilli, G.; Aversa, T.; Stagi, S.; et al. Endocrine disorders in Rett syndrome: A systematic review of the literature. Front. Endocrinol. 2024, 15, 1477227. [Google Scholar] [CrossRef]
- Parmeggiani, L.; Stanzial, F.; Menna, E.; Boni, E.; Manzoni, F.; Benedicenti, F.; Pellegrin, S. Early onset developmental and epileptic encephalopathy and Rett-like phenotype in a 15-year-old girl affected by Cornelia de Lange syndrome type 2 due to a SMC1A gene mutation. Epilepsy Behav. Rep. 2023, 24, 100634. [Google Scholar] [CrossRef]
- Symonds, J.D.; Joss, S.; Metcalfe, K.A.; Somarathi, S.; Cruden, J.; Devlin, A.M.; Donaldson, A.; DiDonato, N.; Fitzpatrick, D.; Kaiser, F.J.; et al. Heterozygous truncation mutations of the SMC1A gene cause a severe early onset epilepsy with cluster seizures in females: Detailed phenotyping of 10 new cases. Epilepsia 2017, 58, 565–575. [Google Scholar] [CrossRef]
- Gibellato, E.; Cianci, P.; Mariani, M.; Parma, B.; Huisman, S.; Smigiel, R.; Bisgaard, A.M.; Massa, V.; Gervasini, C.; Moretti, A.; et al. SMC1A epilepsy syndrome: Clinical data from a large international cohort. Am. J. Med. Genet. A 2024, 194, e63577. [Google Scholar] [CrossRef]
- De Koninck, M.; Losada, A. Cohesin Mutations in Cancer. Cold Spring Harb. Perspect. Med. 2016, 6, a026476. [Google Scholar] [CrossRef]
- Perea-Resa, C.; Wattendorf, L.; Marzouk, S.; Blower, M.D. Cohesin: Behind dynamic genome topology and gene expression reprogramming. Trends Cell Biol. 2021, 31, 760–773. [Google Scholar] [CrossRef]
- Grubert, F.; Srivas, R.; Spacek, D.V.; Kasowski, M.; Ruiz-Velasco, M.; Sinnott-Armstrong, N.; Greenside, P.; Narasimha, A.; Liu, Q.; Geller, B.; et al. Landscape of cohesin-mediated chromatin loops in the human genome. Nature 2020, 583, 737–743. [Google Scholar] [CrossRef]
- Sole-Ferran, M.; Losada, A. Cohesin in 3D: Development, differentiation, and disease. Genes. Dev. 2025, 39, 679–696. [Google Scholar] [CrossRef]
- Horsfield, J.A. Full circle: A brief history of cohesin and the regulation of gene expression. FEBS J. 2023, 290, 1670–1687. [Google Scholar] [CrossRef]
- Mannini, L.; Liu, J.; Krantz, I.D.; Musio, A. Spectrum and consequences of SMC1A mutations: The unexpected involvement of a core component of cohesin in human disease. Hum. Mutat. 2010, 31, 5–10. [Google Scholar] [CrossRef]
- Schar, P.; Fasi, M.; Jessberger, R. SMC1 coordinates DNA double-strand break repair pathways. Nucleic Acids Res. 2004, 32, 3921–3929. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.F.; Jiang, R.; Li, J.D.; Zhang, X.Y.; Zhao, P.; He, M.; Zhang, H.Z.; Sun, L.P.; Shi, D.L.; Zhang, G.X.; et al. SMC1A knockdown induces growth suppression of human lung adenocarcinoma cells through G1/S cell cycle phase arrest and apoptosis pathways in vitro. Oncol. Lett. 2013, 5, 749–755. [Google Scholar] [CrossRef]
- Ma, Z.; Lin, M.; Li, K.; Fu, Y.; Liu, X.; Yang, D.; Zhao, Y.; Zheng, J.; Sun, B. Knocking down SMC1A inhibits growth and leads to G2/M arrest in human glioma cells. Int. J. Clin. Exp. Pathol. 2013, 6, 862–869. [Google Scholar] [PubMed]
- Di Nardo, M.; Astigiano, S.; Baldari, S.; Pallotta, M.M.; Porta, G.; Pigozzi, S.; Antonini, A.; Emionite, L.; Frattini, A.; Valli, R.; et al. The synergism of SMC1A cohesin gene silencing and bevacizumab against colorectal cancer. J. Exp. Clin. Cancer Res. 2024, 43, 49. [Google Scholar] [CrossRef]
- Bozarth, X.L.; Lopez, J.; Fang, H.; Lee-Eng, J.; Duan, Z.; Deng, X. Phenotypes and Genotypes in Patients with SMC1A-Related Developmental and Epileptic Encephalopathy. Genes 2023, 14, 852. [Google Scholar] [CrossRef] [PubMed]
