Investigating the “Dark” Genome: First Report of Partington Syndrome in Cyprus
Abstract
1. Introduction
2. Materials and Methods
2.1. Family Description and Clinical Evaluation
2.2. Cytogenetic and Initial Molecular Testing
2.3. Next-Generation Sequencing
2.4. Reanalysis of “Dark” Genomic Regions
2.5. Sanger Sequencing Validation and Family Segregation Analysis
2.6. Gene and Variant Nomenclature
3. Results
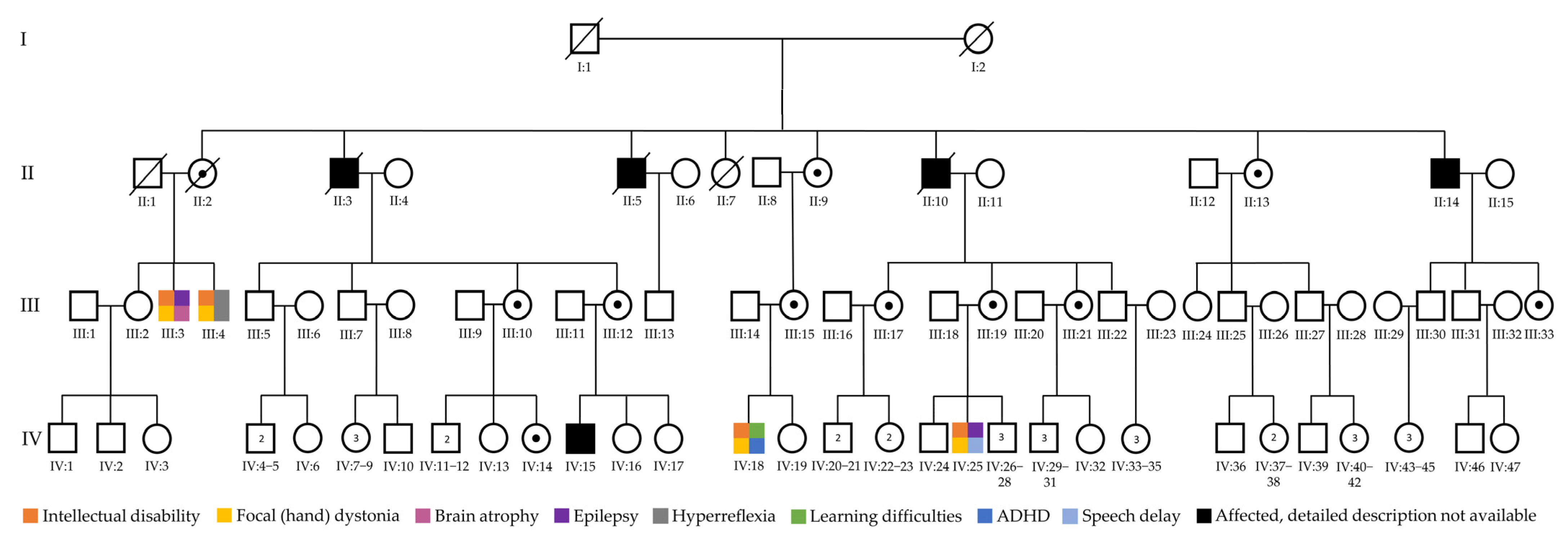
3.1. Clinical Findings and Initial Testing Results
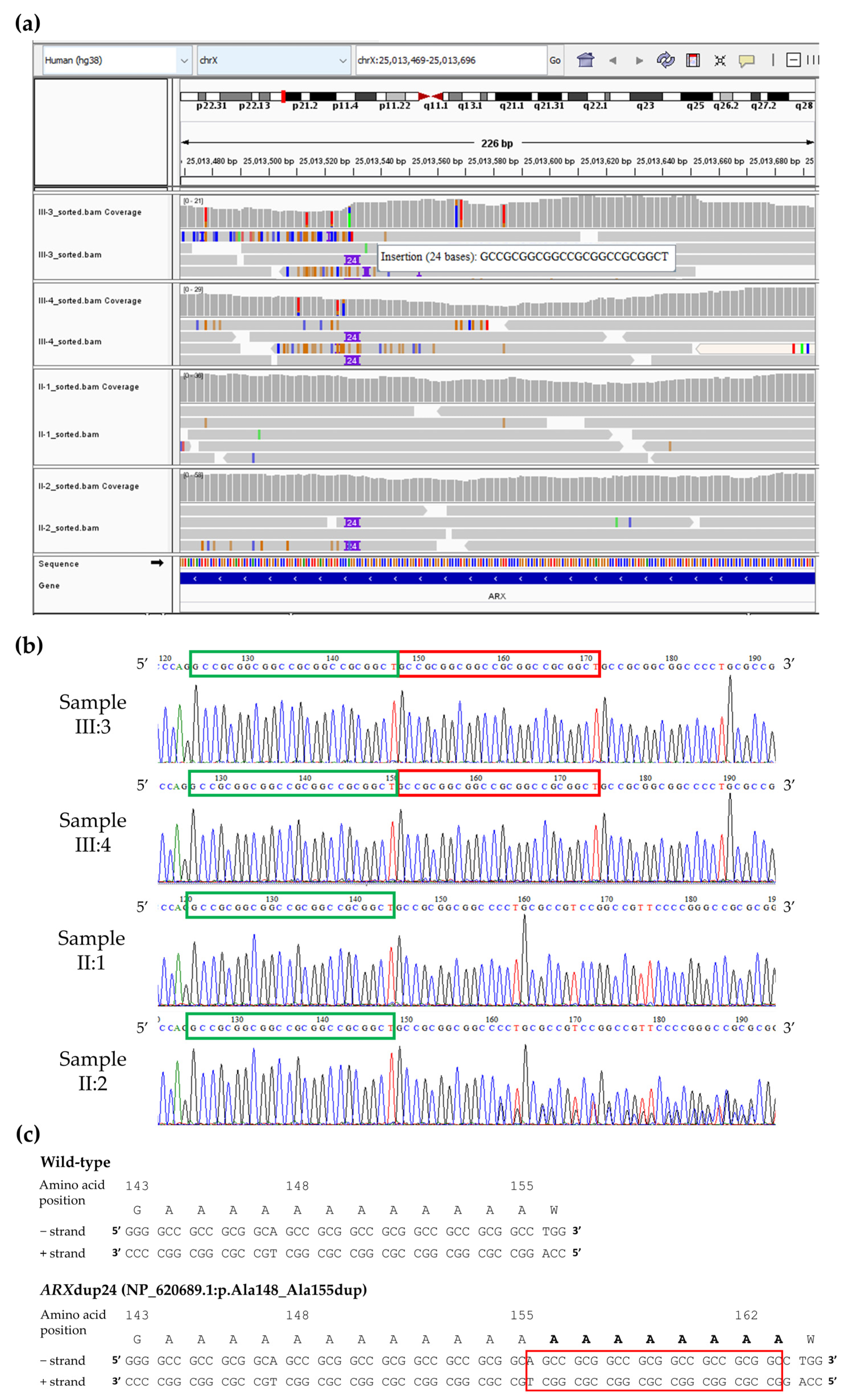
3.2. Unmasking Clinically Relevant “Dark” Genomic Regions
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| ALG13 | ALG13 UDP-N-acetylglucosaminyltransferase subunit |
| array-CGH | Array-Comparative Genomic Hybridization |
| ARX | Aristaless-related homeobox |
| CDS | Coding DNA sequences |
| chr | chromosome |
| CNV | Copy number variant |
| FISH | Fluorescence in situ hybridization |
| FMR1 | Fragile X Messenger Ribonucleoprotein 1 |
| FXS | Fragile X syndrome |
| HGVS | Human genome variation society |
| HPO | Human phenotype ontology |
| ID | Intellectual disability |
| IGV | Integrative genomics viewer |
| indel | Insertion/deletion |
| KDM5C | Lysine-specific demethylase 5C |
| NGS | Next-generation sequencing |
| SMN1 | Survival of motor neuron 1 |
| SMN2 | Survival of motor neuron 2 |
| SNV | Single-nucleotide variant |
| TBX1 | T-box transcription factor 1 |
| WES | Whole-exome sequencing |
| WGS | Whole-genome sequencing |
| XLID | X-linked intellectual disability |
References
- Stevenson, R.E.; Schwartz, C.E. X-linked intellectual disability: Unique vulnerability of the male genome. Dev. Disabil. Res. Rev. 2009, 15, 361–368. [Google Scholar] [CrossRef] [PubMed]
- Lubs, H.A.; Stevenson, R.E.; Schwartz, C.E. Fragile X and X-linked intellectual disability: Four decades of discovery. Am. J. Hum. Genet. 2012, 90, 579–590. [Google Scholar] [CrossRef] [PubMed]
- Schwartz, C.E.; Louie, R.J.; Toutain, A.; Skinner, C.; Friez, M.J.; Stevenson, R.E. X-Linked intellectual disability update 2022. Am. J. Med. Genet. A 2023, 191, 144–159. [Google Scholar] [CrossRef] [PubMed]
- Greenwood Genetic Center. XLID Genetic Research. Available online: https://ggc.org/xlid-genetic-research (accessed on 22 August 2025).
