Recently Integrated Alu Elements in Capuchin Monkeys: A Resource for Cebus/Sapajus Genomics
Abstract
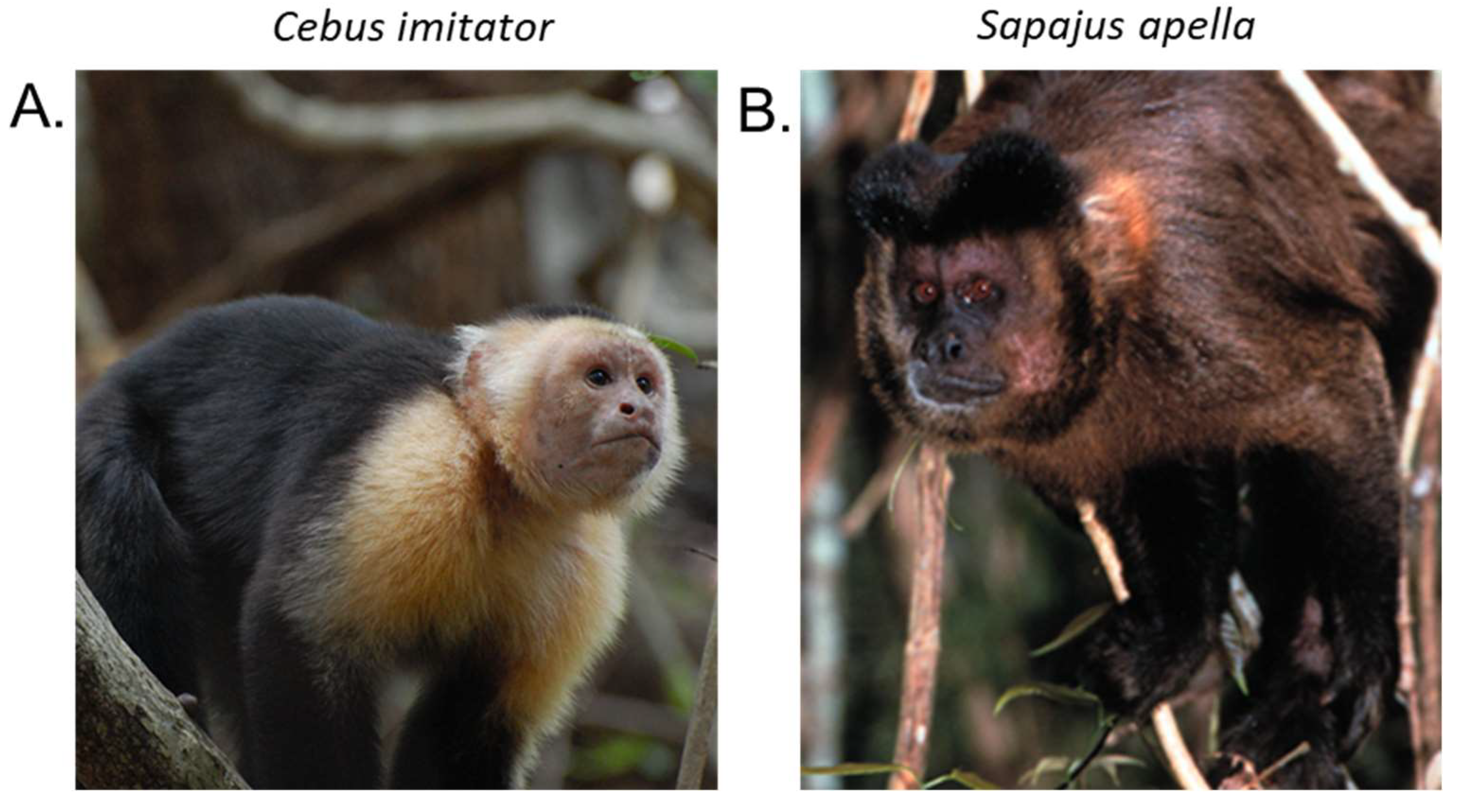
1. Introduction
2. Materials and Methods
2.1. Lineage-Specific Alu Elements
2.2. Oligonucleotide Primer Design
2.3. DNA Samples
2.4. PCR Amplification
2.5. Sanger Chain Termination DNA Sequencing
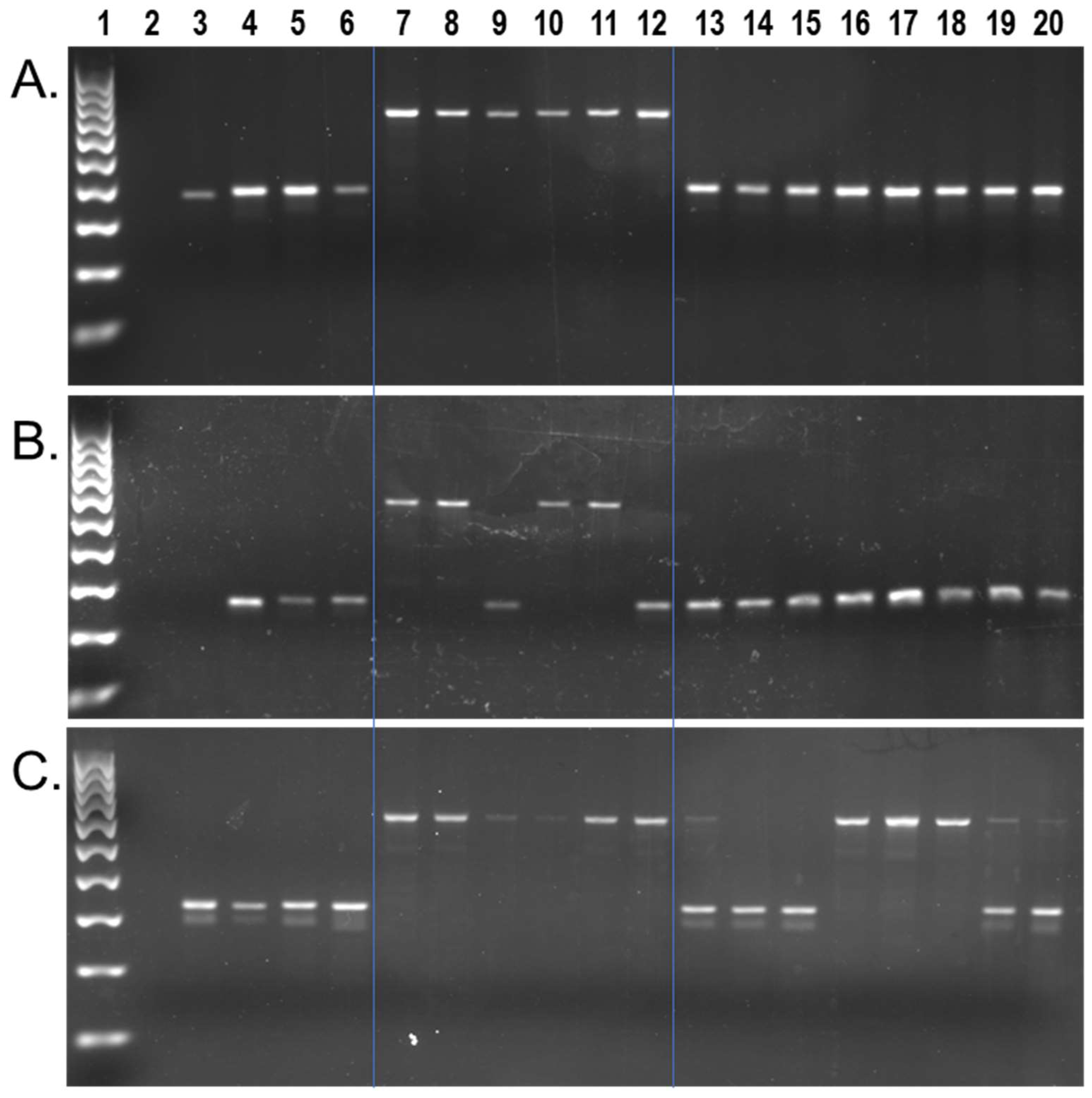
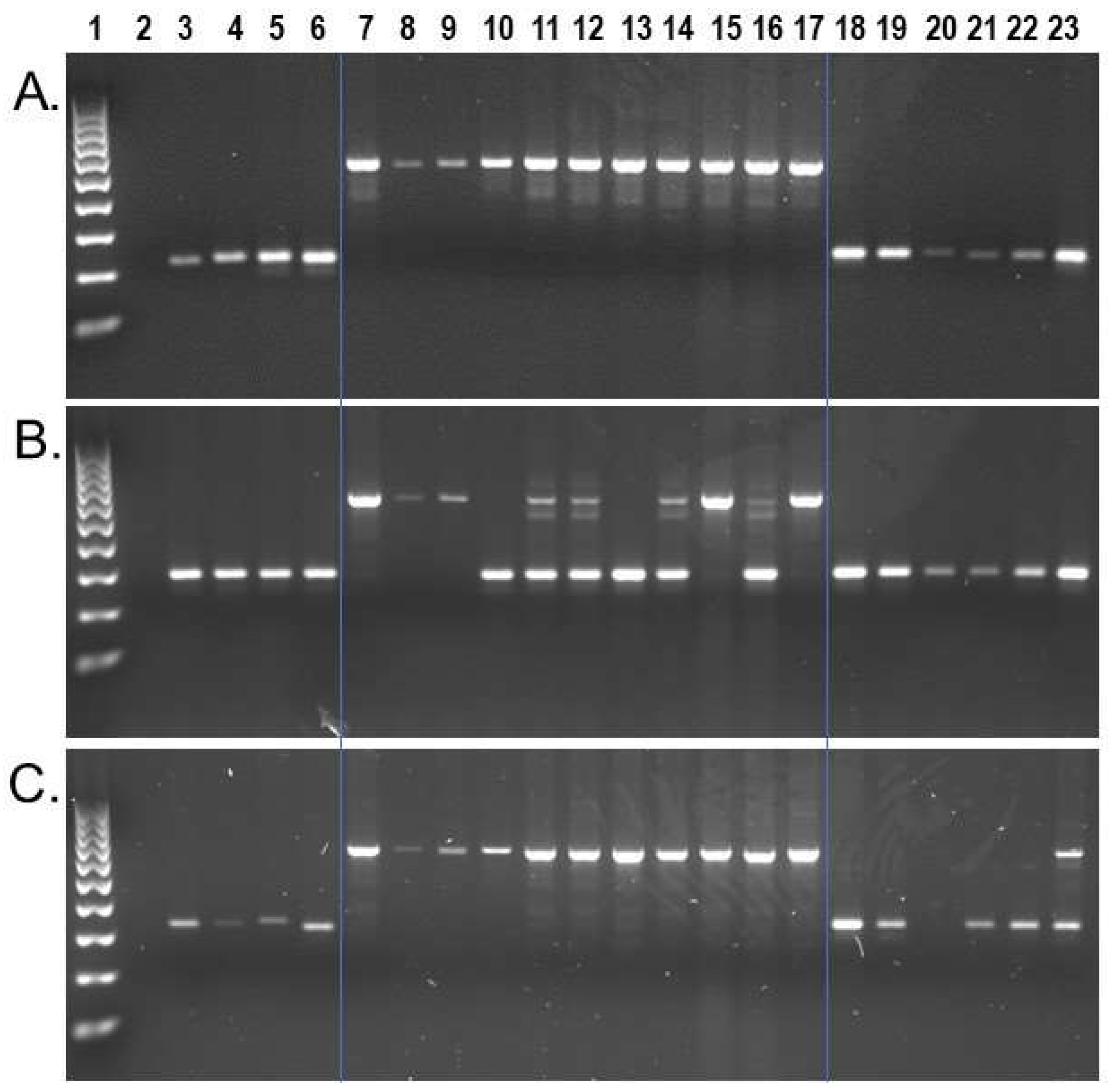
3. Results
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Alfaro, J.W.; Silva, J.D., Jr.; Rylands, A.B. How different are robust and gracile capuchin monkeys? An argument for the use of sapajus and cebus. Am. J. Primatol. 2012, 74, 273–286. [Google Scholar] [CrossRef]
- Martins-Junior, A.M.G.; Carneiro, J.; Sampaio, I.; Ferrari, S.F.; Schneider, H. Phylogenetic relationships among Capuchin (Cebidae, Platyrrhini) lineages: An old event of sympatry explains the current distribution of Cebus and Sapajus. Genet. Mol. Biol. 2018, 41, 699–712. [Google Scholar] [CrossRef] [PubMed]
