Identification of FASN Gene Polymorphisms, Expression and Their Relationship with Body Size Traits in Guizhou White Goat (Capra hircus) with Different Genders
Abstract
1. Introduction
2. Materials and Methods
2.1. Animals and Data Collection
2.2. Primer Design and PCR Amplification
2.3. Measurements and Statistical Analysis
3. Results
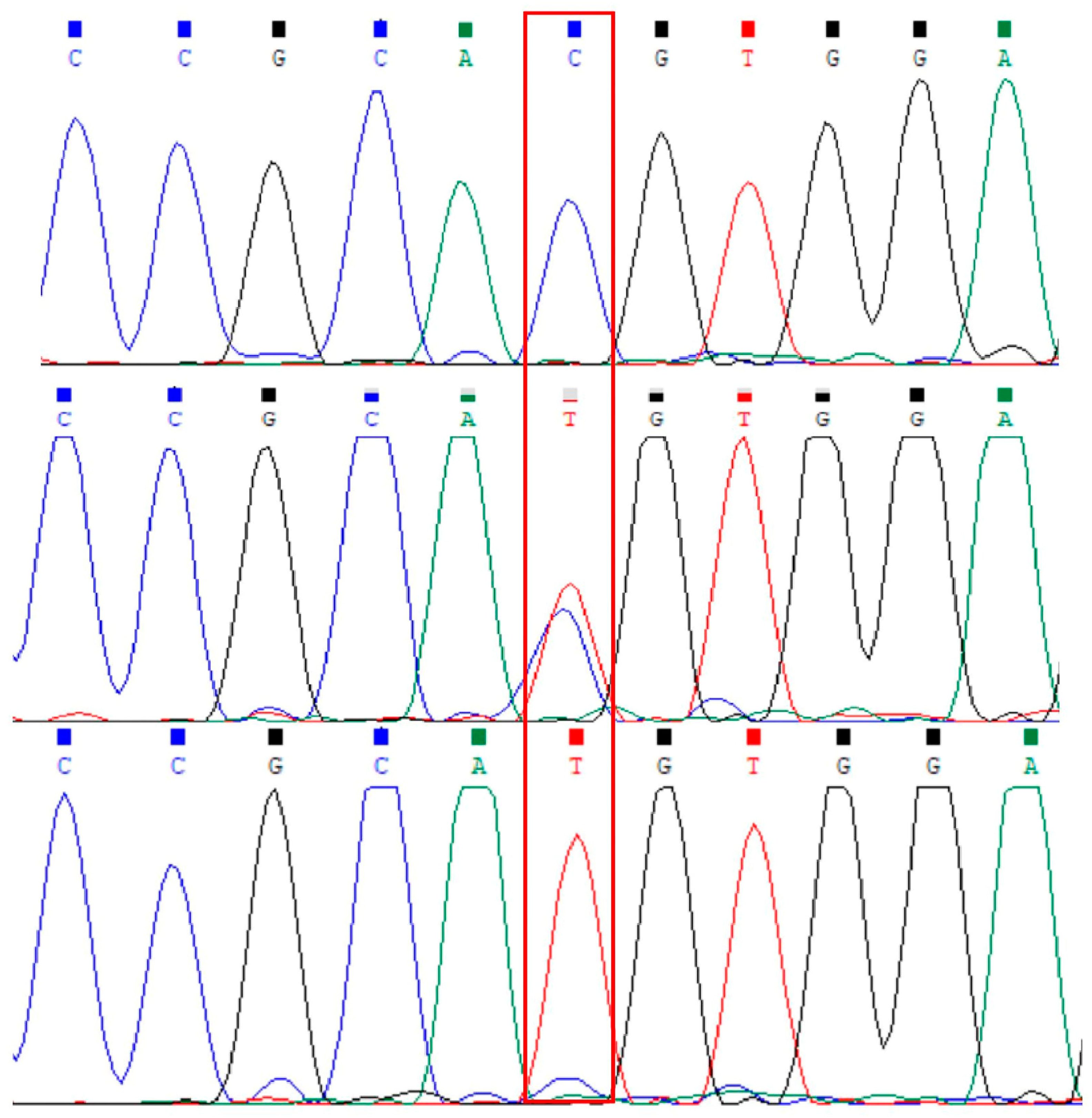
3.1. Analysis of the Polymorphism of the FASN Gene
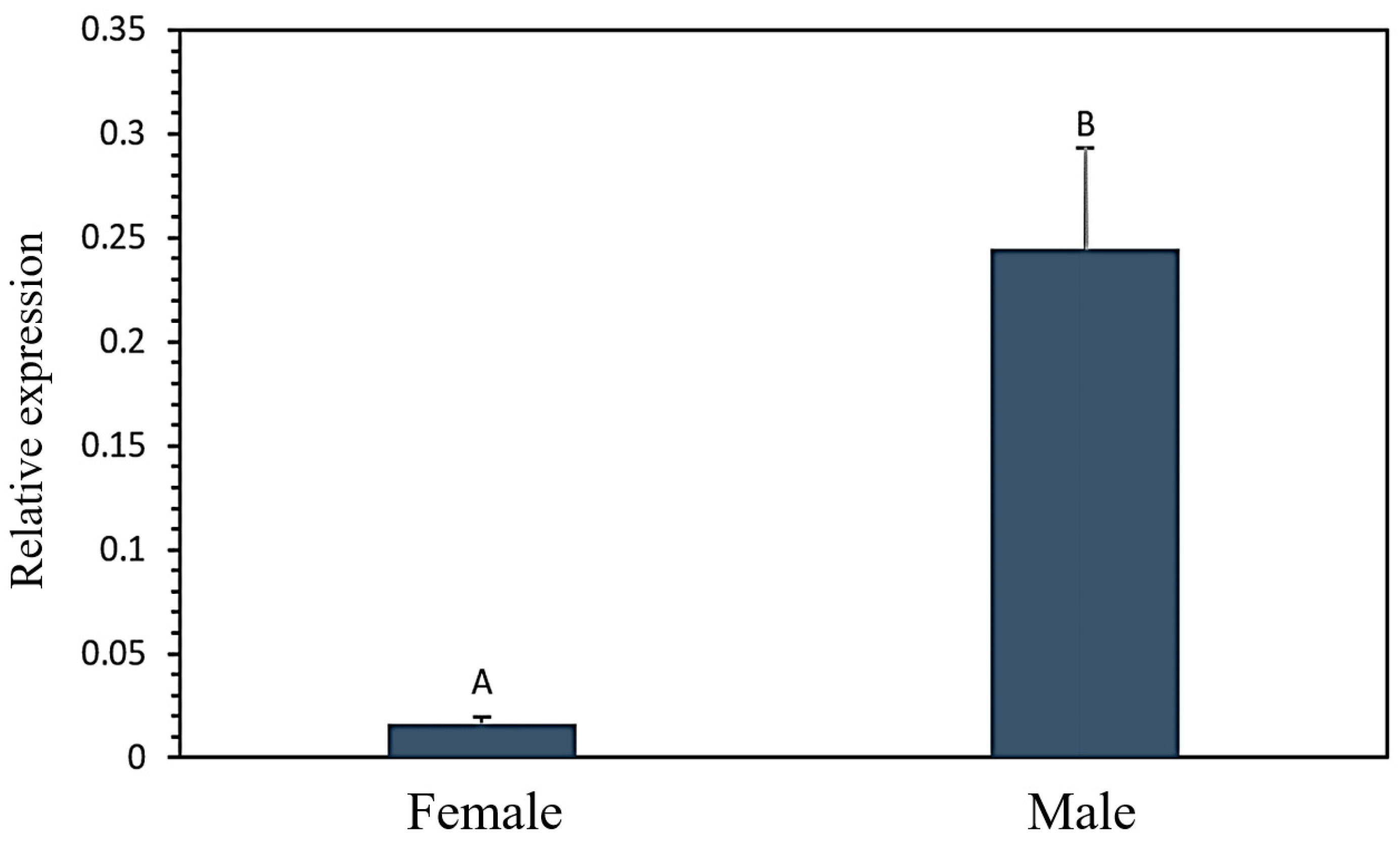
3.2. Analysis of the Differential Expression Level of the FASN Gene in Male and Female Goats
3.3. Effect of Variation in FASN on Body Size Traits
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Conflicts of Interest
References
- Semenkovich, C.F. Regulation of fatty acid synthase (FAS). Prog. Lipid Res. 1997, 36, 43–53. [Google Scholar] [CrossRef] [PubMed]
- Angeles, T.S.; Hudkins, R.L. Recent advances in targeting the fatty acid biosynthetic pathway using fatty acid synthase inhibitors. Expert Opin. Drug Dis. 2016, 11, 1187–1199. [Google Scholar] [CrossRef] [PubMed]
- Smith, S.; Witkowski, A.; Joshi, A.K. Structural and functional organization of the animal fatty acid synthase. Prog. Lipid Res. 2003, 42, 289–317. [Google Scholar] [CrossRef] [PubMed]
- Chirala, S.S.; Jayakumar, A.; Gu, Z.W. Human fatty acid synthase: Role of interdomain in the formation of catalytically active synthase dimer. Proc. Natl. Acad. Sci. USA 2001, 98, 3104–3108. [Google Scholar] [CrossRef] [PubMed]
- Maria, G.; Slawomir, S.; Katarzyna, R.; Maciej, S.; Marek, S. Pig fatness in relation to FASN and INSIG2 genes polymorphism and their transcript level. Mol. Biol. Rep. 2016, 43, 381–389. [Google Scholar]
- Zhao, L.; Li, F.; Liu, T.; Yuan, L.; Zhang, X.; Zhang, D.; Li, X.; Zhang, Y.; Zhao, Y.; Song, Q.; et al. Ovine ELOVL5 and FASN genes polymorphisms and their correlations with sheep tail fat deposition. Gene 2021, 807, 145954. [Google Scholar] [CrossRef] [PubMed]