- Kline, A.D.; Moss, J.F.; Selicorni, A.; Bisgaard, A.M.; Deardorff, M.A.; Gillett, P.M.; Ishman, S.L.; Kerr, L.M.; Levin, A.V.; Mulder, P.A.; et al. Diagnosis and management of Cornelia de Lange syndrome: First international consensus statement. Nat. Rev. Genet. 2018, 19, 649–666. [Google Scholar] [CrossRef]
- Gil-Salvador, M.; Latorre-Pellicer, A.; Lucia-Campos, C.; Arnedo, M.; Darnaude, M.T.; Diaz de Bustamante, A.; Villares, R.; Palma Milla, C.; Puisac, B.; Musio, A.; et al. Case report: A novel case of parental mosaicism in SMC1A gene causes inherited Cornelia de Lange syndrome. Front. Genet. 2022, 13, 993064. [Google Scholar] [CrossRef]
- Chen, J.; Floyd, E.N.; Dawson, D.S.; Rankin, S. Cornelia de Lange Syndrome mutations in SMC1A cause cohesion defects in yeast. Genetics 2023, 225, iyad159. [Google Scholar] [CrossRef]
- Costantino, L.; Hsieh, T.S.; Lamothe, R.; Darzacq, X.; Koshland, D. Cohesin residency determines chromatin loop patterns. eLife 2020, 9, e59889. [Google Scholar] [CrossRef]
- Selivanovskiy, A.V.; Molodova, M.N.; Khrameeva, E.E.; Ulianov, S.V.; Razin, S.V. Liquid condensates: A new barrier to loop extrusion? Cell Mol. Life Sci. 2025, 82, 80. [Google Scholar] [CrossRef]
- Stortz, M.; Presman, D.M.; Levi, V. Transcriptional condensates: A blessing or a curse for gene regulation? Commun. Biol. 2024, 7, 187. [Google Scholar] [CrossRef] [PubMed]
- Morales, C.; Ruiz-Torres, M.; Rodriguez-Acebes, S.; Lafarga, V.; Rodriguez-Corsino, M.; Megias, D.; Cisneros, D.A.; Peters, J.M.; Mendez, J.; Losada, A. PDS5 proteins are required for proper cohesin dynamics and participate in replication fork protection. J. Biol. Chem. 2020, 295, 146–157. [Google Scholar] [CrossRef] [PubMed]
- Alonso-Gil, D.; Losada, A. NIPBL and cohesin: New take on a classic tale. Trends Cell Biol. 2023, 33, 860–871. [Google Scholar] [CrossRef] [PubMed]
- Gause, M.; Webber, H.A.; Misulovin, Z.; Haller, G.; Rollins, R.A.; Eissenberg, J.C.; Bickel, S.E.; Dorsett, D. Functional links between Drosophila Nipped-B and cohesin in somatic and meiotic cells. Chromosoma 2008, 117, 51–66. [Google Scholar] [CrossRef]
- Faure, A.J.; Schmidt, D.; Watt, S.; Schwalie, P.C.; Wilson, M.D.; Xu, H.; Ramsay, R.G.; Odom, D.T.; Flicek, P. Cohesin regulates tissue-specific expression by stabilizing highly occupied cis-regulatory modules. Genome Res. 2012, 22, 2163–2175. [Google Scholar] [CrossRef]
- Brooker, A.S.; Berkowitz, K.M. The roles of cohesins in mitosis, meiosis, and human health and disease. Methods Mol. Biol. 2014, 1170, 229–266. [Google Scholar] [CrossRef] [PubMed]
- Sarogni, P.; Palumbo, O.; Servadio, A.; Astigiano, S.; D’Alessio, B.; Gatti, V.; Cukrov, D.; Baldari, S.; Pallotta, M.M.; Aretini, P.; et al. Overexpression of the cohesin-core subunit SMC1A contributes to colorectal cancer development. J. Exp. Clin. Cancer Res. 2019, 38, 108. [Google Scholar] [CrossRef] [PubMed]
- Remeseiro, S.; Cuadrado, A.; Gomez-Lopez, G.; Pisano, D.G.; Losada, A. A unique role of cohesin-SA1 in gene regulation and development. EMBO J. 2012, 31, 2090–2102. [Google Scholar] [CrossRef]
- Liu, J.; Zhang, Z.; Bando, M.; Itoh, T.; Deardorff, M.A.; Clark, D.; Kaur, M.; Tandy, S.; Kondoh, T.; Rappaport, E.; et al. Transcriptional dysregulation in NIPBL and cohesin mutant human cells. PLoS Biol. 2009, 7, e1000119. [Google Scholar] [CrossRef]
- Izumi, K.; Nakato, R.; Zhang, Z.; Edmondson, A.C.; Noon, S.; Dulik, M.C.; Rajagopalan, R.; Venditti, C.P.; Gripp, K.; Samanich, J.; et al. Germline gain-of-function mutations in AFF4 cause a developmental syndrome functionally linking the super elongation complex and cohesin. Nat. Genet. 2015, 47, 338–344. [Google Scholar] [CrossRef]
- Slavin, T.P.; Lazebnik, N.; Clark, D.M.; Vengoechea, J.; Cohen, L.; Kaur, M.; Konczal, L.; Crowe, C.A.; Corteville, J.E.; Nowaczyk, M.J.; et al. Germline mosaicism in Cornelia de Lange syndrome. Am. J. Med. Genet. A 2012, 158A, 1481–1485. [Google Scholar] [CrossRef]
