Uncovering the Molecular Drivers of NHEJ DNA Repair-Implicated Missense Variants and Their Functional Consequences
Abstract
1. Introduction
2. Methods
2.1. Data Collection
2.2. NHEJ Structural Curation
2.3. Feature Engineering
2.4. Qualitative and Statistical Analysis
2.5. Model Training
3. Results
3.1. Data Curation and Variant Distribution of NHEJ Principal Components
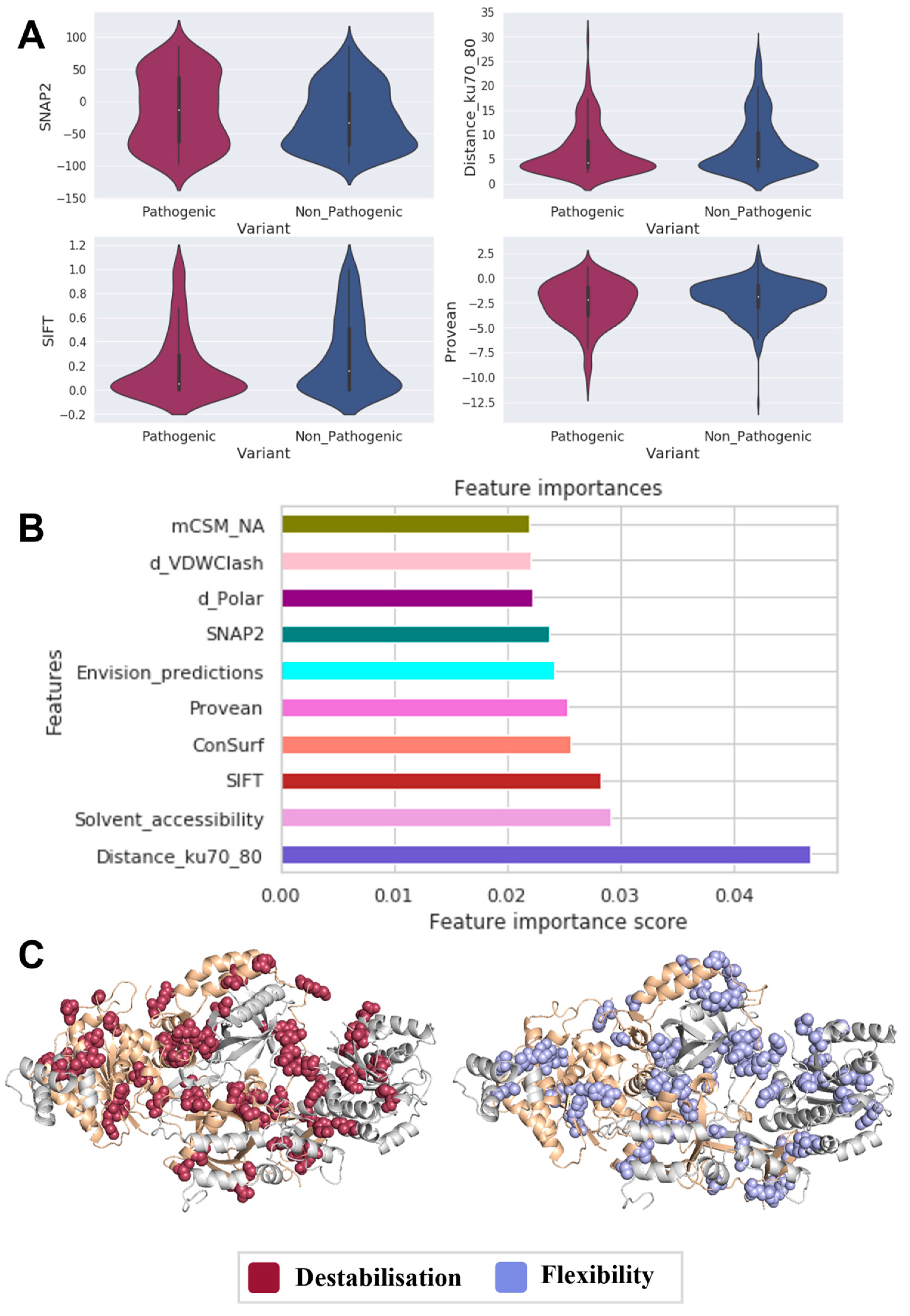
3.2. Identifying Molecular Drivers in Ku70/80 Heterodimer
3.3. Identifying Molecular Drivers of Pathogenicity in DNA-PKcs
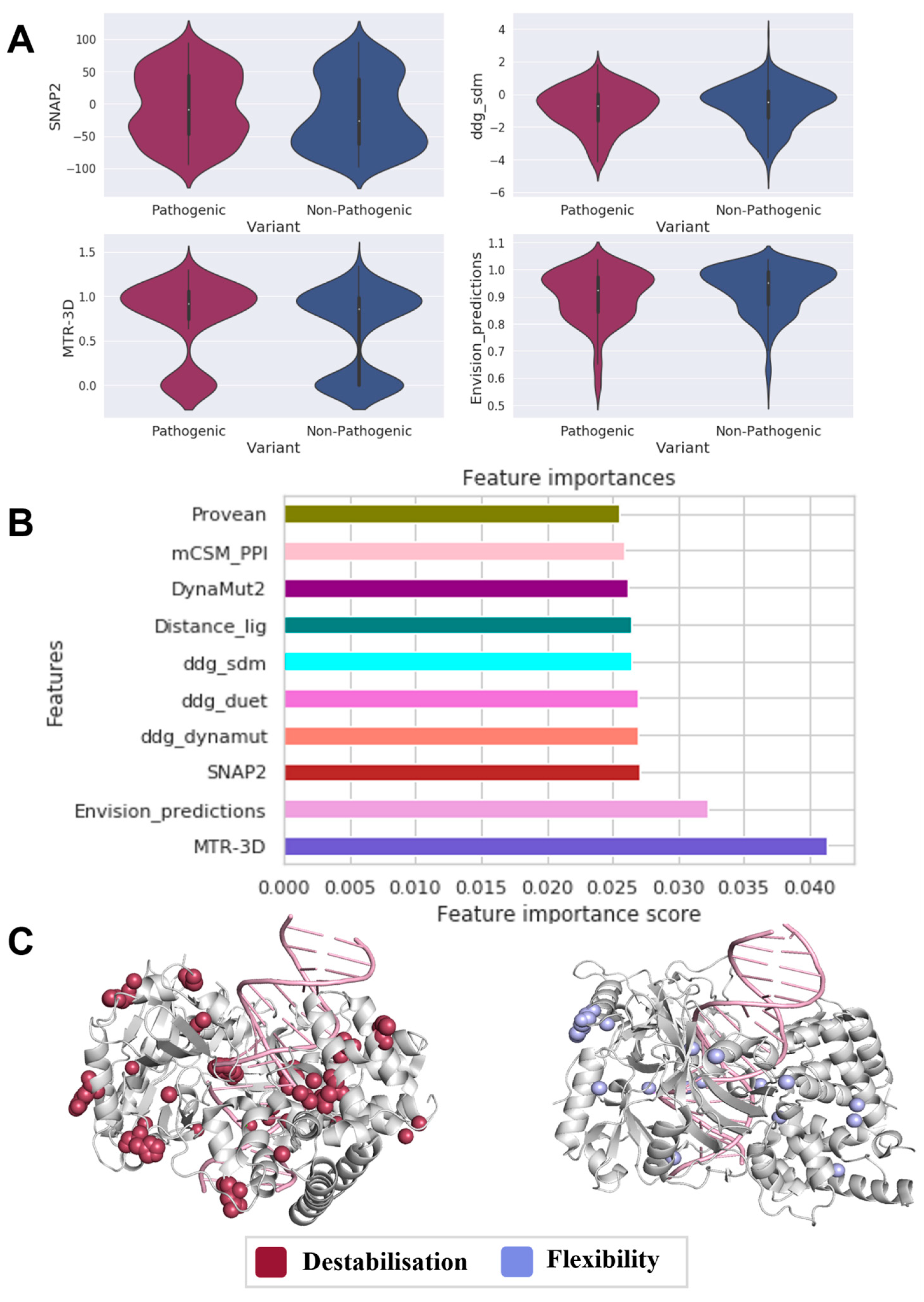
3.4. Uncovering Molecular Drivers in LIG4
3.5. Uncovering Molecular Drivers in XRCC4
4. Discussion
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Yin, M.; Hong, F.; Wang, Q.-E. DNA Damage Response and Cancer Metastasis: Clinical Implications and Therapeutic Opportunities. In Metastasis; Exon Publications: Brisbane, AU, Australia, 2022; pp. 117–136. [Google Scholar]
- Trenner, A.; Sartori, A.A. Harnessing DNA double-strand break repair for cancer treatment. Front. Oncol. 2019, 9, 1388. [Google Scholar] [CrossRef] [PubMed]
- Chang, H.H.; Pannunzio, N.R.; Adachi, N.; Lieber, M.R. Non-homologous DNA end joining and alternative pathways to double-strand break repair. Nat. Rev. Mol. Cell Biol. 2017, 18, 495–506. [Google Scholar] [CrossRef] [PubMed]
- Yano, K.-I.; Morotomi-Yano, K.; Adachi, N.; Akiyama, H. Molecular mechanism of protein assembly on DNA double-strand breaks in the non-homologous end-joining pathway. J. Radiat. Res. 2009, 50, 97–108. [Google Scholar] [CrossRef][Green Version]
- Murray, J.E.; Van Der Burg, M.; IJspeert, H.; Carroll, P.; Wu, Q.; Ochi, T.; Leitch, A.; Miller, E.S.; Kysela, B.; Jawad, A. Mutations in the NHEJ component XRCC4 cause primordial dwarfism. Am. J. Hum. Genet. 2015, 96, 412–424. [Google Scholar] [CrossRef]
- Rosin, N.; Elcioglu, N.H.; Beleggia, F.; Isgüven, P.; Altmüller, J.; Thiele, H.; Steindl, K.; Joset, P.; Rauch, A.; Nürnberg, P. Mutations in XRCC4 cause primary microcephaly, short stature and increased genomic instability. Hum. Mol. Genet. 2015, 24, 3708–3717. [Google Scholar] [CrossRef] [PubMed]
- Gao, Y.; Chaudhuri, J.; Zhu, C.; Davidson, L.; Weaver, D.T.; Alt, F.W. A targeted DNA-PKcs-null mutation reveals DNA-PK-independent functions for KU in V (D) J recombination. Immunity 1998, 9, 367–376. [Google Scholar] [CrossRef]