- Yong, Q.; Zhiguo, H.; Qifa, L.; Zhenshan, L.; Chengli, H.; Guoqing, S.; Rong, D.; Zhuang, X. Developmental changes of the FAS and HSL mRNA expression and their effects on the content of intramuscular fat in Kazak and Xinjiang Sheep. J. Genet. Genom. 2007, 34, 909–917. [Google Scholar]
- Liu, C.; Liu, X.; Yuan, Z.; Zhang, X.; Li, F.; Yue, X. Expression profile of FASN gene and association of its polymorphisms with intramuscular fat content in Hu sheep. Anim. Biotechnol. 2023, 34, 4347–4356. [Google Scholar] [CrossRef]
- Gao, Y.Y.; Cheng, G.; Cheng, Z.X.; Bao, C.; Yamada, T.; Cao, G.F.; Bao, S.Q.; Schreurs, N.M.; Zan, L.S.; Tong, B. Association of variants in FABP4, FASN, SCD, SREBP1 and TCAP genes with intramuscular fat, carcass traits and body size in Chinese Qinchuan cattle. Meat Sci. 2022, 192, 108882. [Google Scholar] [CrossRef]
- Raza, A.H.S.; Gui, L.; Khan, R.; Schreurs, M.N.; Xiaoyu, W.; Wu, S.; Mei, S.; Wang, L.; Ma, X.; Wei, D.; et al. Association between FASN gene polymorphisms ultrasound carcass traits and intramuscular fat in Qinchuan cattle. Gene 2018, 645, 55–59. [Google Scholar] [CrossRef]
- Roy, R.; Taourit, S.; Zaragoza, P.; Eggen, A.; Rodellar, C. Genomic structure and alternative transcript of bovine fatty acid synthase gene (FASN): Comparative analysis of the FASN gene between monogastric and ruminant species. Cytogenet. Genome Res. 2005, 111, 65–73. [Google Scholar] [CrossRef]
- Cesar, A.S.M.; Regitano, L.C.A.; Koltes, J.E.; Fritz-Waters, E.R.; Lanna, D.P.D.; Gasparin, G.; Mourão, G.B.; Oliveira, P.S.N.; Reecy, J.M.; Coutinho, L.L. Putative regulatory factors associated with intramuscular fat content. PLoS ONE 2015, 10, e0128350. [Google Scholar] [CrossRef] [PubMed]