- Boubli, J.P.; Rylands, A.B.; Farias, I.P.; Alfaro, M.E.; Alfaro, J.L. Cebus phylogenetic relationships: A preliminary reassessment of the diversity of the untufted capuchin monkeys. Am. J. Primatol. 2012, 74, 381–393. [Google Scholar] [CrossRef] [PubMed]
- Schoch, C.L.; Ciufo, S.; Domrachev, M.; Hotton, C.L.; Kannan, S.; Khovanskaya, R.; Leipe, D.; McVeigh, R.; O’Neill, K.; Robbertse, B.; et al. NCBI Taxonomy: A comprehensive update on curation, resources and tools. Database J. Biol. Databases Curation 2020, baaa062. [Google Scholar] [CrossRef] [PubMed]
- Lima, M.G.M.; Silva-Júnior, J.S.E.; Černý, D.; Buckner, J.C.; Aleixo, A.; Chang, J.; Zheng, J.; Alfaro, M.E.; Martins, A.; Di Fiore, A.; et al. A phylogenomic perspective on the robust capuchin monkey (Sapajus) radiation: First evidence for extensive population admixture across South America. Mol. Phylogenet. Evol. 2018, 124, 137–150. [Google Scholar] [CrossRef]
- Lynch Alfaro, J.W.; Boubli, J.P.; Olson, L.E.; Di Fiore, A.; Wilson, B.; Gutiérrez-Espeleta, G.A.; Chiou, K.L.; Schulte, M.; Neitzel, S.; Ross, V.; et al. Explosive Pleistocene range expansion leads to widespread Amazonian sympatry between robust and gracile capuchin monkeys. J. Biogeogr. 2012, 39, 272–288. [Google Scholar] [CrossRef]