- Martin, J.P.; Bell, J. A Pedigree of Mental Defect Showing Sex-Linkage. J. Neurol. Psychiatry 1943, 6, 154–157. [Google Scholar] [CrossRef]
- Arveiler, B.; Alembik, Y.; Hanauer, A.; Jacobs, P.; Tranebjaerg, L.; Mikkelsen, M.; Puissant, H.; Piet, L.L.; Mandel, J.L. Linkage analysis suggests at least two loci for X-linked non-specific mental retardation. Am. J. Med. Genet. 1988, 30, 473–483. [Google Scholar] [CrossRef]
- Verkerk, A.J.; Pieretti, M.; Sutcliffe, J.S.; Fu, Y.H.; Kuhl, D.P.; Pizzuti, A.; Reiner, O.; Richards, S.; Victoria, M.F.; Zhang, F.P.; et al. Identification of a gene (FMR-1) containing a CGG repeat coincident with a breakpoint cluster region exhibiting length variation in fragile X syndrome. Cell 1991, 65, 905–914. [Google Scholar] [CrossRef]
- Bauters, M.; Van Esch, H.; Marynen, P.; Froyen, G. X chromosome array-CGH for the identification of novel X-linked mental retardation genes. Eur. J. Med. Genet. 2005, 48, 263–275. [Google Scholar] [CrossRef]
- Bashiardes, S.; Kousoulidou, L.; van Bokhoven, H.; Ropers, H.H.; Chelly, J.; Moraine, C.; de Brouwer, A.P.; Van Esch, H.; Froyen, G.; Patsalis, P.C. A new chromosome x exon-specific microarray platform for screening of patients with X-linked disorders. J. Mol. Diagn. 2009, 11, 562–568. [Google Scholar] [CrossRef]
- Whibley, A.C.; Plagnol, V.; Tarpey, P.S.; Abidi, F.; Fullston, T.; Choma, M.K.; Boucher, C.A.; Shepherd, L.; Willatt, L.; Parkin, G.; et al. Fine-scale survey of X chromosome copy number variants and indels underlying intellectual disability. Am. J. Hum. Genet. 2010, 87, 173–188. [Google Scholar] [CrossRef]
- Moyses-Oliveira, M.; Guilherme, R.S.; Meloni, V.A.; Di Battista, A.; de Mello, C.B.; Bragagnolo, S.; Moretti-Ferreira, D.; Kosyakova, N.; Liehr, T.; Carvalheira, G.M.; et al. X-linked intellectual disability related genes disrupted by balanced X-autosome translocations. Am. J. Med. Genet. B Neuropsychiatr. Genet. 2015, 168, 669–677. [Google Scholar] [CrossRef]
- Neri, G.; Schwartz, C.E.; Lubs, H.A.; Stevenson, R.E. X-linked intellectual disability update 2017. Am. J. Med. Genet A 2018, 176, 1375–1388. [Google Scholar] [CrossRef]