- Hausman, G.J.; Dodson, M.V.; Ajuwon, K.; Azain, M.; Barnes, K.M.; Guan, L.L.; Jiang, Z.; Poulos, S.P.; Sainz, R.D.; Smith, S.; et al. Boardinvited review: The biology and regulation of preadipocytes and adipocytes in meat animals. J. Anim. Sci. 2009, 87, 1218–1246. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.N.; Qi, Z.; Li, X.L.; Xiang, J.Z. Nutritional regulation and molecular mechanism of intramuscular fat deposition. Chin. J. Anim. Nutr. 2020, 32, 2947–2958. [Google Scholar]
- Li, T.; Meng, W.; Liu, T.C.; Wang, Y.Z.; Zhang, M. Sex differences in FASN protein concentrations in urinary exosomes related to serum triglycerides levels in healthy adults. Lipids Health Dis. 2023, 22, 176. [Google Scholar] [CrossRef] [PubMed]
- Ji, S.; Yang, R.; Lu, C.; Qiu, Z.; Yan, C.; Zhao, Z. Differential expression of PPAR gamma, FASN, and ACADM genes in various adipose tissues and longissimus dorsi muscle from Yanbian Yellow cattle and Yan Yellow cattle. Asian Australas. J. Anim. Sci. 2014, 27, 10–18. [Google Scholar] [CrossRef]
- Shi, B.; Jiang, Y.; Chen, Y.; Zhao, Z.; Zhou, H.; Luo, Y.; Hu, J.; Hickford, J.G.H. Variation in the Fatty Acid Synthase gene (FASN) and its association with milk traits in Gannan yaks. Animals 2019, 9, 613. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.; Yang, X.J.; Tong, H.; Chen, J.; Zhao, R.Q. Expressions of FAS and HSL mRNA in longissimus dorsi muscle and their relation to intramescular fat contents in pig. Chin. J. Agric. Biotechnol. 2004, 12, 422–426. [Google Scholar]
- Almaamory, Y.A.; Al-Anbari, N.N. Relationship of FASN Gene Polymorphism in Growth Performance and Wool Traits of Awassi Sheep. IOP Conf. Ser. Earth Environ. Sci. 2023, 1214, 012032. [Google Scholar] [CrossRef]
- Martina, Z.; Diana, L.; Paolo, Z.; Marcello, M.; Andrea, S.; Leonardo, N.C.; Roberta, D. Association study between backfat fatty acid composition and SNPs in candidate genes highlights the effect of FASN polymorphism in large white pigs. Meat Sci. 2019, 156, 75–84. [Google Scholar]