- Ruiz-Garcia, M.; Castillo, M.I.; Ledezma, A.; Leguizamon, N.; Sánchez, R.; Chinchilla, M.; Gutierrez-Espeleta, G.A. Molecular systematics and phylogeography of Cebus capucinus (Cebidae, Primates) in Colombia and Costa Rica by means of the mitochondrial COII gene. Am. J. Primatol. 2012, 74, 366–380. [Google Scholar] [CrossRef] [PubMed]
- Ruiz-García, M.; Castillo, M.I.; Lichilín-Ortiz, N.; Pinedo-Castro, M. Molecular relationships and classification of several tufted capuchin lineages (Cebus apella, Cebus xanthosternos and Cebus nigritus, Cebidae), by means of mitochondrial cytochrome oxidase II gene sequences. Folia Primatol. Int. J. Primatol. 2012, 83, 100–125. [Google Scholar] [CrossRef] [PubMed]
- Lynch Alfaro, J.W.; Izar, P.; Ferreira, R.G. Capuchin monkey research priorities and urgent issues. Am. J. Primatol. 2014, 76, 705–720. [Google Scholar] [CrossRef]
- Martins, W.P.; de Melo, F.R.; Kierulff, M.C.M.; Mittermeier, R.A.; Lynch Alfaro, J.W.; Jerusalinsky, L. Sapajus robustus (amended version of 2019 assessment). IUCN Red List Threat. Species 2021, e.T42697A192592444. [Google Scholar] [CrossRef]
