Spatial Colinear but Broken Temporal Expression of Duplicated ParaHox Genes in Asexually Reproducing Annelids, Nais communis and Pristina longiseta
Abstract
1. Introduction
2. Materials and Methods
2.1. Animal Material and Fixation
2.2. Sequence Retrieval and Phylogenetic Analysis
2.3. Gene Cloning
2.4. Whole-Mount In Situ Hybridization
2.5. Immunohistochemistry
2.6. Data Visualization
3. Results
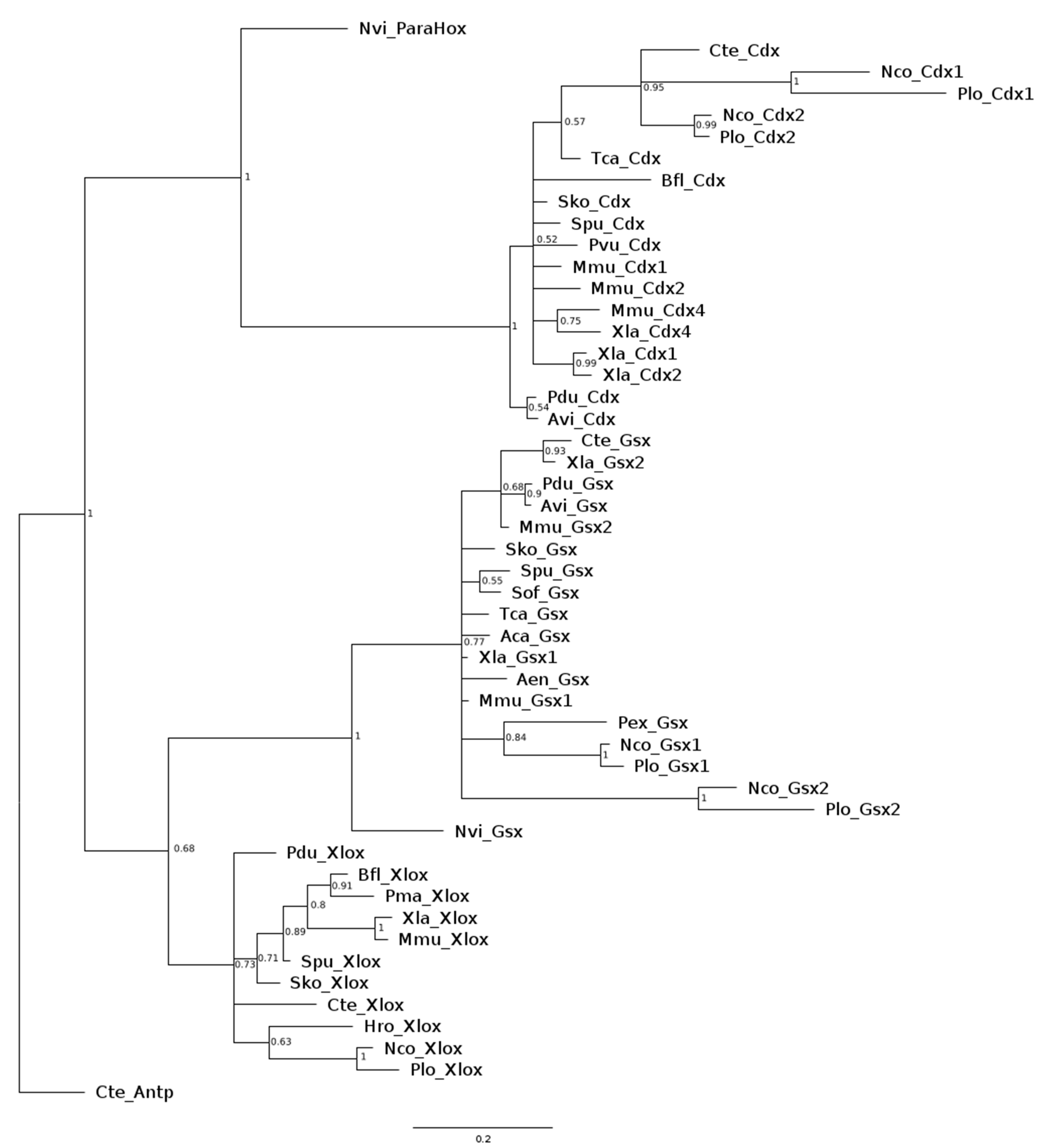
3.1. Sequence Analysis
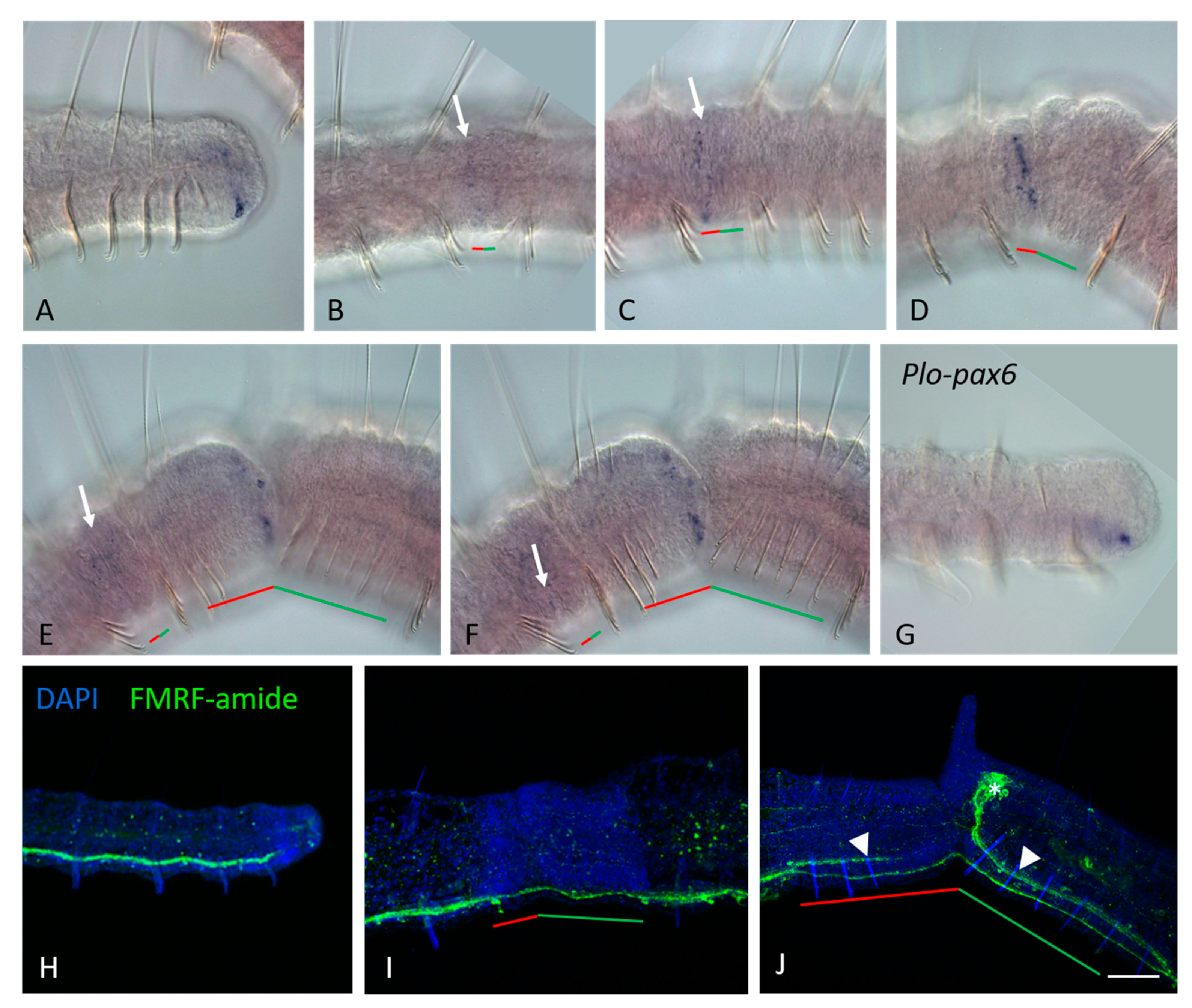
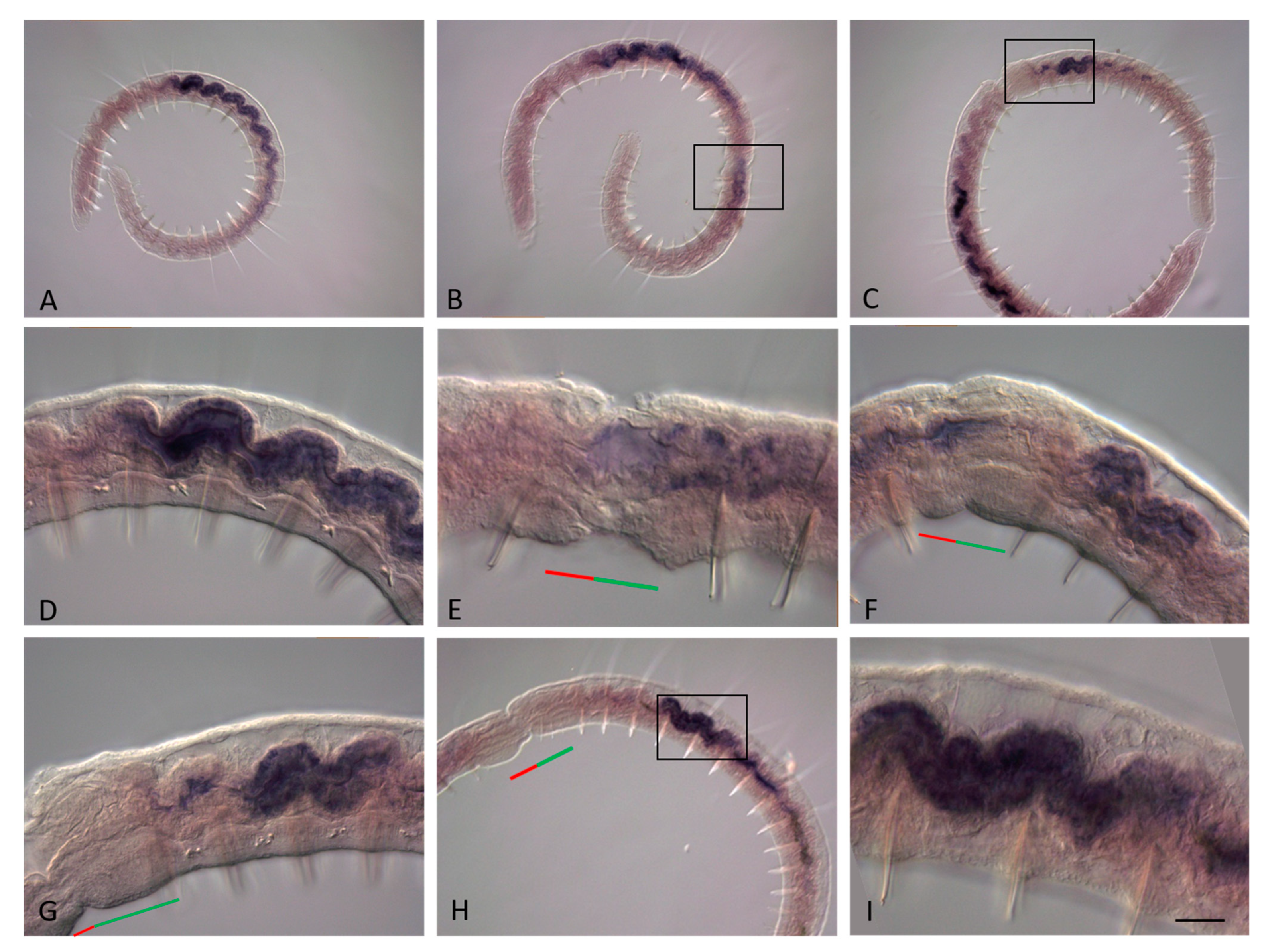
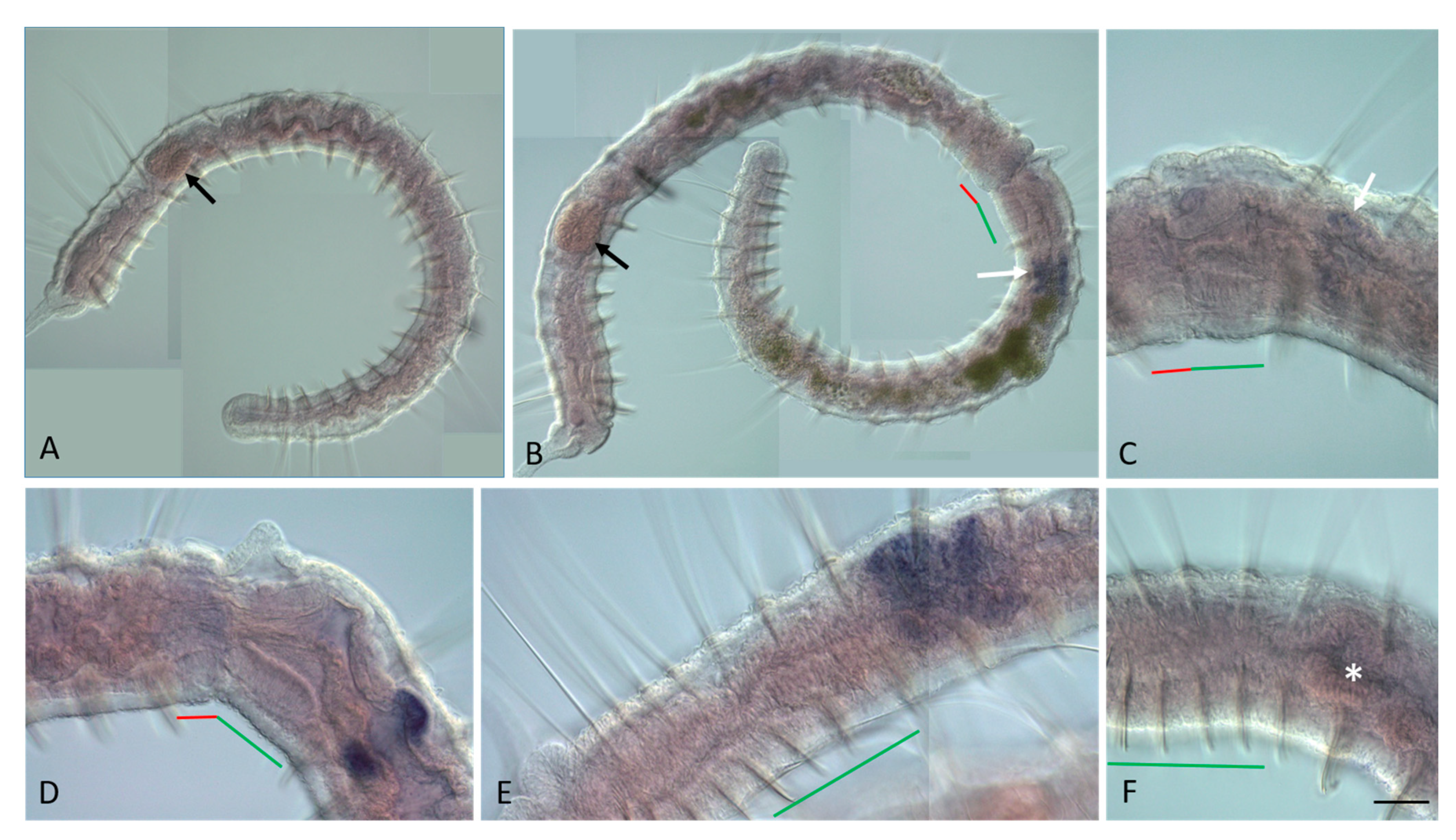
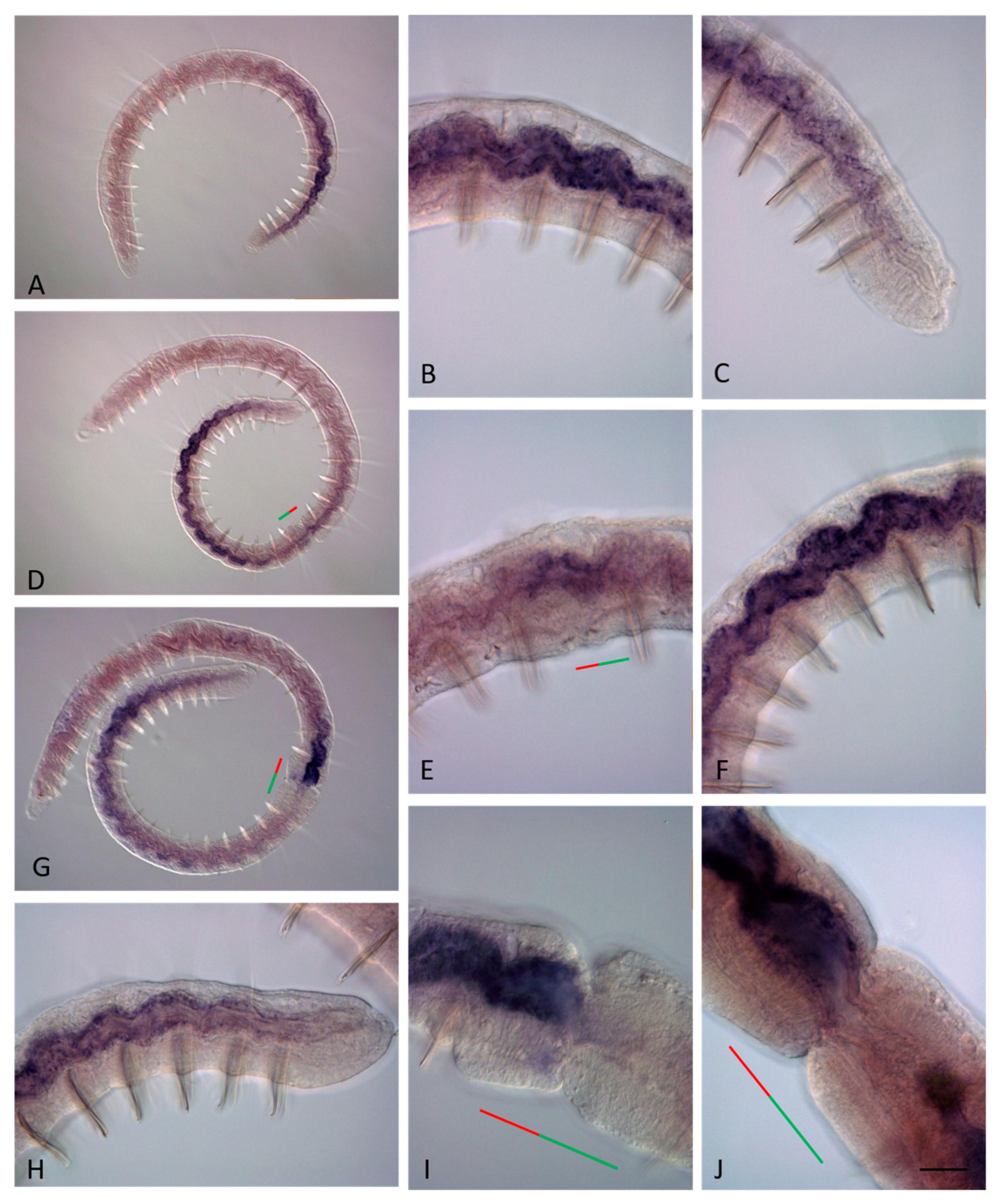
3.2. ParaHox Gene Expression in Growing Adult N. communis and P. longiseta
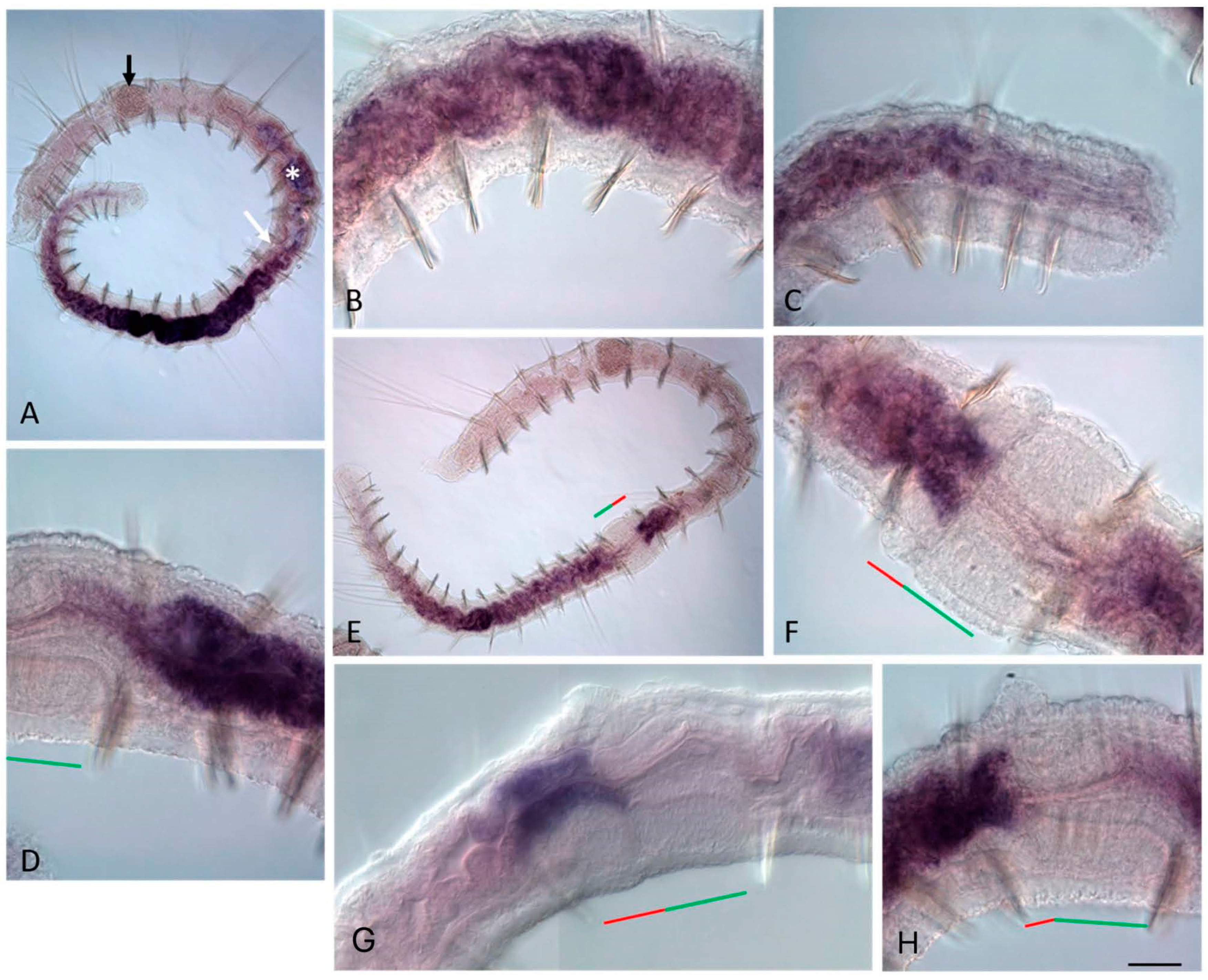
3.3. ParaHox Gene Expression in the Asexually Reproducing Animals, N. communis and P. longiseta
4. Discussion
4.1. ParaHox Genes in Naidids N. communis and P. longiseta
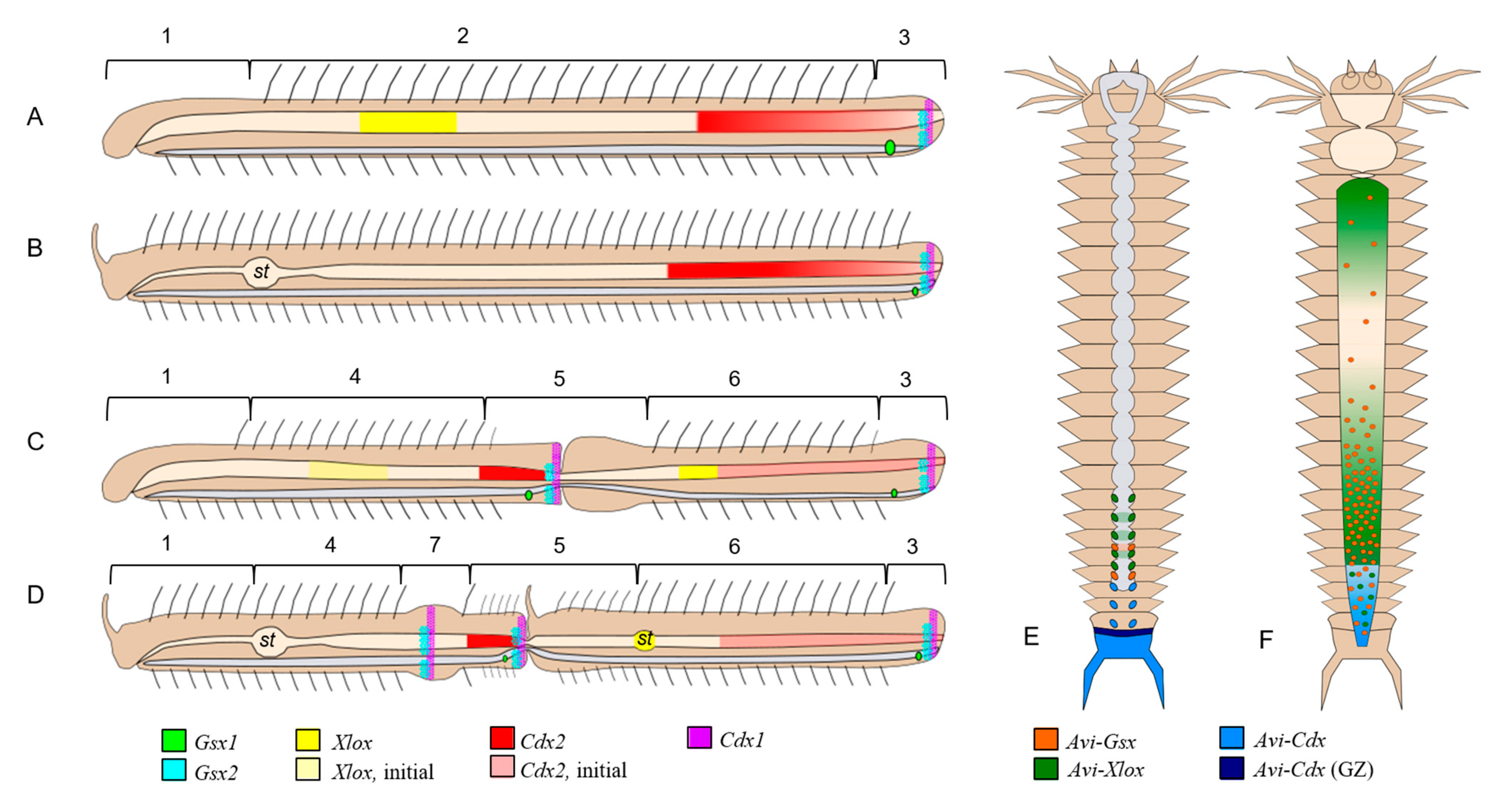
4.2. Expression of ParaHox Genes in Growing adult N. communis and P. longiseta
4.3. Expression of ParaHox Genes in Asexually Reproducing N. communis and P. longiseta
4.4. Signs of Colinearity in the Expression of ParaHox Genes in Growing and Asexually Reproducing N. communis and P. longiseta
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Brooke, N.M.; Garcia-Fernàndez, J.; Holland, P.W.H. The ParaHox gene cluster is an evolutionary sister of the Hox gene cluster. Nature 1998, 392, 920–922. [Google Scholar] [CrossRef]
- Garcia-Fernàndez, J. The genesis and evolution of homeobox gene clusters. Nat. Rev. Genet. 2005, 6, 881–892. [Google Scholar] [CrossRef] [PubMed]
- Ferrier, D.E.K.; Dewar, K.; Cook, A.; Chang, J.L.; Hill-Force, A.; Amemiya, C. The chordate ParaHox cluster. Curr. Biol. 2005, 15, 820–822. [Google Scholar] [CrossRef]
- Ferrier, D.E.K. The origin of the Hox/ParaHox genes, the Ghost Locus hypothesis and the complexity of the first animal. Brief. Funct. Genom. 2016, 15, 333–341. [Google Scholar] [CrossRef] [PubMed]
- Holland, P.W. Beyond the Hox: How widespread is homeobox gene clustering? J. Anat. 2001, 199, 13–23. [Google Scholar] [CrossRef] [PubMed]