- Gardner, R.M.; Sutherland, G.R.; Shaffer, L.G. Chromosome Abnormalities and Genetic Counseling; Oxford University Press (OUP): New York, NY, USA, 2012. [Google Scholar]
- Jansen, S.; Kleefstra, T.; Willemsen, M.H.; de Vries, P.; Pfundt, R.; Hehir-Kwa, J.Y.; Gilissen, C.; Veltman, J.A.; de Vries, B.B.; Vissers, L.E. De novo loss-of-function mutations in X-linked SMC1A cause severe ID and therapy-resistant epilepsy in females: Expanding the phenotypic spectrum. Clin. Genet. 2016, 90, 413–419. [Google Scholar] [CrossRef]
- Lebrun, N.; Lebon, S.; Jeannet, P.Y.; Jacquemont, S.; Billuart, P.; Bienvenu, T. Early-onset encephalopathy with epilepsy associated with a novel splice site mutation in SMC1A. Am. J. Med. Genet. A 2015, 167A, 3076–3081. [Google Scholar] [CrossRef] [PubMed]
- Ye, Y.Z.; Duan, J.; Hu, Z.Q.; Cao, D.Z.; Liao, J.X.; Chen, L. Developmental and epileptic encephalopathy 85 caused by SMC1A gene truncating variation: 4 cases report and literature review. Zhonghua Er Ke Za Zhi 2022, 60, 583–587. [Google Scholar] [CrossRef] [PubMed]
- Generoso, S.F.; Neguembor, M.V.; Hershberg, E.A.; Sadreyev, R.I.; Kurimoto, K.; Yabuta, Y.; Ricci, R.; Audergon, P.; Bauer, M.; Saitou, M.; et al. Cohesin controls X chromosome structure remodeling and X-reactivation during mouse iPSC-reprogramming. Proc. Natl. Acad. Sci. USA 2023, 120, e2213810120. [Google Scholar] [CrossRef]
- Schierding, W.; Vickers, M.H.; O’Sullivan, J.M.; Cutfield, W.S. 9-Epigenetics. In Fetal and Neonatal Physiology, 5th ed.; Polin, R.A., Abman, S.H., Rowitch, D.H., Benitz, W.E., Fox, W.W., Eds.; Elsevier: Amsterdam, The Netherlands, 2017; pp. 89–100.e103. [Google Scholar]
- Brand, B.A.; Blesson, A.E.; Smith-Hicks, C.L. The Impact of X-Chromosome Inactivation on Phenotypic Expression of X-Linked Neurodevelopmental Disorders. Brain Sci. 2021, 11, 904. [Google Scholar] [CrossRef] [PubMed]
- Borck, G.; Zarhrate, M.; Bonnefont, J.P.; Munnich, A.; Cormier-Daire, V.; Colleaux, L. Incidence and clinical features of X-linked Cornelia de Lange syndrome due to SMC1L1 mutations. Hum. Mutat. 2007, 28, 205–206. [Google Scholar] [CrossRef]
- Pettersen, E.F.; Goddard, T.D.; Huang, C.C.; Meng, E.C.; Couch, G.S.; Croll, T.I.; Morris, J.H.; Ferrin, T.E. UCSF ChimeraX: Structure visualization for researchers, educators, and developers. Protein Sci. 2021, 30, 70–82. [Google Scholar] [CrossRef]
- Moss, J.; Penhallow, J.; Ansari, M.; Barton, S.; Bourn, D.; FitzPatrick, D.R.; Goodship, J.; Hammond, P.; Roberts, C.; Welham, A.; et al. Genotype-phenotype correlations in Cornelia de Lange syndrome: Behavioral characteristics and changes with age. Am. J. Med. Genet. A 2017, 173, 1566–1574. [Google Scholar] [CrossRef] [PubMed]
- Moncayo, J.A.; Ayala, I.N.; Argudo, J.M.; Aguirre, A.S.; Parwani, J.; Pachano, A.; Ojeda, D.; Cordova, S.; Mora, M.G.; Tapia, C.M.; et al. Understanding Protein Protocadherin-19 (PCDH19) Syndrome: A Literature Review of the Pathophysiology. Cureus 2022, 14, e25808. [Google Scholar] [CrossRef]
- Tehrani Fateh, S.; Mohammad Zadeh, N.; Salehpour, S.; Hashemi-Gorji, F.; Omidi, A.; Sadeghi, H.; Mirfakhraie, R.; Moghimi, P.; Keyvanfar, S.; Mohammadi Sarvaleh, S.; et al. Comprehensive review and expanding the genetic landscape of Cornelia-de-Lange spectrum: Insights from novel mutations and skin biopsy in exome-negative cases. BMC Med. Genom. 2024, 17, 20. [Google Scholar] [CrossRef]
- Spagnoli, C.; Fusco, C.; Pisani, F. Rett Syndrome Spectrum in Monogenic Developmental-Epileptic Encephalopathies and Epilepsies: A Review. Genes 2021, 12, 1157. [Google Scholar] [CrossRef] [PubMed]