- Fialho, M.S.; Jerusalinsky, L.; Moura, E.F.; Ravetta, A.L.; Laroque, P.O.; de Queiroz, H.L.; Boubli, J.P.; Lynch Alfaro, J.W. Cebus kaapori (amended version of 2020 assessment). IUCN Red List Threat. Species 2021, e.T40019A191704766. [Google Scholar] [CrossRef]
- Martins, A.B.; Fialho, M.S.; Jerusalinsky, L.; Valença-Montenegro, M.M.; Bezerra, B.M.; Laroque, P.O.; de Melo, F.R.; Lynch Alfaro, J.W. Sapajus libidinosus (amended version of 2019 assessment). IUCN Red List Threat. Species 2021, e.T136346A192593226. [Google Scholar] [CrossRef]
- De Moraes, B.L.C.; Borges, D.B.; Souza-Alves, J.P.; Boubli, J.P.; Bezerra, B. Microsatellite Markers for Bearded Capuchins (Sapajus libidinosus): Transferability and Characterization. An. Acad. Bras. Cienc. 2021, 93, e20190802. [Google Scholar] [CrossRef]
- Gron, K.J. April 17. Primate Factsheets: Tufted capuchin (Cebus apella) Taxonomy, Morphology, & Ecology. 2009. Available online: https://primate.wisc.edu/primate-info-net/pin-factsheets/pin-factsheet-tufted-capuchin/Link (accessed on 2 March 2022).