- Nie, Y.; Li, Y.; Li, X.; Wilson, A.F.; Pang, Q. The non-homologous end-joining activity is required for Fanconi anemia fetal HSC maintenance. Stem Cell Res. Ther. 2019, 10, 114. [Google Scholar] [CrossRef] [PubMed]
- Woodbine, L.; Gennery, A.R.; Jeggo, P.A. The clinical impact of deficiency in DNA non-homologous end-joining. DNA Repair 2014, 16, 84–96. [Google Scholar] [CrossRef]
- Bau, D.-T.; Fu, Y.-P.; Chen, S.-T.; Cheng, T.-C.; Yu, J.-C.; Wu, P.-E.; Shen, C.-Y. Breast cancer risk and the DNA double-strand break end-joining capacity of nonhomologous end-joining genes are affected by BRCA1. Cancer Res. 2004, 64, 5013–5019. [Google Scholar] [CrossRef]
- Caracciolo, D.; Riillo, C.; Di Martino, M.T.; Tagliaferri, P.; Tassone, P. Alternative Non-Homologous End-Joining: Error-Prone DNA Repair as Cancer’s Achilles’ Heel. Cancers 2021, 13, 1392. [Google Scholar] [CrossRef]
- Sishc, B.J.; Davis, A.J. The role of the core non-homologous end joining factors in carcinogenesis and cancer. Cancers 2017, 9, 81. [Google Scholar] [CrossRef] [PubMed]
- Ferguson, D.O.; Sekiguchi, J.M.; Chang, S.; Frank, K.M.; Gao, Y.; DePinho, R.A.; Alt, F.W. The nonhomologous end-joining pathway of DNA repair is required for genomic stability and the suppression of translocations. Proc. Natl. Acad. Sci. USA 2000, 97, 6630–6633. [Google Scholar] [CrossRef] [PubMed]
- Gu, Y.; Jin, S.; Gao, Y.; Weaver, D.T.; Alt, F.W. Ku70-deficient embryonic stem cells have increased ionizing radiosensitivity, defective DNA end-binding activity, and inability to support V (D) J recombination. Proc. Natl. Acad. Sci. USA 1997, 94, 8076–8081. [Google Scholar] [CrossRef] [PubMed]
- Nussenzweig, A.; Sokol, K.; Burgman, P.; Li, L.; Li, G.C. Hypersensitivity of Ku80-deficient cell lines and mice to DNA damage: The effects of ionizing radiation on growth, survival, and development. Proc. Natl. Acad. Sci. USA 1997, 94, 13588–13593. [Google Scholar] [CrossRef]
- Portelli, S.; Phelan, J.E.; Ascher, D.B.; Clark, T.G.; Furnham, N. Understanding molecular consequences of putative drug resistant mutations in Mycobacterium tuberculosis. Sci. Rep. 2018, 8, 15356. [Google Scholar] [CrossRef]