- Goldstein, J.H.; Tim-Aroon, T.; Shieh, J.; Merrill, M.; Deeb, K.K.; Zhang, S.; Bass, N.E.; Bedoyan, J.K. Novel SMC1A frameshift mutations in children with developmental delay and epilepsy. Eur. J. Med. Genet. 2015, 58, 562–568. [Google Scholar] [CrossRef] [PubMed]
- Fang, H.; Deng, X.; Disteche, C.M. X-factors in human disease: Impact of gene content and dosage regulation. Hum. Mol. Genet. 2021, 30, R285–R295. [Google Scholar] [CrossRef]
- Berletch, J.B.; Ma, W.; Yang, F.; Shendure, J.; Noble, W.S.; Disteche, C.M.; Deng, X. Escape from X inactivation varies in mouse tissues. PLoS Genet. 2015, 11, e1005079. [Google Scholar] [CrossRef]
- Musio, A.; Selicorni, A.; Focarelli, M.L.; Gervasini, C.; Milani, D.; Russo, S.; Vezzoni, P.; Larizza, L. X-linked Cornelia de Lange syndrome owing to SMC1L1 mutations. Nat. Genet. 2006, 38, 528–530. [Google Scholar] [CrossRef]
- Deardorff, M.A.; Noon, S.E.; Krantz, I.D. Cornelia de Lange Syndrome; University of Washington: Seattle, WA, USA, 2020. [Google Scholar]
- Vetri, L.; Cali, F.; Saccone, S.; Vinci, M.; Chiavetta, N.V.; Carotenuto, M.; Roccella, M.; Costanza, C.; Elia, M. Whole Exome Sequencing as a First-Line Molecular Genetic Test in Developmental and Epileptic Encephalopathies. Int. J. Mol. Sci. 2024, 25, 1146. [Google Scholar] [CrossRef]
- Gonzalez Garcia, A.; Malone, J.; Li, H. A novel mosaic variant on SMC1A reported in buccal mucosa cells, albeit not in blood, of a patient with Cornelia de Lange-like presentation. Cold Spring Harb. Mol. Case Stud. 2020, 6, a005322. [Google Scholar] [CrossRef]
- Gil-Salvador, M.; Trujillano, L.; Lucia-Campos, C.; Del Rincon, J.; Pamplona, P.; Arnedo, M.; Puisac, B.; Marcos-Alcalde, I.; Gomez-Puertas, P.; Ramos, F.J.; et al. Postzygotic mosaicism in SMC1A and the first reported case of a female with Cornelia de Lange syndrome. Sci. Rep. 2025, 15, 20772. [Google Scholar] [CrossRef]
- Simkin, D.; Kiskinis, E. Modeling Pediatric Epilepsy Through iPSC-Based Technologies. Epilepsy Curr. 2018, 18, 240–245. [Google Scholar] [CrossRef]
- Zou, H.; Zhang, Q.; Liao, J.; Zou, D.; Hu, Z.; Li, B.; Chen, L.; Wen, J.; Zhao, X.; Zhang, V.W.; et al. Diagnostic efficiency of exome-based sequencing in pediatric patients with epilepsy. Front. Genet. 2024, 15, 1496411. [Google Scholar] [CrossRef] [PubMed]
- Crowgey, E.L.; Mahajan, N.; Wong, W.H.; Gopalakrishnapillai, A.; Barwe, S.P.; Kolb, E.A.; Druley, T.E. Error-corrected sequencing strategies enable comprehensive detection of leukemic mutations relevant for diagnosis and minimal residual disease monitoring. BMC Med. Genom. 2020, 13, 32. [Google Scholar] [CrossRef] [PubMed]
- Clement, K.; Farouni, R.; Bauer, D.E.; Pinello, L. AmpUMI: Design and analysis of unique molecular identifiers for deep amplicon sequencing. Bioinformatics 2018, 34, i202–i210. [Google Scholar] [CrossRef] [PubMed]
- Royer-Bertrand, B.; Cisarova, K.; Niel-Butschi, F.; Mittaz-Crettol, L.; Fodstad, H.; Superti-Furga, A. CNV Detection from Exome Sequencing Data in Routine Diagnostics of Rare Genetic Disorders: Opportunities and Limitations. Genes 2021, 12, 1427. [Google Scholar] [CrossRef]
- Moreno-Cabrera, J.M.; Del Valle, J.; Castellanos, E.; Feliubadalo, L.; Pineda, M.; Brunet, J.; Serra, E.; Capella, G.; Lazaro, C.; Gel, B. Evaluation of CNV detection tools for NGS panel data in genetic diagnostics. Eur. J. Hum. Genet. 2020, 28, 1645–1655. [Google Scholar] [CrossRef]
- Zhang, X.; Cai, M.; Ma, Y.; Chen, J.; Huang, S.; Cai, M.; Ding, Y.; Ma, D.; Gao, Q.; Hu, X.; et al. Minigene Assay as an Effective Molecular Diagnostic Strategy in Determining the Pathogenicity of Noncanonical Splice-Site Variants in FLCN. J. Mol. Diagn. 2023, 25, 110–120. [Google Scholar] [CrossRef]