- Hui, J.H.L.; Raible, F.; Korchagina, N.; Dray, N.; Samain, S.; Magdelenat, G.; Jubin, C.; Segurens, B.; Balavoine, G.; Arendt, D.; et al. Features of the ancestral bilaterian inferred from Platynereis dumerilii ParaHox genes. BMC Biol. 2009, 7, 43. [Google Scholar] [CrossRef]
- Wollesen, T.; Rodríguez Monje, S.V.; McDougall, C.; Degnan, B.M.; Wanninger, A. The ParaHox gene Gsx patterns the apical organ and central nervous system but not the foregut in scaphopod and cephalopod mollusks. EvoDevo 2015, 6, 41. [Google Scholar] [CrossRef]
- Arnone, M.I.; Rizzo, F.; Annunciata, R.; Cameron, R.A.; Peterson, K.J.; Martínez, P. Genetic organization and embryonic expression of the ParaHox genes in the sea urchin S. purpuratus: Insights into the relationship between clustering and colinearity. Dev. Biol. 2006, 300, 63–73. [Google Scholar] [CrossRef]
- Ferrier, D.E.K.; Holland, P.W.H. Ciona intestinalis ParaHox genes: Evolution of Hox/ParaHox cluster integrity, developmental mode, and temporal colinearity. Mol. Phylogenetics Evol. 2002, 24, 412–417. [Google Scholar] [CrossRef]
- Mulley, J.F.; Chiu, C.H.; Holland, P.W.H. Breakup of a homeobox cluster after genome duplication in teleosts. Proc. Natl. Acad. Sci. USA 2006, 103, 10369–10372. [Google Scholar] [CrossRef]
- Annunziata, R.; Martinez, P.; Arnone, M.I. Intact cluster and chordate-like expression of ParaHox genes in a sea star. BMC Biol. 2013, 11, 68. [Google Scholar] [CrossRef]
- Ikuta, T.; Chen, Y.C.; Annunziata, R.; Ting, H.C.; Tung, C.H.; Koyanagi, R.; Tagawa, K.; Humphreys, T.; Fujiyama, A.; Saiga, H.; et al. Identification of an intact ParaHox cluster with temporal colinearity but altered spatial colinearity in the hemichordate Ptychodera flava. BMC Evol. Biol. 2013, 13, 129. [Google Scholar] [CrossRef]
- Martín-Durán, J.M.; Romero, R. Evolutionary implications of morphogenesis and molecular patterning of the blind gut in the planarian Schmidtea polychroa. Dev. Biol. 2011, 352, 164–176. [Google Scholar] [CrossRef]
- Hudson, C.; Lemaire, P. Induction of anterior neural fates in the ascidian Ciona intestinalis. Mech. Dev. 2001, 100, 189–203. [Google Scholar] [CrossRef]