- Portelli, S.; Barr, L.; de Sá, A.G.; Pires, D.E.; Ascher, D.B. Distinguishing between PTEN clinical phenotypes through mutation analysis. Comput. Struct. Biotechnol. J. 2021, 19, 3097–3109. [Google Scholar] [CrossRef]
- Airey, E.; Portelli, S.; Xavier, J.S.; Myung, Y.C.; Silk, M.; Karmakar, M.; Velloso, J.P.; Rodrigues, C.H.; Parate, H.H.; Garg, A. Artificial Neural Networks; Springer: Berlin/Heidelberg, Germany, 2021; pp. 1–32. [Google Scholar]
- Tate, J.G.; Bamford, S.; Jubb, H.C.; Sondka, Z.; Beare, D.M.; Bindal, N.; Boutselakis, H.; Cole, C.G.; Creatore, C.; Dawson, E. COSMIC: The catalogue of somatic mutations in cancer. Nucleic Acids Res. 2019, 47, D941–D947. [Google Scholar] [CrossRef]
- Karczewski, K.J.; Francioli, L.C.; Tiao, G.; Cummings, B.B.; Alföldi, J.; Wang, Q.; Collins, R.L.; Laricchia, K.M.; Ganna, A.; Birnbaum, D.P. The mutational constraint spectrum quantified from variation in 141,456 humans. Nature 2020, 581, 434–443. [Google Scholar] [CrossRef]
- McLaren, W.; Gil, L.; Hunt, S.E.; Riat, H.S.; Ritchie, G.R.; Thormann, A.; Flicek, P.; Cunningham, F. The ensembl variant effect predictor. Genome Biol. 2016, 17, 122. [Google Scholar] [CrossRef]
- Nemoz, C.; Ropars, V.; Frit, P.; Gontier, A.; Drevet, P.; Yu, J.; Guerois, R.; Pitois, A.; Comte, A.; Delteil, C. XLF and APLF bind Ku80 at two remote sites to ensure DNA repair by non-homologous end joining. Nat. Struct. Mol. Biol. 2018, 25, 971–980. [Google Scholar] [CrossRef]
- Yin, X.; Liu, M.; Tian, Y.; Wang, J.; Xu, Y. Cryo-EM structure of human DNA-PK holoenzyme. Cell Res. 2017, 27, 1341–1350. [Google Scholar] [CrossRef] [PubMed]
- Jumper, J.; Evans, R.; Pritzel, A.; Green, T.; Figurnov, M.; Ronneberger, O.; Tunyasuvunakool, K.; Bates, R.; Žídek, A.; Potapenko, A. Highly accurate protein structure prediction with AlphaFold. Nature 2021, 596, 583–589. [Google Scholar] [CrossRef] [PubMed]
- Ashkenazy, H.; Abadi, S.; Martz, E.; Chay, O.; Mayrose, I.; Pupko, T.; Ben-Tal, N. ConSurf 2016: An improved methodology to estimate and visualize evolutionary conservation in macromolecules. Nucleic Acids Res. 2016, 44, W344–W350. [Google Scholar] [CrossRef]
- Adzhubei, I.; Jordan, D.; Sunyaev, S. Predicting functional effect of human missense mutations using PolyPhen-2. In Current Protocols in Human Genetics; Wiley: Hoboken, NJ, USA, 2013; Chapter 7, Unit 7.20. [Google Scholar]