- Weng, O.Y.; Li, Y.; Wang, L.Y. Modeling Epilepsy Using Human Induced Pluripotent Stem Cells-Derived Neuronal Cultures Carrying Mutations in Ion Channels and the Mechanistic Target of Rapamycin Pathway. Front. Mol. Neurosci. 2022, 15, 810081. [Google Scholar] [CrossRef]
- Francis, D.I.; Stark, Z.; Scheffer, I.E.; Tan, T.Y.; Murali, K.; Gallacher, L.; Amor, D.J.; Goel, H.; Downie, L.; Stutterd, C.A.; et al. Comparing saliva and blood for the detection of mosaic genomic abnormalities that cause syndromic intellectual disability. Eur. J. Hum. Genet. 2023, 31, 521–525. [Google Scholar] [CrossRef]
- Kim, S.H.; Kwon, S.S.; Park, M.R.; Lee, H.A.; Kim, J.H.; Cha, J.; Kim, S.; Baek, S.T.; Kim, S.H.; Lee, J.S.; et al. Detecting Low-Variant Allele Frequency Mosaic Pathogenic Variants of NF1, TSC2, and AKT3 Genes from Blood in Patients with Neurodevelopmental Disorders. J. Mol. Diagn. 2023, 25, 583–591. [Google Scholar] [CrossRef]
- Rehm, H.L.; Bale, S.J.; Bayrak-Toydemir, P.; Berg, J.S.; Brown, K.K.; Deignan, J.L.; Friez, M.J.; Funke, B.H.; Hegde, M.R.; Lyon, E.; et al. ACMG clinical laboratory standards for next-generation sequencing. Genet. Med. 2013, 15, 733–747. [Google Scholar] [CrossRef]
- Barisic, I.; Tokic, V.; Loane, M.; Bianchi, F.; Calzolari, E.; Garne, E.; Wellesley, D.; Dolk, H.; Group, E.W. Descriptive epidemiology of Cornelia de Lange syndrome in Europe. Am. J. Med. Genet. A 2008, 146A, 51–59. [Google Scholar] [CrossRef] [PubMed]
- Gil-Rodriguez, M.C.; Deardorff, M.A.; Ansari, M.; Tan, C.A.; Parenti, I.; Baquero-Montoya, C.; Ousager, L.B.; Puisac, B.; Hernandez-Marcos, M.; Teresa-Rodrigo, M.E.; et al. De novo heterozygous mutations in SMC3 cause a range of Cornelia de Lange syndrome-overlapping phenotypes. Hum. Mutat. 2015, 36, 454–462. [Google Scholar] [CrossRef] [PubMed]
- Balciuniene, J.; DeChene, E.T.; Akgumus, G.; Romasko, E.J.; Cao, K.; Dubbs, H.A.; Mulchandani, S.; Spinner, N.B.; Conlin, L.K.; Marsh, E.D.; et al. Use of a Dynamic Genetic Testing Approach for Childhood-Onset Epilepsy. JAMA Netw. Open 2019, 2, e192129. [Google Scholar] [CrossRef] [PubMed]
- Braunholz, D.; Obieglo, C.; Parenti, I.; Pozojevic, J.; Eckhold, J.; Reiz, B.; Braenne, I.; Wendt, K.S.; Watrin, E.; Vodopiutz, J.; et al. Hidden mutations in Cornelia de Lange syndrome limitations of sanger sequencing in molecular diagnostics. Hum. Mutat. 2015, 36, 26–29. [Google Scholar] [CrossRef]
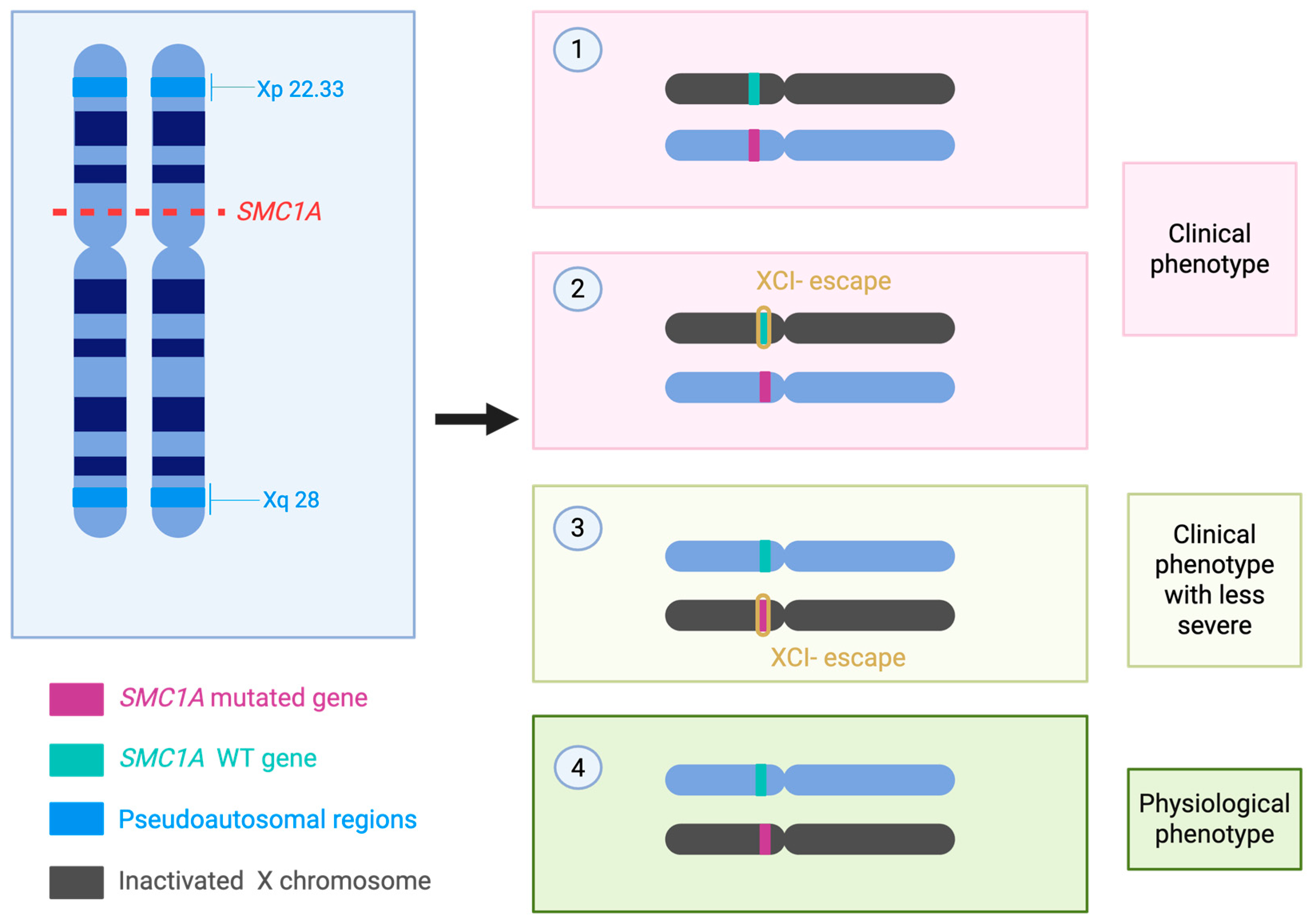
| Biological Pathway | Effect of SMC1A Mutation | References |
|---|---|---|
| Chromatid cohesion | Chromosomal instability and segregation errors | [25] |
| DNA damage repair (Homologous recombination HR, Non homologous end joining NHEJ) | Reduced repair fidelity, genomic instability | [26] |
| Gene transcription and chromatin | Misregulation of developmental genes and epigenetic control | [8] |
| Cell cycle control | G1/S or G2/M arrest, reduced proliferation, increased apoptosis | [27,28,29] |
| Neural development | Epilepsy, intellectual disability, Rett-like features in females | [30] |
| # | Nucleotide Changes | Amino Acid Changes | Type | XCI | Age of Seizure Onset | Speech | ID | Walking | MRI |
|---|---|---|---|---|---|---|---|---|---|
| 1 | c.20_23del | Ile7Argfs * 42 | Frameshift | Skewed 81:19 | 15 mo | None | Yes | No | N/A |
| 2 | c.31A > T | Asn11Tyr | Missense | N/A | 2.5 mo | N/A | Yes Yes | N/A | Normal |
| 3 | c.140T > G | Phe47Cys | Missense | Random 7:26 | 3 mo | None | Yes Yes | No | N/A |
| 4 | c.157dup | Thr53AsnfsX34 | Frameshift | N/A | 5 mo | N/A | Yes Yes | N/A | Cerebral volume loss |
| 5 | c.287G > C | Arg96Pro | Missense | Random 7:26 | 18 mo | N/A | Yes Yes | N/A | N/A |
| 6 | c.421G > A | Glu141Lys | Missense | Highly skewed 100:0 | 2.5 mo | None | Yes Yes | N/A | N/A |