- Kulakova, M.A.; Cook, C.E.; Andreeva, T.F. ParaHox gene expression in larval and postlarval development of the polychaete Nereis virens (Annelida, Lophotrochozoa). BMC Dev. Biol. 2008, 8, 61. [Google Scholar] [CrossRef] [PubMed]
- Fröbius, A.C.; Seaver, E.C. ParaHox gene expression in the polychaete annelid Capitella sp. I. Dev. Genes Evol. 2006, 216, 81–88. [Google Scholar] [CrossRef]
- Samadi, L.; Steiner, G. Conservation of ParaHox genes’ function in patterning of the digestive tract of the marine gastropod Gibbula varia. BMC Dev. Biol. 2010, 10, 74. [Google Scholar] [CrossRef] [PubMed]
- Wright, C.V.; Cho, K.W.; Oliver, G.; De Robertis, E.M. Vertebrate homeodomain proteins: Families of region-specific transcription factors. Trends Biochem. Sci. 1989, 14, 52–56. [Google Scholar] [CrossRef]
- Jonsson, J.; Carlsson, L.; Edlund, T.; Edlund, H. Insulin-promoter-factor 1 is required for pancreas development in mice. Nature 1994, 371, 606–609. [Google Scholar] [CrossRef]
- Perez-Villamil, B.; Schwartz, P.T.; Vallejo, M. The pancreatic homeodomain transcription factor IDX1/IPF1 is expressed in neural cells during brain development. Endocrinology 1999, 140, 3857–3860. [Google Scholar] [CrossRef] [PubMed]
- Wysocka-Diller, J.; Aisemberg, G.O.; Macagno, E.R. A novel homeobox cluster expressed in repeated structures of the midgut. Dev. Biol. 1995, 171, 439–447. [Google Scholar] [CrossRef]
- Beck, F.; Stringer, E.J. The role of Cdx genes in the gut and in axial development. Biochem. Soc. Trans. 2010, 38, 353–357. [Google Scholar] [CrossRef] [PubMed]
- Jedrusik, A.; Parfitt, D.-E.; Guo, G.; Skamagki, M.; Grabarek, J.B.; Johnson, M.H.; Robson, P.; Zernicka-Goetz, M. Role of Cdx2 and cell polarity in cell allocation and specification of trophectoderm and inner cell mass in the mouse embryo. Genes Dev. 2008, 22, 2692–2706. [Google Scholar] [CrossRef] [PubMed]
- Neijts, R.; Amin, S.; van Rooijen, C.; Deschamps, J. Cdx is crucial for the timing mechanism driving colinear Hox activation and defines a trunk segment in the Hox cluster topology. Dev. Biol. 2017, 422, 146–154. [Google Scholar] [CrossRef] [PubMed]
- Copf, T.; Schröder, R.; Averof, M. Ancestral role of caudal genes in axis elongation and segmentation. Proc. Natl. Acad. Sci. USA 2004, 101, 17711–17715. [Google Scholar] [CrossRef]