- de Ligt, J.; Willemsen, M.H.; van Bon, B.W.; Kleefstra, T.; Yntema, H.G.; Kroes, T.; Vulto-van Silfhout, A.T.; Koolen, D.A.; de Vries, P.; Gilissen, C.; et al. Diagnostic exome sequencing in persons with severe intellectual disability. N. Engl. J. Med. 2012, 367, 1921–1929. [Google Scholar] [CrossRef]
- Tzschach, A.; Grasshoff, U.; Beck-Woedl, S.; Dufke, C.; Bauer, C.; Kehrer, M.; Evers, C.; Moog, U.; Oehl-Jaschkowitz, B.; Di Donato, N.; et al. Next-generation sequencing in X-linked intellectual disability. Eur. J. Hum. Genet. 2015, 23, 1513–1518. [Google Scholar] [CrossRef] [PubMed]
- Hu, H.; Haas, S.A.; Chelly, J.; Van Esch, H.; Raynaud, M.; de Brouwer, A.P.; Weinert, S.; Froyen, G.; Frints, S.G.; Laumonnier, F.; et al. X-exome sequencing of 405 unresolved families identifies seven novel intellectual disability genes. Mol. Psychiatry 2016, 21, 133–148. [Google Scholar] [CrossRef] [PubMed]
- Field, M.J.; Kumar, R.; Hackett, A.; Kayumi, S.; Shoubridge, C.A.; Ewans, L.J.; Ivancevic, A.M.; Dudding-Byth, T.; Carroll, R.; Kroes, T.; et al. Different types of disease-causing noncoding variants revealed by genomic and gene expression analyses in families with X-linked intellectual disability. Hum. Mutat. 2021, 42, 835–847. [Google Scholar] [CrossRef] [PubMed]
- Wojcik, M.H.; Reuter, C.M.; Marwaha, S.; Mahmoud, M.; Duyzend, M.H.; Barseghyan, H.; Yuan, B.; Boone, P.M.; Groopman, E.E.; Delot, E.C.; et al. Beyond the exome: What’s next in diagnostic testing for Mendelian conditions. Am. J. Hum. Genet. 2023, 110, 1229–1248. [Google Scholar] [CrossRef]
- Mantere, T.; Kersten, S.; Hoischen, A. Long-Read Sequencing Emerging in Medical Genetics. Front. Genet. 2019, 10, 426. [Google Scholar] [CrossRef]
- Mantere, T.; Neveling, K.; Pebrel-Richard, C.; Benoist, M.; van der Zande, G.; Kater-Baats, E.; Baatout, I.; van Beek, R.; Yammine, T.; Oorsprong, M.; et al. Optical genome mapping enables constitutional chromosomal aberration detection. Am. J. Hum. Genet. 2021, 108, 1409–1422. [Google Scholar] [CrossRef]
- Ebbert, M.T.W.; Jensen, T.D.; Jansen-West, K.; Sens, J.P.; Reddy, J.S.; Ridge, P.G.; Kauwe, J.S.K.; Belzil, V.; Pregent, L.; Carrasquillo, M.M.; et al. Systematic analysis of dark and camouflaged genes reveals disease-relevant genes hiding in plain sight. Genome Biol. 2019, 20, 97. [Google Scholar] [CrossRef]