- Baker, J.N.; Walker, J.A.; Denham, M.W.; Loupe, C.D., 3rd; Batzer, M.A. Recently integrated Alu insertions in the squirrel monkey (Saimiri) lineage and application for population analyses. Mob DNA 2018, 9, 9. [Google Scholar] [CrossRef] [PubMed]
- Batzer, M.A.; Deininger, P.L. Alu repeats and human genomic diversity. Nat. Rev. Genet. 2002, 3, 370–379. [Google Scholar] [CrossRef]
- Li, J.; Han, K.; Xing, J.; Kim, H.S.; Rogers, J.; Ryder, O.A.; Disotell, T.; Yue, B.; Batzer, M.A. Phylogeny of the macaques (Cercopithecidae: Macaca) based on Alu elements. Gene 2009, 448, 242–249. [Google Scholar] [CrossRef] [PubMed]
- McLain, A.T.; Meyer, T.J.; Faulk, C.; Herke, S.W.; Oldenburg, J.M.; Bourgeois, M.G.; Abshire, C.F.; Roos, C.; Batzer, M.A. An alu-based phylogeny of lemurs (infraorder: Lemuriformes). PLoS ONE 2012, 7, e44035. [Google Scholar] [CrossRef]
- Meyer, T.J.; McLain, A.T.; Oldenburg, J.M.; Faulk, C.; Bourgeois, M.G.; Conlin, E.M.; Mootnick, A.R.; de Jong, P.J.; Roos, C.; Carbone, L.; et al. An Alu-based phylogeny of gibbons (hylobatidae). Mol. Biol. Evol. 2012, 29, 3441–3450. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Perna, N.T.; Batzer, M.A.; Deininger, P.L.; Stoneking, M. Alu insertion polymorphism: A new type of marker for human population studies. Hum. Biol. 1992, 64, 641–648. [Google Scholar]
- Ray, D.A.; Batzer, M.A. Tracking Alu evolution in New World primates. BMC Evol. Biol. 2005, 5, 51. [Google Scholar] [CrossRef][Green Version]
- Salem, A.H.; Ray, D.A.; Xing, J.; Callinan, P.A.; Myers, J.S.; Hedges, D.J.; Garber, R.K.; Witherspoon, D.J.; Jorde, L.B.; Batzer, M.A. Alu elements and hominid phylogenetics. Proc. Natl. Acad. Sci. USA 2003, 100, 12787–12791. [Google Scholar] [CrossRef]
- Steely, C.J.; Walker, J.A.; Jordan, V.E.; Beckstrom, T.O.; McDaniel, C.L.; St Romain, C.P.; Bennett, E.C.; Robichaux, A.; Clement, B.N.; Raveendran, M.; et al. Alu Insertion Polymorphisms as Evidence for Population Structure in Baboons. Genome Biol. Evol. 2017, 9, 2418–2427. [Google Scholar] [CrossRef] [PubMed]
- Stewart, C.; Kural, D.; Stromberg, M.P.; Walker, J.A.; Konkel, M.K.; Stutz, A.M.; Urban, A.E.; Grubert, F.; Lam, H.Y.; Lee, W.P.; et al. A comprehensive map of mobile element insertion polymorphisms in humans. PLoS Genet. 2011, 7, e1002236. [Google Scholar] [CrossRef]
- Xing, J.; Wang, H.; Han, K.; Ray, D.A.; Huang, C.H.; Chemnick, L.G.; Stewart, C.B.; Disotell, T.R.; Ryder, O.A.; Batzer, M.A. A mobile element based phylogeny of Old World monkeys. Mol. Phylogenet. Evol. 2005, 37, 872–880. [Google Scholar] [CrossRef]
- Xing, J.; Wang, H.; Zhang, Y.; Ray, D.A.; Tosi, A.J.; Disotell, T.R.; Batzer, M.A. A mobile element-based evolutionary history of guenons (tribe Cercopithecini). BMC Biol. 2007, 5, 5. [Google Scholar] [CrossRef] [PubMed]
- Roos, C.; Zinner, D.; Kubatko, L.S.; Schwarz, C.; Yang, M.; Meyer, D.; Nash, S.D.; Xing, J.; Batzer, M.A.; Brameier, M.; et al. Nuclear versus mitochondrial DNA: Evidence for hybridization in colobine monkeys. BMC Evol. Biol. 2011, 11, 77. [Google Scholar] [CrossRef] [PubMed]