- De Rosa, R.; Prud’homme, B.; Balavoine, G. Caudal and even-skipped in the annelid Platynereis dumerilii and the ancestry of posterior growth. Evol. Dev. 2005, 7, 574–587. [Google Scholar] [CrossRef]
- Edgar, L.G.; Carr, S.; Wang, H.; Wood, W.B. Zygotic expression of the caudal homolog pal-1 is required for posterior patterning in Caenorhabditis elegans embryogenesis. Dev. Biol. 2001, 229, 71–88. [Google Scholar] [CrossRef]
- Moreno, E.; Morata, G. Caudal is the Hox gene that specifies the most posterior Drosophile segment. Nature 1999, 400, 873–877. [Google Scholar] [CrossRef]
- Matsuo, K.; Yoshida, H.; Shimizu, T. Differential expression of caudal and dorsal genes in the teloblast lineages of the oligochaete annelid Tubifex tubifex. Dev. Genes Evol. 2005, 215, 238–247. [Google Scholar] [CrossRef]
- Le Gouar, M.; Lartillot, N.; Adoutte, A.; Vervoort, M. The expression of caudal homologue in mollusc, Patella vulgata. Gene Expr. Patt. 2003, 3, 35–37. [Google Scholar] [CrossRef]
- Johnson, A.B.; Lambert, J.D. The Caudal ParaHox gene is required for hindgut development in the mollusc Tritia (a.k.a. Ilyanassa). Dev. Biol. 2021, 470, 1–9. [Google Scholar] [CrossRef]
- Perry, K.J.; Lyons, D.C.; Truchado-Garcia, M.; Fischer, A.H.L.; Helfrich, L.W.; Johansson, K.B.; Diamond, J.C.; Grande, C.; Henry, J.Q. Deployment of regulatory genes during gastrulation and germ layer specification in a model spiralian mollusc Crepidula. Deve. Dyn. 2015, 244, 1215–1248. [Google Scholar] [CrossRef] [PubMed]
- Bely, A.E. Decoupling of fission and regenerative capabilities in an asexual oligochaete. Hydrobiologia 1999, 406, 243–251. [Google Scholar] [CrossRef]
- Kharin, A.V.; Zagainova, I.V.; Kostyuchenko, R.P. Formation of the paratomic fission zone in freshwater oligochaetes. Russ. J. Dev. Biol. 2006, 37, 354–365. [Google Scholar] [CrossRef]
- Kostyuchenko, R.P.; Kozin, V.V.; Kupriashova, E.E. Regeneration and asexual reproduction in annelids: Cells, genes, and evolution. Biol. Bull. 2016, 43, 185–194. [Google Scholar] [CrossRef]