- Kritioti, E.; Theodosiou, A.; Parpaite, T.; Alexandrou, A.; Nicolaou, N.; Papaevripidou, I.; Séjourné, N.; Coste, B.; Christophidou-Anastasiadou, V.; Tanteles, G.A.; et al. Unravelling the genetic causes of multiple malformation syndromes: A whole exome sequencing study of the Cypriot population. PLoS ONE 2021, 16, e0253562. [Google Scholar] [CrossRef]
- Aristidou, C.; Theodosiou, A.; Alexandrou, A.; Papaevripidou, I.; Evangelidou, P.; Kosmaidou-Aravidou, Z.; Behjati, F.; Christophidou-Anastasiadou, V.; Tanteles, G.A.; Sismani, C. Exploring the Genetic Causality of Discordant Phenotypes in Familial Apparently Balanced Translocation Cases Using Whole Exome Sequencing. Genes 2022, 14, 82. [Google Scholar] [CrossRef]
- Krumm, N.; Sudmant, P.H.; Ko, A.; O’Roak, B.J.; Malig, M.; Coe, B.P.; Project, N.E.S.; Quinlan, A.R.; Nickerson, D.A.; Eichler, E.E. Copy number variation detection and genotyping from exome sequence data. Genome Res. 2012, 22, 1525–1532. [Google Scholar] [CrossRef]
- Raczy, C.; Petrovski, R.; Saunders, C.T.; Chorny, I.; Kruglyak, S.; Margulies, E.H.; Chuang, H.Y.; Kallberg, M.; Kumar, S.A.; Liao, A.; et al. Isaac: Ultra-fast whole-genome secondary analysis on Illumina sequencing platforms. Bioinformatics 2013, 29, 2041–2043. [Google Scholar] [CrossRef]
- Kim, S.; Scheffler, K.; Halpern, A.L.; Bekritsky, M.A.; Noh, E.; Kallberg, M.; Chen, X.; Kim, Y.; Beyter, D.; Krusche, P.; et al. Strelka2: Fast and accurate calling of germline and somatic variants. Nat. Methods 2018, 15, 591–594. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.; Schulz-Trieglaff, O.; Shaw, R.; Barnes, B.; Schlesinger, F.; Kallberg, M.; Cox, A.J.; Kruglyak, S.; Saunders, C.T. Manta: Rapid detection of structural variants and indels for germline and cancer sequencing applications. Bioinformatics 2016, 32, 1220–1222. [Google Scholar] [CrossRef] [PubMed]
- Boeva, V.; Popova, T.; Bleakley, K.; Chiche, P.; Cappo, J.; Schleiermacher, G.; Janoueix-Lerosey, I.; Delattre, O.; Barillot, E. Control-FREEC: A tool for assessing copy number and allelic content using next-generation sequencing data. Bioinformatics 2012, 28, 423–425. [Google Scholar] [CrossRef] [PubMed]