- Hartig, G.; Churakov, G.; Warren, W.C.; Brosius, J.; Makałowski, W.; Schmitz, J. Retrophylogenomics place tarsiers on the evolutionary branch of anthropoids. Sci. Rep. 2013, 3, 1756. [Google Scholar] [CrossRef]
- Liu, G.E.; Alkan, C.; Jiang, L.; Zhao, S.; Eichler, E.E. Comparative analysis of Alu repeats in primate genomes. Genome Res. 2009, 19, 876–885. [Google Scholar] [CrossRef]
- Schmitz, J.; Ohme, M.; Zischler, H. SINE insertions in cladistic analyses and the phylogenetic affiliations of Tarsius bancanus to other primates. Genetics 2001, 157, 777–784. [Google Scholar] [CrossRef]
- Singer, S.S.; Schmitz, J.; Schwiegk, C.; Zischler, H. Molecular cladistic markers in New World monkey phylogeny (Platyrrhini, Primates). Mol. Phylogenet. Evol. 2003, 26, 490–501. [Google Scholar] [CrossRef]
- Luan, D.D.; Korman, M.H.; Jakubczak, J.L.; Eickbush, T.H. Reverse transcription of R2Bm RNA is primed by a nick at the chromosomal target site: A mechanism for non-LTR retrotransposition. Cell 1993, 72, 595–605. [Google Scholar] [CrossRef]
- Batzer, M.A.; Deininger, P.L. A human-specific subfamily of Alu sequences. Genomics 1991, 9, 481–487. [Google Scholar] [CrossRef]
- Batzer, M.A.; Deininger, P.L.; Hellmann-Blumberg, U.; Jurka, J.; Labuda, D.; Rubin, C.M.; Schmid, C.W.; Zietkiewicz, E.; Zuckerkandl, E. Standardized nomenclature for Alu repeats. J. Mol. Evol. 1996, 42, 3–6. [Google Scholar] [CrossRef]
- Jurka, J. Sequence patterns indicate an enzymatic involvement in integration of mammalian retroposons. Proc. Natl. Acad. Sci. USA 1997, 94, 1872–1877. [Google Scholar] [CrossRef] [PubMed]
- Kapitonov, V.; Jurka, J. The age of Alu subfamilies. J. Mol. Evol. 1996, 42, 59–65. [Google Scholar] [CrossRef]
- Konkel, M.K.; Walker, J.A.; Batzer, M.A. LINEs and SINEs of Primate Evolution. Evol. Anthropol. 2010, 19, 236–249. [Google Scholar] [CrossRef]
- Churakov, G.; Grundmann, N.; Kuritzin, A.; Brosius, J.; Makałowski, W.; Schmitz, J. A novel web-based TinT application and the chronology of the Primate Alu retroposon activity. BMC Evol. Biol. 2010, 10, 376. [Google Scholar] [CrossRef]
- Worley, K.C.; Warren, W.C.; Rogers, J.; Locke, D.; Muzny, D.M.; Mardis, E.R.; Weinstock, G.M.; Tardif, S.D. The common marmoset genome provides insight into primate biology and evolution. Nat. Genet. 2014, 46, 850–857. [Google Scholar] [CrossRef]
- Baker, J.N.; Walker, J.A.; Vanchiere, J.A.; Phillippe, K.R.; St Romain, C.P.; Gonzalez-Quiroga, P.; Denham, M.W.; Mierl, J.R.; Konkel, M.K.; Batzer, M.A. Evolution of Alu Subfamily Structure in the Saimiri Lineage of New World Monkeys. Genome Biol. Evol. 2017, 9, 2365–2376. [Google Scholar] [CrossRef] [PubMed]
- Storer, J. Characterization and Amplification of Retrotransposable Elements Platy-1 and Alu in the Cebidae Lineage of Platyrrhine Primates. LSU Doctoral Dissertations. 5053. 2019. Available online: https://digitalcommons.lsu.edu/gradschool_dissertations/5053 (accessed on 2 March 2022).