- Zattara, E.E.; Bely, A.E. Phylogenetic distribution of regeneration and asexual reproduction in Annelida: Regeneration is ancestral and fission evolves in regenerative clades. Invert. Biol. 2016, 135, 400–414. [Google Scholar] [CrossRef]
- Nikanorova, D.D.; Kupriashova, E.E.; Kostyuchenko, R.P. Regeneration in Annelids: Cell Sources, Tissue Remodeling, and Differential Gene Expression. Russ. J. Dev. Biol. 2020, 51, 148–161. [Google Scholar] [CrossRef]
- Zattara, E.E.; Bely, A.E. Evolution of a Novel Developmental Trajectory: Fission Is Distinct from Regeneration in the Annelid Pristina leidyi. Evol. Dev. 2011, 13, 80–95. [Google Scholar] [CrossRef] [PubMed]
- Kostyuchenko, R.P.; Kozin, V.V. Comparative Aspects of Annelid Regeneration: Towards Understanding the Mechanisms of Regeneration. Genes 2021, 12, 1148. [Google Scholar] [CrossRef]
- Kostyuchenko, R.P.; Kozin, V.V. Morphallaxis versus Epimorphosis? Cellular and Molecular Aspects of Regeneration and Asexual Reproduction in Annelids. Biol. Bull. 2020, 47, 237–246. [Google Scholar] [CrossRef]
- Steinmetz, P.R.H.; Urbach, R.; Posnien, N.; Eriksson, J.; Kostyuchenko, R.P.; Brena, C.; Guy, K.; Akam, M.; Bucher, G.; Arendt, D. Six3 demarcates the anterior-most developing brain region in bilaterian animals. EvoDevo 2010, 1, 14. [Google Scholar] [CrossRef] [PubMed]
- Bely, A.E.; Wray, G.A. Evolution of regeneration and fission in annelids: Insights from engrailed- and orthodenticle-class gene expression. Development 2001, 128, 2781–2791. [Google Scholar] [CrossRef] [PubMed]
- Özpolat, B.D.; Bely, A.E. Gonad Establishment during Asexual Reproduction in the Annelid Pristina leidyi. Dev. Biol. 2015, 405, 123–136. [Google Scholar] [CrossRef]
- Arnold, C.P.; Lozano, A.M.; Mann, F.G.; Nowotarski, S.H.; Haug, J.O.; Langet, J.J.; Seidel, C.W.; Alvarado, A.S. Hox genes regulate asexual reproductive behavior and tissue segmentation in adult animals. Nat. Commun. 2021, 12, 6706. [Google Scholar] [CrossRef]
- del Olmo, I.; Verdes, A.; Álvarez-Campos, P. Distinct patterns of gene expression during regeneration and asexual reproduction in the annelid Pristina leidyi. J. Exp. Zool. P. B Mol. Dev. Evol. 2022, 338, 405. [Google Scholar] [CrossRef]