- Cingolani, P.; Platts, A.; Wang, L.L.; Coon, M.; Nguyen, T.; Wang, L.; Land, S.J.; Lu, X.; Ruden, D.M. A program for annotating and predicting the effects of single nucleotide polymorphisms, SnpEff: SNPs in the genome of Drosophila melanogaster strain w1118; iso-2; iso-3. Fly 2012, 6, 80–92. [Google Scholar] [CrossRef]
- Köhler, S.; Schulz, M.H.; Krawitz, P.; Bauer, S.; Dölken, S.; Ott, C.E.; Mundlos, C.; Horn, D.; Mundlos, S.; Robinson, P.N. Clinical Diagnostics in Human Genetics with Semantic Similarity Searches in Ontologies. Am. J. Hum. Genet. 2009, 85, 457–464. [Google Scholar] [CrossRef]
- Gargano, M.A.; Matentzoglu, N.; Coleman, B.; Addo-Lartey, E.B.; Anagnostopoulos, A.V.; Anderton, J.; Avillach, P.; Bagley, A.M.; Bakstein, E.; Balhoff, J.P.; et al. The Human Phenotype Ontology in 2024: Phenotypes around the world. Nucleic Acids Res. 2024, 52, D1333–D1346. [Google Scholar] [CrossRef]
- Untergasser, A.; Cutcutache, I.; Koressaar, T.; Ye, J.; Faircloth, B.C.; Remm, M.; Rozen, S.G. Primer3—New capabilities and interfaces. Nucleic Acids Res. 2012, 40, e115. [Google Scholar] [CrossRef]
- Perez, G.; Barber, G.P.; Benet-Pages, A.; Casper, J.; Clawson, H.; Diekhans, M.; Fischer, C.; Gonzalez, J.N.; Hinrichs, A.S.; Lee, C.M.; et al. The UCSC Genome Browser database: 2025 update. Nucleic Acids Res. 2025, 53, D1243–D1249. [Google Scholar] [CrossRef]
- Seal, R.L.; Braschi, B.; Gray, K.; Jones, T.E.M.; Tweedie, S.; Haim-Vilmovsky, L.; Bruford, E.A. Genenames.org: The HGNC resources in 2023. Nucleic Acids Res. 2023, 51, D1003–D1009. [Google Scholar] [CrossRef] [PubMed]
- Hart, R.K.; Fokkema, I.; DiStefano, M.; Hastings, R.; Laros, J.F.J.; Taylor, R.; Wagner, A.H.; den Dunnen, J.T. HGVS Nomenclature 2024: Improvements to community engagement, usability, and computability. Genome Med. 2024, 16, 149. [Google Scholar] [CrossRef] [PubMed]
- Ryan, N.M.; Corvin, A. Investigating the dark-side of the genome: A barrier to human disease variant discovery? Biol. Res. 2023, 56, 42. [Google Scholar] [CrossRef] [PubMed]
- Strømme, P.; Mangelsdorf, M.E.; Shaw, M.A.; Lower, K.M.; Lewis, S.M.E.; Bruyere, H.; Lütcherath, V.; Gedeon, Á.K.; Wallace, R.H.; Scheffer, I.E.; et al. Mutations in the human ortholog of Aristaless cause X-linked mental retardation and epilepsy. Nat. Genet. 2002, 30, 441–445. [Google Scholar] [CrossRef]