- Storer, J.M.; Walker, J.A.; Jordan, V.E.; Batzer, M.A. Sensitivity of the polyDetect computational pipeline for phylogenetic analyses. Anal. Biochem. 2020, 593, 113516. [Google Scholar] [CrossRef] [PubMed]
- Osterholz, M.; Vermeer, J.; Walter, L.; Roos, C. A PCR-based marker to simply identify Saimiri sciureus and S. boliviensis boliviensis. Am. J. Primatol. 2008, 70, 1177–1180. [Google Scholar] [CrossRef] [PubMed]
- Osterholz, M.; Walter, L.; Roos, C. Retropositional events consolidate the branching order among New World monkey genera. Mol. Phylogenet. Evol. 2009, 50, 507–513. [Google Scholar] [CrossRef]
- Ray, D.A.; Xing, J.; Hedges, D.J.; Hall, M.A.; Laborde, M.E.; Anders, B.A.; White, B.R.; Stoilova, N.; Fowlkes, J.D.; Landry, K.E.; et al. Alu insertion loci and platyrrhine primate phylogeny. Mol. Phylogenet. Evol. 2005, 35, 117–126. [Google Scholar] [CrossRef]
- Martins, A.M., Jr.; Amorim, N.; Carneiro, J.C.; de Mello Affonso, P.R.; Sampaio, I.; Schneider, H. Alu elements and the phylogeny of capuchin (Cebus and Sapajus) monkeys. Am. J. Primatol. 2015, 77, 368–375. [Google Scholar] [CrossRef]
- Orkin, J.D.; Montague, M.J.; Tejada-Martinez, D.; de Manuel, M.; Del Campo, J.; Cheves Hernandez, S.; Di Fiore, A.; Fontsere, C.; Hodgson, J.A.; Janiak, M.C.; et al. The genomics of ecological flexibility, large brains, and long lives in capuchin monkeys revealed with fecalFACS. Proc. Natl. Acad. Sci. USA 2021, 118, e2010632118. [Google Scholar] [CrossRef]
- Smit, A.F.A.; Hubley, R.; Green, P. 2013–2015, RepeatMasker Open-4.0. 2015. Available online: http://www.repeatmasker.org (accessed on 4 March 2022).