- Piórkowska, K.; Małopolska, M.; Ropka-Molik, K.; Szyndler-Nędza, M.; Wiechniak, A.; Żukowski, K.; Lambert, B.; Tyra, M. Evaluation of SCD, ACACA and FASN Mutations: Effects on Pork Quality and Other Production Traits in Pigs Selected Based on RNA-Seq Results. Animals 2020, 10, 123. [Google Scholar] [CrossRef] [PubMed]
- Vohra, V.; Kumar, M.; Chopra, A.; Niranjan, S.K.; Mishra, A.K.; Kataria, R.S. Polymorphism in Exon-40 of FASN Gene in Lesser known Buffalo breeds of India. J. Anim. Res. 2015, 5, 325–328. [Google Scholar]

| Primer | Primer Sequences (5′ → 3′) | Tm (°C) | Product Length (bp) |
|---|---|---|---|
| FASN-P1 | F:CTCTGGAAGGACAACTGGGT | 57 °C | 638 bp |
| R:AAGAGGGGCGAAATCTGTCA | |||
| FASN-P2 | F:CCGACGTGGTAATGAACAGG | 57 °C | 361 bp |
| R:CCCCTCAGAAGACATCAGCT | |||
| FASN-β (RT-qPCR) | F:CTTGACACGGCTCAACTCG | 58 °C | 118 bp |
| R:ATAGGTGGGGATGCTGAGC | |||
| GAPDH (RT-qPCR) | F: GGCCTCCAAGGAGTAAGGTC | 58 °C | 124 bp |
| R: CGGGAGATTCTCAGTGTGGT |
| Genotype | Genotype Frequency | Allele | Allele Frequency | Ho | He | Ne | χ2 | PIC |
|---|---|---|---|---|---|---|---|---|
| CC | 43.08% | C | 64.1% | 0.5415 | 0.4585 | 0.4573 | 1.3437 (p = 0.25) | 0.35 |
| TT | 14.87% | T | 35.9% | |||||
| CT | 42.05% |
| Body Size Trait | Allele | Present | Absent | p-Value |
|---|---|---|---|---|
| Body weight (kg) | C | 25.48 ± 0.73 | 32.71 ± 1.44 | <0.01 |
| T | 28.30 ± 0.87 | 24.82 ± 1.04 | <0.01 | |
| Heart girth (cm) | C | 69.19 ± 0.78 | 73.95 ± 1.48 | <0.01 |
| T | 71.33 ± 0.91 | 68.56 ± 1.07 | 0.05 | |
| Wither height (cm) | C | 57.23 ± 0.70 | 60.12 ± 1.24 | 0.04 |
| T | 57.81 ± 0.78 | 58.10 ± 1.04 | 0.82 | |
| Body length (cm) | C | 51.14 ± 1.09 | 54.08 ± 2.04 | 0.19 |
| T | 51.45 ± 1.23 | 52.21 ± 1.60 | 0.70 | |
| Circumference of cannon bone (cm) | C | 7.55 ± 0.18 | 7.90 ± 0.35 | 0.37 |
| T | 7.69 ± 0.20 | 7.51 ± 0.25 | 0.57 |
| Body Size Traits | Genotype | Mean ± Standard Error | p-Value |
|---|---|---|---|