- Ohira, R.; Zhang, Y.H.; Guo, W.; Dipple, K.; Shih, S.L.; Doerr, J.; Huang, B.L.; Fu, L.J.; Abu-Khalil, A.; Geschwind, D.; et al. Human ARX gene: Genomic characterization and expression. Mol. Genet. Metab. 2002, 77, 179–188. [Google Scholar] [CrossRef]
- Bienvenu, T.; Poirier, K.; Friocourt, G.; Bahi, N.; Beaumont, D.; Fauchereau, F.; Jeema, L.B.; Zemni, R.; Vinet, M.C.; Francis, F.; et al. ARX, a novel Prd-class-homeobox gene highly expressed in the telencephalon, is mutated in X-linked mental retardation. Hum. Mol. Genet. 2002, 11, 981–991. [Google Scholar] [CrossRef]
- Seufert, D.W.; Prescott, N.L.; El-Hodiri, H.M. Xenopus aristaless-related homeobox (xARX) gene product functions as both a transcriptional activator and repressor in forebrain development. Dev. Dyn. 2005, 232, 313–324. [Google Scholar] [CrossRef]
- McKenzie, O.; Ponte, I.; Mangelsdorf, M.; Finnis, M.; Colasante, G.; Shoubridge, C.; Stifani, S.; Gecz, J.; Broccoli, V. Aristaless-related homeobox gene, the gene responsible for West syndrome and related disorders, is a Groucho/transducin-like enhancer of split dependent transcriptional repressor. Neuroscience 2007, 146, 236–247. [Google Scholar] [CrossRef]
- Gecz, J.; Cloosterman, D.; Partington, M. ARX: A gene for all seasons. Curr. Opin. Genet. Dev. 2006, 16, 308–316. [Google Scholar] [CrossRef]
- Poirier, K.; Lacombe, D.; Gilbert-Dussardier, B.; Raynaud, M.; Desportes, V.; de Brouwer, A.P.; Moraine, C.; Fryns, J.P.; Ropers, H.H.; Beldjord, C.; et al. Screening of ARX in mental retardation families: Consequences for the strategy of molecular diagnosis. Neurogenetics 2006, 7, 39–46. [Google Scholar] [CrossRef]
- Shoubridge, C.; Fullston, T.; Gecz, J. ARX spectrum disorders: Making inroads into the molecular pathology. Hum. Mutat. 2010, 31, 889–900. [Google Scholar] [CrossRef]
- Mahmoud, M.; Harting, J.; Corbitt, H.; Chen, X.; Jhangiani, S.N.; Doddapaneni, H.; Meng, Q.; Han, T.; Lambert, C.; Zhang, S.; et al. Closing the gap: Solving complex medically relevant genes at scale. medRxiv 2024. [Google Scholar] [CrossRef] [PubMed]
- ACGS Best Practice Guidelines for Variant Classification in Rare Disease 2024. Available online: https://www.genomicseducation.hee.nhs.uk/wp-content/uploads/2024/08/ACGS-2024_UK-practice-guidelines-for-variant-classification.pdf (accessed on 30 July 2025).