- Hecht, M.; Bromberg, Y.; Rost, B. Better prediction of functional effects for sequence variants. BMC Genom. 2015, 16 (Suppl. 8), S1. [Google Scholar] [CrossRef] [PubMed]
- Choi, Y.; Chan, A.P. PROVEAN web server: A tool to predict the functional effect of amino acid substitutions and indels. Bioinformatics 2015, 31, 2745–2747. [Google Scholar] [CrossRef] [PubMed]
- Silk, M.; Pires, D.E.; Rodrigues, C.H.; D’Souza, E.N.; Olshansky, M.; Thorne, N.; Ascher, D.B. MTR3D: Identifying regions within protein tertiary structures under purifying selection. Nucleic Acids Res. 2021, 49, W438–W445. [Google Scholar] [CrossRef] [PubMed]
- Gray, V.E.; Hause, R.J.; Luebeck, J.; Shendure, J.; Fowler, D.M. Quantitative Missense Variant Effect Prediction Using Large-Scale Mutagenesis Data. Cell Syst. 2018, 6, 116–124.e3. [Google Scholar] [CrossRef]
- Ng, P.C.; Henikoff, S. SIFT: Predicting amino acid changes that affect protein function. Nucleic Acids Res. 2003, 31, 3812–3814. [Google Scholar] [CrossRef]
- Pires, D.E.; Ascher, D.B.; Blundell, T.L. mCSM: Predicting the effects of mutations in proteins using graph-based signatures. Bioinformatics 2014, 30, 335–342. [Google Scholar] [CrossRef]
- Rodrigues, C.H.M.; Pires, D.E.V.; Ascher, D.B. DynaMut2: Assessing changes in stability and flexibility upon single and multiple point missense mutations. Protein Sci. 2021, 30, 60–69. [Google Scholar] [CrossRef]
- Rodrigues, C.H.; Pires, D.E.; Ascher, D.B. DynaMut: Predicting the impact of mutations on protein conformation, flexibility and stability. Nucleic Acids Res. 2018, 46, W350–W355. [Google Scholar] [CrossRef] [PubMed]
- Pandurangan, A.P.; Ochoa-Montaño, B.; Ascher, D.B.; Blundell, T.L. SDM: A server for predicting effects of mutations on protein stability. Nucleic Acids Res. 2017, 45, W229–W235. [Google Scholar] [CrossRef] [PubMed]
- Pires, D.E.; Ascher, D.B.; Blundell, T.L. DUET: A server for predicting effects of mutations on protein stability using an integrated computational approach. Nucleic Acids Res. 2014, 42, W314–W319. [Google Scholar] [CrossRef]
- Rodrigues, C.H.; Pires, D.E.; Ascher, D.B. mmCSM-PPI: Predicting the effects of multiple point mutations on protein–protein interactions. Nucleic Acids Res. 2021, 49, W417–W424. [Google Scholar] [CrossRef] [PubMed]