- Katoh, M.; Kuma, M. MAFFT: A novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Res. 2002, 30, 3059–3066. [Google Scholar] [CrossRef] [PubMed]
- Okonechnikov, K.; Golosova, O.; Fursov, M.; UGENE team. Unipro UGENE: A unified bioinformatics toolkit. Bioinformatics 2012, 28, 1166–1167. [Google Scholar] [CrossRef]
- Miller, M.A.; Pfeiffer, W.; Schwartz, T. Creating the CIPRES Science Gateway for inference of large phylogenetic trees. In Proceedings of the Gateway Computing Environments Workshop (GCE), New Orleans, LA, USA, 14 November 2010; pp. 1–8. [Google Scholar] [CrossRef]
- Tamura, K.; Stecher, G.; Kumar, S. MEGA11: Molecular Evolutionary Genetics Analysis version 11. Mol. Biol. Evol. 2021, 38, 3022–3027. [Google Scholar] [CrossRef]
- Kostyuchenko, R.P.; Kozin, V.V.; Filippova, N.A.; Sorokina, E.V. FoxA expression pattern in two polychaete species, Alitta virens and Platynereis dumerilii: Examination of the conserved key regulator of the gut development from cleavage through larval life, postlarval growth, and regeneration. Dev. Dyn. 2019, 248, 728–743. [Google Scholar] [CrossRef]
- Kostyuchenko, R.P. Nanos Is Expressed in Somatic and Germline Tissue during Larval and Post-Larval Development of the Annelid Alitta virens. Genes 2022, 13, 270. [Google Scholar] [CrossRef]
- Zwarycz, A.S.; Nossa, C.W.; Putnam, N.H.; Ryan, J.F. Timing and scope of genomic expansion within annelida: Evidence from homeoboxes in the genome of the earthworm Eisenia fetida. Genome Biol. Evol. 2016, 8, 271–281. [Google Scholar] [CrossRef]
- Simakov, O.; Marletaz, F.; Cho, S.J.; Edsinger-Gonzales, E.; Havlak, P.; Hellsten, U.; Kuo, D.H.; Larsson, T.; Lv, J.; Arendt, D.; et al. Insights into bilaterian evolution from three spiralian genomes. Nature 2013, 493, 526–531. [Google Scholar] [CrossRef]
- Holland, P.W.H. Evolution of homeobox genes. Wiley Interdiscip. Rev. Dev. Biol. 2013, 2, 31–45. [Google Scholar] [CrossRef]
- Nong, W.; Cao, J.; Li, Y.; Qu, Z.; Sun, J.; Swale, T.; Yip, H.Y.; Qian, P.Y.; Qiu, J.W.; Kwan, H.S.; et al. Jellyfish genomes reveal distinct homeobox gene clusters and conservation of small RNA processing. Nat. Commun. 2020, 11, 3051. [Google Scholar] [CrossRef]