- Poeta, L.; Fusco, F.; Drongitis, D.; Shoubridge, C.; Manganelli, G.; Filosa, S.; Paciolla, M.; Courtney, M.; Collombat, P.; Lioi, M.B.; et al. A regulatory path associated with X-linked intellectual disability and epilepsy links KDM5C to the polyalanine expansions in ARX. Am. J. Hum. Genet. 2013, 92, 114–125. [Google Scholar] [CrossRef] [PubMed]
- Stromme, P.; Mangelsdorf, M.E.; Scheffer, I.E.; Gecz, J. Infantile spasms, dystonia, and other X-linked phenotypes caused by mutations in Aristaless related homeobox gene, ARX. Brain Dev. 2002, 24, 266–268. [Google Scholar] [CrossRef] [PubMed]
- Kato, M.; Das, S.; Petras, K.; Kitamura, K.; Morohashi, K.I.; Abuelo, D.N.; Barr, M.; Bonneau, D.; Brady, A.F.; Carpenter, N.J.; et al. Mutations of ARX Are Associated with Striking Pleiotropy and Consistent Genotype-Phenotype Correlation. Hum. Mutat. 2004, 23, 147–159. [Google Scholar] [CrossRef]
- Stepp, M.L.; Cason, A.L.; Finnis, M.; Mangelsdorf, M.; Holinski-Feder, E.; Macgregor, D.; MacMillan, A.; Holden, J.J.; Gecz, J.; Stevenson, R.E.; et al. XLMR in MRX families 29, 32, 33 and 38 results from the dup24 mutation in the ARX (Aristaless related homeobox) gene. BMC Med. Genet. 2005, 6, 16. [Google Scholar] [CrossRef]
- Kato, M.; Das, S.; Petras, K.; Sawaishi, Y.; Dobyns, W.B. Polyalanine expansion of ARX associated with cryptogenic West syndrome. Neurology 2003, 61, 267–276. [Google Scholar] [CrossRef]
- Partington, M.W.; Mulley, J.C.; Sutherland, G.R.; Hockey, A.; Thode, A.; Turner, G. X-linked mental retardation with dystonic movements of the hands. Am. J. Med. Genet. 1988, 30, 251–262. [Google Scholar] [CrossRef]
- Partington, M.W.; Turner, G.; Boyle, J.; Gecz, J. Three new families with X-linked mental retardation caused by the 428–451dup(24bp) mutation in ARX. Clin. Genet. 2004, 66, 39–45. [Google Scholar] [CrossRef]
- Gedeon, A.; Partington, M.; Mulley, J. X-linked mental retardation with dystonic movements of the hands (PRTS): Revisited. Am. J. Med. Genet. 1994, 51, 565–568. [Google Scholar] [CrossRef]
- Turner, G.; Partington, M.; Kerr, B.; Mangelsdorf, M.; Gecz, J. Variable expression of mental retardation, autism, seizures, and dystonic hand movements in two families with an identical ARX gene mutation. Am. J. Med. Genet. 2002, 112, 405–411. [Google Scholar] [CrossRef]
- Frints, S.G.; Froyen, G.; Marynen, P.; Willekens, D.; Legius, E.; Fryns, J.P. Re-evaluation of MRX36 family after discovery of an ARX gene mutation reveals mild neurological features of Partington syndrome. Am. J. Med. Genet. 2002, 112, 427–428. [Google Scholar] [CrossRef]
- Frints, S.G.; Borghgraef, M.; Froyen, G.; Marynen, P.; Fryns, J.P. Clinical study and haplotype analysis in two brothers with Partington syndrome. Am. J. Med. Genet. 2002, 112, 361–368. [Google Scholar] [CrossRef]
- Gras, M.; Heide, S.; Keren, B.; Valence, S.; Garel, C.; Whalen, S.; Jansen, A.C.; Keymolen, K.; Stouffs, K.; Jennesson, M.; et al. Further characterisation of ARX-related disorders in females due to inherited or de novo variants. J. Med. Genet. 2024, 61, 103–108. [Google Scholar] [CrossRef]