| Body weight (kg) | CC | 24.79 ± 1.0 a | <0.01 |
| CT | 26.18 ± 1.01 a | ||
| TT | 32.72 ± 1.44 b | ||
| Heart girth (cm) | CC | 68.54 ± 1.06 a | 0.01 |
| CT | 69.89 ± 1.11 ab | ||
| TT | 73.97 ± 1.48 b | ||
| Wither height (cm) | CC | 58.09 ± 1.01 | 0.07 |
| CT | 56.48 ± 0.95 | ||
| TT | 60.11 ± 1.24 | ||
| Body length (cm) | CC | 52.34 ± 1.58 | 0.25 |
| CT | 50.13 ± 1.45 | ||
| TT | 54.16 ± 2.04 | ||
| Circumference of cannon bone (cm) | CC | 7.51 ± 0.25 | 0.65 |
| CT | 7.59 ± 0.25 | ||
| TT | 7.90 ± 0.36 |
| Body Size Trait | Allele | Male | Female | ||||
|---|---|---|---|---|---|---|---|
| Present | Absent | p-Value | Present | Absent | p-Value | ||
| Body weight (kg) | C | 30.41 ± 1.59 | 30.19 ± 2.11 | 0.93 | 28.42 ± 0.58 | 33.16 ± 1.24 | <0.01 |
| T | 30.47 ± 1.46 | 30.08 ± 1.80 | 0.84 | 30.55 ± 0.68 | 27.29 ± 0.86 | <0.01 | |
| Heart girth (cm) | C | 73.34 ± 2.89 | 71.35 ± 3.56 | 0.65 | 71.94 ± 0.63 | 75.20 ± 1.28 | 0.02 |
| T | 73.20 ± 2.57 | 70.99 ± 3.59 | 0.57 | 75.27 ± 0.70 | 71.18 ± 0.96 | 0.08 | |
| Wither height (cm) | C | 61.52 ± 2.10 | 57.96 ± 2.58 | 0.27 | 56.90 ± 0.64 | 61.51 ± 1.18 | <0.01 |
| T | 58.98 ± 1.84 | 63.12 ± 2.57 | 0.15 | 58.01 ± 0.75 | 57.67 ± 1.07 | 0.79 | |
| Body length (cm) | C | 54.98 ± 1.70 | 53.81 ± 2.09 | 0.66 | 50.25 ± 1.27 | 58.50 ± 3.12 | 0.03 |
| T | 54.01 ± 1.50 | 55.82 ± 2.10 | 0.43 | 50.90 ± 1.71 | 52.75 ± 2.70 | 0.57 | |
| Circumference of cannon bone (cm) | C | 8.17 ± 0.29 | 7.96 ± 0.36 | 0.63 | 7.32 ± 0.18 | 8.05 ± 0.43 | 0.15 |
| T | 8.11 ± 0.26 | 8.04 ± 0.36 | 0.87 | 7.41 ± 0.21 | 7.45 ± 0.34 | 0.92 | |
| Body Size Trait | Genotype | Male | Female | ||
|---|---|---|---|---|---|
| Mean ± Standard Error | p-Value | Mean ± Standard Error | p-Value | ||
| Body weight (kg) | CC | 30.16 ± 1.88 | 0.97 | 27.29 ± 0.84 a | <0.01 |
| CT | 30.71 ± 1.98 | 29.45 ± 0.80 a | |||
| TT | 30.19 ± 2.13 | 33.16 ± 1.23 b | |||