- Rodrigues, C.H.; Myung, Y.; Pires, D.E.; Ascher, D.B. mCSM-PPI2: Predicting the effects of mutations on protein–protein interactions. Nucleic Acids Res. 2019, 47, W338–W344. [Google Scholar] [CrossRef] [PubMed]
- Pires, D.E.V.; Ascher, D.B. MCSM-NA: Predicting the effects of mutations on protein-nucleic acids interactions. Nucleic Acids Res. 2017, 45, W241–W246. [Google Scholar] [CrossRef]
- Jubb, H.C.; Higueruelo, A.P.; Ochoa-Montaño, B.; Pitt, W.R.; Ascher, D.B.; Blundell, T.L. Arpeggio: A web server for calculating and visualising interatomic interactions in protein structures. J. Mol. Biol. 2017, 429, 365–371. [Google Scholar] [CrossRef]
- Pedregosa, F.; Varoquaux, G.; Gramfort, A.; Michel, V.; Thirion, B.; Grisel, O.; Blondel, M.; Prettenhofer, P.; Weiss, R.; Dubourg, V. Scikit-learn: Machine learning in Python. J. Mach. Learn. Res. 2011, 12, 2825–2830. [Google Scholar]
- Doherty, A.J.; Jackson, S.P. DNA repair: How Ku makes ends meet. Curr. Biol. 2001, 11, R920–R924. [Google Scholar] [CrossRef]

| Protein | Class | n |
|---|---|---|
| Ku70/80 heterodimer | Pathogenic | 346 |
| Nonpathogenic | 380 | |
| DNA-PKcs | Pathogenic | 654 |
| Nonpathogenic | 1483 | |
| DNA Ligase IV | Pathogenic | 259 |
| Nonpathogenic | 444 | |
| XRCC4 | Pathogenic | 67 |
| Nonpathogenic | 83 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Al-Jarf, R.; Karmakar, M.; Myung, Y.; Ascher, D.B. Uncovering the Molecular Drivers of NHEJ DNA Repair-Implicated Missense Variants and Their Functional Consequences. Genes 2023, 14, 1890. https://doi.org/10.3390/genes14101890
Al-Jarf R, Karmakar M, Myung Y, Ascher DB. Uncovering the Molecular Drivers of NHEJ DNA Repair-Implicated Missense Variants and Their Functional Consequences. Genes. 2023; 14(10):1890. https://doi.org/10.3390/genes14101890
Chicago/Turabian StyleAl-Jarf, Raghad, Malancha Karmakar, Yoochan Myung, and David B. Ascher. 2023. "Uncovering the Molecular Drivers of NHEJ DNA Repair-Implicated Missense Variants and Their Functional Consequences" Genes 14, no. 10: 1890. https://doi.org/10.3390/genes14101890
APA StyleAl-Jarf, R., Karmakar, M., Myung, Y., & Ascher, D. B. (2023). Uncovering the Molecular Drivers of NHEJ DNA Repair-Implicated Missense Variants and Their Functional Consequences. Genes, 14(10), 1890. https://doi.org/10.3390/genes14101890