- Landrum, M.J.; Chitipiralla, S.; Brown, G.R.; Chen, C.; Gu, B.; Hart, J.; Hoffman, D.; Jang, W.; Kaur, K.; Liu, C.; et al. ClinVar: Improvements to accessing data. Nucleic Acids Res. 2020, 48, D835–D844. [Google Scholar] [CrossRef]
| Phenotype/ Initial Test | Case III:3 | Case III:4 |
|---|---|---|
| Abnormality of mental function | Mild-moderate intellectual disability, psychiatric abnormalities, irritability and anger tendencies. | Mild-moderate intellectual disability. |
| Abnormal brain morphology | Brain atrophy (signs of bilateral hippocampal atrophy) evidenced at the age of 56. | NA 1 |
| Seizures | Yes, generalised tonic–clonic (grand mal) seizures initiating at the age of 3. Last reported seizure at the age of 22. Seizures were controlled with antiepileptic drugs. | No |
| Abnormality of movement | Focal (hand) dystonia and gait disturbance. | Focal (hand) dystonia and hyperreflexia. |
| Abnormal speech pattern | Dysarthria | NA 1 |
| Dysmorphic features | Relatively large ears, wide nasal base with broad nasal tip. | Protruding large ears, pinched nose. |
| Chromosomal analysis | Apparently normal, 46,XY | Apparently normal, 46,XY |
| chrX-specific array/ Array-CGH | Apparently normal constitution | Apparently normal constitution |
| Fragile X testing | Negative (30 CGG repeats) | Negative (21 CGG repeats) |
| WES/WGS | Negative | Negative |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Aristidou, C.; Theodosiou, A.; Antoniou, P.; Alexandrou, A.; Papaevripidou, I.; Kousoulidou, L.; Koutsou, P.; Georghiou, A.; Delikurt, T.; Spanou, E.; et al. Investigating the “Dark” Genome: First Report of Partington Syndrome in Cyprus. Genes 2025, 16, 1224. https://doi.org/10.3390/genes16101224
Aristidou C, Theodosiou A, Antoniou P, Alexandrou A, Papaevripidou I, Kousoulidou L, Koutsou P, Georghiou A, Delikurt T, Spanou E, et al. Investigating the “Dark” Genome: First Report of Partington Syndrome in Cyprus. Genes. 2025; 16(10):1224. https://doi.org/10.3390/genes16101224
Chicago/Turabian StyleAristidou, Constantia, Athina Theodosiou, Pavlos Antoniou, Angelos Alexandrou, Ioannis Papaevripidou, Ludmila Kousoulidou, Pantelitsa Koutsou, Anthi Georghiou, Türem Delikurt, Elena Spanou, and et al. 2025. "Investigating the “Dark” Genome: First Report of Partington Syndrome in Cyprus" Genes 16, no. 10: 1224. https://doi.org/10.3390/genes16101224
APA StyleAristidou, C., Theodosiou, A., Antoniou, P., Alexandrou, A., Papaevripidou, I., Kousoulidou, L., Koutsou, P., Georghiou, A., Delikurt, T., Spanou, E., Salameh, N., Evangelidou, P., Christodoulou, K., Verloes, A., Christophidou-Anastasiadou, V., Tanteles, G. A., & Sismani, C. (2025). Investigating the “Dark” Genome: First Report of Partington Syndrome in Cyprus. Genes, 16(10), 1224. https://doi.org/10.3390/genes16101224