| Heart girth (cm) | CC | 71.45 ± 3.66 | 0.63 | 71.23 ± 0.95 a | 0.05 |
| CT | 75.08 ± 3.55 | 72.48 ± 0.82 ab | |||
| TT | 71.29 ± 3.57 | 75.17 ± 1.28 b | |||
| Wither height (cm) | CC | 63.25 ± 2.64 | 0.31 | 57.70 ± 0.97 a | <0.01 |
| CT | 59.94 ± 2.56 | 56.34 ± 0.82 a | |||
| TT | 58.01 ± 2.58 | 61.52 ± 1.18 b | |||
| Body length (cm) | CC | 55.86 ± 2.16 | 0.73 | 52.75 ± 2.10 a | 0.04 |
| CT | 54.18 ± 2.10 | 49.00 ± 1.49 ab | |||
| TT | 53.84 ± 2.11 | 58.50 ± 2.97 b | |||
| Circumference of cannon bone (cm) | CC | 8.08 ± 0.37 | 0.82 | 7.45 ± 0.32 | 0.32 |
| CT | 8.26 ± 0.36 | 7.25 ± 0.22 | |||
| TT | 7.95 ± 0.36 | 8.05 ± 0.45 | |||
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
An, Q.; Zeng, L.; Wang, W.; Yang, J.; Meng, J.; Zhao, Y.; Song, X. Identification of FASN Gene Polymorphisms, Expression and Their Relationship with Body Size Traits in Guizhou White Goat (Capra hircus) with Different Genders. Genes 2024, 15, 656. https://doi.org/10.3390/genes15060656
An Q, Zeng L, Wang W, Yang J, Meng J, Zhao Y, Song X. Identification of FASN Gene Polymorphisms, Expression and Their Relationship with Body Size Traits in Guizhou White Goat (Capra hircus) with Different Genders. Genes. 2024; 15(6):656. https://doi.org/10.3390/genes15060656
Chicago/Turabian StyleAn, Qingming, Lingli Zeng, Wenying Wang, Jiangyu Yang, Jinzhu Meng, Yuanyuan Zhao, and Xingchao Song. 2024. "Identification of FASN Gene Polymorphisms, Expression and Their Relationship with Body Size Traits in Guizhou White Goat (Capra hircus) with Different Genders" Genes 15, no. 6: 656. https://doi.org/10.3390/genes15060656
APA StyleAn, Q., Zeng, L., Wang, W., Yang, J., Meng, J., Zhao, Y., & Song, X. (2024). Identification of FASN Gene Polymorphisms, Expression and Their Relationship with Body Size Traits in Guizhou White Goat (Capra hircus) with Different Genders. Genes, 15(6), 656. https://doi.org/10.3390/genes15060656