| 7 | c.511C > T | Arg171Ter | Nonsense | Random | 4 wk | None | Yes Yes | No | Normal |
| 8 | c.615G > A | Glu2055 * | Splice-site | N/A | P3 (18) 4 mo; P12 (22): 13 mo | None | Yes Yes | No | P3: small hemorrhage along with posterior falx and tentorium |
| 9 | c.615G + 1G > C | Glu2055 * | Splice-site | Moderately skewed 83:17 | 1 mo | N/A | Yes Yes | No | N/A |
| 10 | c.615 + 5G > A | Glu2055 * | Splice-site | Random | 4 mo | None | Yes | No | Normal |
| 11 | c.694G > T | Glu232Ter | Nonsense | N/A | 4 mo | N/A | Yes | N/A | N/A |
| 12 | c.1113 + 1G > A | Gln371fs * | Splice-site | N/A | 7 wk | None | Yes | N/A | N/A |
| 13 | c.1114delG | Val372Ter | Nonsense | N/A | 7 wk | None | Yes | N/A | N/A |
| 14 | c.1487G > A | Arg496His | Missense | Skewed in patient P3 | 35 mo | limited | Yes | Yes | N/A |
| 15 | c.1489C > T | Arg497Ter | Nonsense | N/A | 4 mo | None | Yes | N/A | N/A |
| 16 | c.1495C > T | Arg499Ter | Nonsense | N/A | 40 mo | None | Yes | Yes | Microform of HPE, ventricular ectasia |
| 17 | c.1591C > T | Gln531Ter | Nonsense | N/A | 15 mo | None | Yes | Yes | Normal |
| 18 | c.1609delG | Val537Phefs * 42 | Frameshift | N/A | 5 mo | None | Yes | No | N/A |
| 19 | c.1636_1638delATT | Ile546del | In-frame | Moderately skewed 86:14, Mosaic | 2 mo | None | Yes | No | N/A |
| 20 | c.1900C > T | Gln634Ter | Nonsense | N/A | 3 mo | N/A | Yes | N/A | N/A |
| 21 | c.1911 + 1G > T | Thr638Valfs * 48 | Splice-site | N/A | Neonate | None | Yes | no | Small frontal lobe, thin CC |
| 22 | c.2197G > T | Glu733Ter | Nonsense | Random | 5 mo | None | Yes | No | Volume loss |
| 23 | c.236del | Asn788Lysfs * 10 | Frameshift | Random | 9 mo | None | Yes | No | Slightly enlarged ventricles, hypoplastic cerebellar vermis |
| 24 | c.2394delA | Lys798Asnfs * 31 | Frameshift | N/A | N/A | N/A | Yes | N/A | Semi-lobar HPE |
| 25 | c.2394dupA | Arg799fs | Frameshift | N/A | 4 mo | None | Yes | N/A | N/A |
| 26 | c.2421_2562del | Leu808Argfs * 6 | Frameshift | Moderately skewed 85:15 | 2 mo | None | Yes | N/A | Mild periventricular white matter abnormalities |
| 27 | c.2477delA | p825fs | Frameshift | N/A | 28 mo in P8; <1 mo in P9 | P8: None | P9: None | P8: Yes | P9: normal; P8: hemi-lobar HPE |
| 28 | c.2663C > G | Arg898Gly | Missense | Skewed; P1/P2 | 25 mo; P1 | None | Yes | P1: Yes; P2: normal until onset | P2: cerebellar atrophy |
| 29 | c.2769dupC | Ser924Glnfs * 2 | Frameshift | N/A | 24 mo | N/A | Yes | N/A | N/A |
| 30 | c.2834delG | Gly945Lysfs * 19 | Frameshift | N/A | N/A | N/A | Yes | N/A | Semi-lobar HPE |
| 31 | c.2853_2856delTCAG | Ser951Argfs * 12 | Frameshift | Skewed | 4 mo | N/A | severe | no | Mild ventriculomegaly |
| 32 | c.2873delA | Gln958Argfs * 6 | Frameshift | Random | 3 mo | None | Yes | No | N/A |
| 33 | c.2923C > T | Arg975Ter | Nonsense | 5 mo | Moderate to severe | no | Yes | Normal | |
| 34 | c.3046_3048delGTCinsG | Val1016Alafs * 28 | Frameshift | N/A | Neonate | None | Yes | No | Thin abnormal CC and minimal cerebellar atrophy |
| 35 | c.3115C > T | Gln1039Ter | Nonsense | Random 76:24 | 2 mo | None | Yes | No | N/A |
| 36 | c.3145C > T | Arg1049Ter | Nonsense | N/A | 5–6 wk | None | Yes | no | Cerebral volume loss |
| 37 | c.3241A > T | Ile1081Phe | Missense | N/A | 4 yr | None | Yes | Yes | Mildly prominent lateral ventricles |
| 38 | c.3285 + 1G > C | p1095 | Splice-site | N/A | Not reported | N/A | Yes | N/A | Middle interhemispheric variant, HPE |
| 39 | c.3312C > A | Tyr1107Ter | Nonsense | N/A | 12 yr | Normal before SE | Yes | Yes | Normal |
| 40 | c.3326_3330delATGGCinsC | Asp1109Alafs * 102 | Frameshift | N/A | 6 mo | None | Yes | no | Small cavum septum vergae |