- Iwanoff, P.P. Die Entwiklung der Larvalsegmente bei den Annelide. Z. Morph. Oekol. Tiere. 1928, 10, 62–161. [Google Scholar] [CrossRef]
- Kozin, V.V.; Kostyuchenko, R.P. Vasa, PL10, and Piwi gene expression during caudal regeneration of the polychaete annelid Alitta virens. Dev. Genes Evol. 2015, 225, 129–138. [Google Scholar] [CrossRef] [PubMed]
- Kostyuchenko, R.P.; Smirnova, N.P. Vasa, Piwi, and Pl10 expression during sexual maturation and asexual reproduction in the annelid Pristina longiseta . J. Dev. Biol. 2023, 11. [Google Scholar]
- Balavoine, G. Segment formation in annelids: Patterns, processes and evolution. Int. J. Dev. Biol. 2014, 58, 469–483. [Google Scholar] [CrossRef] [PubMed]
- Pfeifer, K.; Dorresteijn, A.W.C.; Fröbius, A.C. Activation of Hox Genes during Caudal Regeneration of the Polychaete Annelid Platynereis Dumerilii. Dev. Genes Evol. 2012, 222, 165–179. [Google Scholar] [CrossRef] [PubMed]
- Novikova, E.L.; Bakalenko, N.I.; Nesterenko, A.Y.; Kulakova, M.A. Expression of Hox Genes during Regeneration of Nereid Polychaete Alitta (Nereis) virens (Annelida, Lophotrochozoa). EvoDevo 2013, 4, 14. [Google Scholar] [CrossRef]
- Planques, A.; Malem, J.; Parapar, J.; Vervoort, M.; Gazave, E. Morphological, Cellular and Molecular Characterization of Posterior Regeneration in the Marine Annelid Platynereis dumerilii. Dev. Biol. 2019, 445, 189–210. [Google Scholar] [CrossRef]
- Weigert, A.; Bleidorn, C. Current status of annelid phylogeny. Org. Divers. Evol. 2016, 16, 345–362. [Google Scholar] [CrossRef]
- Takeo, M.; Yoshida-Noro, C.; Tochinai, S. Morphallactic regeneration as revealed by region-specific gene expression in the digestive tract of Enchytraeus japonensis (Oligochaeta, Annelida). Dev. Dyn. 2008, 237, 1284–1294. [Google Scholar] [CrossRef]
- Žídek, R.; Machoň, O.; Kozmik, Z. Wnt/β-catenin signalling is necessary for gut differentiation in a marine annelid, Platynereis dumerilii. EvoDevo 2018, 9, 14. [Google Scholar] [CrossRef]
- Annunziata, R.; Arnone, M.I. A dynamic regulatory network explains ParaHox gene control of gut patterning in the sea urchin. Development 2014, 141, 2462–2472. [Google Scholar] [CrossRef] [PubMed]