| 41 | c.3549_3552dupGGCC | Ile1185glyfs * 23 | Frameshift | Random in P1 (59)-N/A-P3-(22) | 17 mo P2-; 16 (59)mo P13-(22) | N/A-Goldstein; P13-Baranano-limited | Yes | N/A-; P13(59)(22)-Yes | P2 (59): mild enlarged extra-axial spaces and slight thinning of CC |
| 42 | c.2320G > A | Asp774Asn | Missense | N/A | N/A | delay | Yes | At 4 yr | N/A |
| 43 | c.3103C > T | Arg1035Ter | Nonsense | ||||||
| 44 | c.1342_1348del | Ser448Lysfs * 6 | Frameshift/Truncating | ||||||
| 44 | c.967C > T | Gln323Ter | Nonsense | ||||||
| 45 | c.2368del | Arg790Glyfs * 8 | Frameshift/Truncating | ||||||
| 46 | c.866del | Ser289Ter | Nonsense | ||||||
| 47 | 3428bp Deletion | - | Microdeletion | ||||||
| 48 | c.2110C > T | Gln704Ter | Nonsense | ||||||
| 49 | c.3557T > C | Val1186Ala | Missense | ||||||
| 50 | c.358del | Glu120Asnfs * 2 | Frameshift/Truncating | ||||||
| 51 | c.72del | Gln25Argfs * 25 | Frameshift/Truncating | ||||||
| 52 | c.3165dup | Lys1056Insfs * 13 | Frameshift/Truncating | ||||||
| 53 | c.353dup | Ser118Argfs * 2 | Frameshift/Truncating | ||||||
| 54 | c.3037C > T | Gln1013Ter | Nonsense | ||||||
| 55 | c.2314G > T | Val772Leu | Missense | ||||||
| 56 | c.2948A > G | Tyr983Cys | Missense | ||||||
| 57 | Ex1 del | - | Microdeletion | ||||||
| 58 | c.95G > A | Gly326Glu | Missense | ||||||
| 59 | c.2607_2608del | Gln869Hisfs * 17 | Frameshift/Truncating | ||||||
| 60 | c.547C > T | Gln183Ter | Nonsense | ||||||
| 61 | c.2493_2496del | Asp831Glu/Gln832del | Missense/Del In-frame | ||||||
| 62 | c.1478A > C | Glu493Ala | Missense | ||||||
| 63 | c.3254A > G | Tyr1085Cys | Missense | ||||||
| 64 | c.587G > A | Arg196His | Missense |
| Primary Clinical Presentation | Multisystem Developmental Disorder | Severe Neurodevelopmental Disorder with Epilepsy |
|---|---|---|
| Neurological features | Mild-to-moderate intellectual disability; variable motor delay | Profound developmental impairment; hypotonia; regression possible |
| Seizures | Rare or absent | Early-onset, often refractory epilepsy |
| Neurodevelopmental course | Stable developmental impairment | Progressive decline related to epileptic activity |
| Facial dysmorphism | Distinctive CdLS facial features (arched eyebrows, long eyelashes, short nose, thin upper lip) | Absent or subtle facial dysmorphism |
| Growth parameters | Prenatal and postnatal growth retardation | Normal growth parameters in many cases |
| Other systemic involvement | Possible limb anomalies, gastrointestinal and cardiac defects | Less frequent systemic malformations |
| Genetic cause (SMC1A context) | Pathogenic variants in SMC1A (often missense or in-frame changes) | Pathogenic variants in SMC1A (often LoF, splice-site, or truncating) |
| Onset | Congenital | Infancy (usually within the first year) |
| Method | Advantages | Limitations | Specimen/Practical Notes | References |
|---|---|---|---|---|
| Multigene Next generation sequencing (NGS) panel (epilepsy/CdLS panels) | High diagnostic yield when phenotype is suggestive; targeted content increases interpretability; panels can combine single-nucleotide variant (SNV) and copy number variation (CNV) detection. | May miss very low-level mosaic variants if sequencing depth is limited; gene content and sensitivity vary between laboratories. | DNA from peripheral blood; collect additional mosaic-enriched tissue (buccal swab, uncultured skin fibroblasts) if mosaicism is suspected. Panels should include validated CNV detection. | [64] |
| Trio whole-exome sequencing (WES)/whole-genome sequencing (WGS) | Broad coverage for atypical presentations; facilitates de novo variant detection and parental phasing. | Standard WES/WGS pipelines often lack sensitivity for very low variant allele frequency (VAF) mosaicism; cost and analytic complexity are higher. | DNA from proband + parents (trio) recommended. Follow-up targeted testing or deep sequencing may be required for mosaic detection. | [65,69] |