- Garstang, M.G.; Osborne, P.W.; Ferrier, D.E.K. TCF/Lef regulates the Gsx ParaHox gene in central nervous system development in chordates. BMC Evol. Biol. 2016, 16, 57. [Google Scholar] [CrossRef]
- Osborne, P.W.; Benoit, G.; Laudet, V.; Schubert, M.; Ferrier, D.E.K. Differential regulation of ParaHox genes by retinoic acid in the invertebrate chordate amphioxus (Branchiostoma floridae). Dev. Biol. 2009, 327, 252–262. [Google Scholar] [CrossRef] [PubMed]
- Zhong, Y.; Herrera-Úbeda, C.; Herrera-Úbeda, C.; Garcia-Fernàndez, J.; Li, G.; Holland, P.W.H. Mutation of amphioxus Pdx and Cdx demonstrates conserved roles for ParaHox genes in gut, anus and tail patterning. BMC Biol. 2020, 18, 68. [Google Scholar] [CrossRef]
- Kang, D.; Huang, F.; Li, D.; Shankland, M.; Gaffield, W.; Weisblat, D.A. A hedgehog homolog regulates gut formation in leech (Helobdella). Development 2003, 130, 1645–1657. [Google Scholar] [CrossRef]
- Seaver, E.C.; Kaneshige, L.M. Expression of ‘segmentation’ genes during larval and juvenile development in the polychaetes Capitella sp. I and H. elegans. Dev. Biol. 2006, 289, 179–194. [Google Scholar] [CrossRef]





Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kostyuchenko, R.P.; Amosov, A.V. Spatial Colinear but Broken Temporal Expression of Duplicated ParaHox Genes in Asexually Reproducing Annelids, Nais communis and Pristina longiseta. Genes 2023, 14, 1501. https://doi.org/10.3390/genes14071501
Kostyuchenko RP, Amosov AV. Spatial Colinear but Broken Temporal Expression of Duplicated ParaHox Genes in Asexually Reproducing Annelids, Nais communis and Pristina longiseta. Genes. 2023; 14(7):1501. https://doi.org/10.3390/genes14071501
Chicago/Turabian StyleKostyuchenko, Roman P., and Artem V. Amosov. 2023. "Spatial Colinear but Broken Temporal Expression of Duplicated ParaHox Genes in Asexually Reproducing Annelids, Nais communis and Pristina longiseta" Genes 14, no. 7: 1501. https://doi.org/10.3390/genes14071501
APA StyleKostyuchenko, R. P., & Amosov, A. V. (2023). Spatial Colinear but Broken Temporal Expression of Duplicated ParaHox Genes in Asexually Reproducing Annelids, Nais communis and Pristina longiseta. Genes, 14(7), 1501. https://doi.org/10.3390/genes14071501