| Ultra-deep targeted amplicon NGS | Sensitive detection of low VAF mosaic variants (with appropriate error-correction); ideal for confirmatory testing. | Requires locus-specific assay design, rigorous error-correction and validation; not genome-wide. | Multi-tissue sampling recommended (blood, buccal, fibroblasts). Typical target depths ≥ 500–1000× with validated limit of detection. | [70,71] |
| Copy-number analysis (Array comparative genomic hybridization (a-CGH)/Multiple ligation-dependent probe (MLPA)/NGS-based CNV calling) | Detects intragenic deletions/duplications not identified by SNV calling. | May have reduced sensitivity for low-level mosaic CNVs and cannot detect single-nucleotide variants. | DNA from blood or alternative tissues when mosaicism is suspected. Integrate with sequencing results. | [72,73] |
| RNA-based assays (Reverse transcription (RT-PCR), RNA-seq, minigene splicing assays) | Provide functional evidence for splice-altering variants and quantify transcript consequences; aid reclassification of variant of uncertain significance (VUS). | Require expression of SMC1A in the sampled tissue; patient RNA may be unavailable or low expressing. | Use tissue with relevant expression; if unavailable, deploy minigene constructs or fibroblast RNA where feasible. | [74] |
| Functional cellular models (Induced pluripotent stem cells iPSC-derived neurons, gene-edited cellular assays) | Directly assess cellular/neuronal consequences of variants and support pathogenicity assignments. | Resource-intensive, time-consuming, and generally confined to research or specialized diagnostic labs. | Derive iPSCs from patient fibroblasts or blood; used for mechanistic studies and advanced validation. | [68,75] |
| X-inactivation/allele-specific expression assays | Critical for interpreting variant effect in heterozygous females given X-linkage and variable X-escape; refines genotype–phenotype correlation. | Interpretation can be complex due to tissue-specific X-chromosome inactivation (XCI) patterns and mosaicism. | Perform XCI assays on multiple tissues when possible; combine with allele-specific expression and phenotypic data. | [66] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Astorino, M.F.; Speranza, D.; Luppino, G.; La Rosa, M.A.; Briuglia, S.; Calabrò, M. Linking Genotype to Clinical Features in SMC1A-Related Phenotypes: From Cornelia de Lange Syndrome to Developmental and Epileptic Encephalopathy, a Comprehensive Review. Genes 2025, 16, 1196. https://doi.org/10.3390/genes16101196
Astorino MF, Speranza D, Luppino G, La Rosa MA, Briuglia S, Calabrò M. Linking Genotype to Clinical Features in SMC1A-Related Phenotypes: From Cornelia de Lange Syndrome to Developmental and Epileptic Encephalopathy, a Comprehensive Review. Genes. 2025; 16(10):1196. https://doi.org/10.3390/genes16101196
Chicago/Turabian StyleAstorino, Maria Francesca, Desirèe Speranza, Giovanni Luppino, Maria Angela La Rosa, Silvana Briuglia, and Marco Calabrò. 2025. "Linking Genotype to Clinical Features in SMC1A-Related Phenotypes: From Cornelia de Lange Syndrome to Developmental and Epileptic Encephalopathy, a Comprehensive Review" Genes 16, no. 10: 1196. https://doi.org/10.3390/genes16101196
APA StyleAstorino, M. F., Speranza, D., Luppino, G., La Rosa, M. A., Briuglia, S., & Calabrò, M. (2025). Linking Genotype to Clinical Features in SMC1A-Related Phenotypes: From Cornelia de Lange Syndrome to Developmental and Epileptic Encephalopathy, a Comprehensive Review. Genes, 16(10), 1196. https://doi.org/10.3390/genes16101196