- Kent, W.J. BLAT—the BLAST-like alignment tool. Genome Res. 2002, 12, 656–664. [Google Scholar] [CrossRef]
- Hall, T.A. BioEdit: A user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp. Ser. 1999, 41, 95–98. [Google Scholar]
- Untergasser, A.; Cutcutache, I.; Koressaar, T.; Ye, J.; Faircloth, B.C.; Remm, M.; Rozen, S.G. Primer3--new capabilities and interfaces. Nucleic Acids Res. 2012, 40, e115. [Google Scholar] [CrossRef]
- Ye, J.; Coulouris, G.; Zaretskaya, I.; Cutcutache, I.; Rozen, S.; Madden, T.L. Primer-BLAST: A tool to design target-specific primers for polymerase chain reaction. BMC Bioinform. 2012, 13, 134. [Google Scholar] [CrossRef] [PubMed]
- Phillips, K.A.; Thompson, C.R. Hand preference for tool-use in capuchin monkeys (Cebus apella) is associated with asymmetry of the primary motor cortex. Am. J. Primatol. 2013, 75, 435–440. [Google Scholar] [CrossRef] [PubMed]
- Renner, E.; Abramo, A.M.; Karen Hambright, M.; Phillips, K.A. Insightful problem solving and emulation in brown capuchin monkeys. Anim. Cogn. 2017, 20, 531–536. [Google Scholar] [CrossRef]
- Sanger, F.; Nicklen, S.; Coulson, A.R. DNA sequencing with chain-terminating inhibitors. Proc. Natl. Acad. Sci. USA 1977, 74, 5463–5467. [Google Scholar] [CrossRef]
- Konkel, M.K.; Walker, J.A.; Hotard, A.B.; Ranck, M.C.; Fontenot, C.C.; Storer, J.; Stewart, C.; Marth, G.T.; Batzer, M.A. Sequence Analysis and Characterization of Active Human Alu Subfamilies Based on the 1000 Genomes Pilot Project. Genome Biol. Evol. 2015, 7, 2608–2622. [Google Scholar] [CrossRef] [PubMed]
- Faircloth, B.C.; McCormack, J.E.; Crawford, N.G.; Harvey, M.G.; Brumfield, R.T.; Glenn, T.C. Ultraconserved elements anchor thousands of genetic markers spanning multiple evolutionary timescales. Syst. Biol. 2012, 61, 717–726. [Google Scholar] [CrossRef] [PubMed]
- Kuritzin, A.; Kischka, T.; Schmitz, J.; Churakov, G. Incomplete Lineage Sorting and Hybridization Statistics for Large-Scale Retroposon Insertion Data. PLoS Comput. Biol. 2016, 12, e1004812. [Google Scholar] [CrossRef] [PubMed]
- Ray, D.A.; Xing, J.; Salem, A.H.; Batzer, M.A. SINEs of a nearly perfect character. Syst. Biol. 2006, 55, 928–935. [Google Scholar] [CrossRef] [PubMed]
- Martins, W.P.; Lynch Alfaro, J.; Rylands, A.B. Reduced range of the endangered crested capuchin monkey (Sapajus robustus) and a possible hybrid zone with Sapajus nigritus. Am. J. Primatol. 2017, 79, e22696. [Google Scholar] [CrossRef]
- Nieves, M.; Mendez, G.; Ortiz, A.; Mühlmann, M.; Mudry, M.D. Karyological diagnosis of Cebus (Primates, Platyrrhini) in captivity: Detection of hybrids and management program applications. Anim. Reprod. Sci. 2008, 108, 66–78. [Google Scholar] [CrossRef]
- Lander, E.S.; Linton, L.M.; Birren, B.; Nusbaum, C.; Zody, M.C.; Baldwin, J.; Devon, K.; Dewar, K.; Doyle, M.; FitzHugh, W.; et al. Initial sequencing and analysis of the human genome. Nature 2001, 409, 860–921. [Google Scholar] [CrossRef] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Storer, J.M.; Walker, J.A.; Rockwell, C.E.; Mores, G.; Beckstrom, T.O.; Orkin, J.D.; Melin, A.D.; Phillips, K.A.; Roos, C.; Batzer, M.A. Recently Integrated Alu Elements in Capuchin Monkeys: A Resource for Cebus/Sapajus Genomics. Genes 2022, 13, 572. https://doi.org/10.3390/genes13040572
Storer JM, Walker JA, Rockwell CE, Mores G, Beckstrom TO, Orkin JD, Melin AD, Phillips KA, Roos C, Batzer MA. Recently Integrated Alu Elements in Capuchin Monkeys: A Resource for Cebus/Sapajus Genomics. Genes. 2022; 13(4):572. https://doi.org/10.3390/genes13040572
Chicago/Turabian StyleStorer, Jessica M., Jerilyn A. Walker, Catherine E. Rockwell, Grayce Mores, Thomas O. Beckstrom, Joseph D. Orkin, Amanda D. Melin, Kimberley A. Phillips, Christian Roos, and Mark A. Batzer. 2022. "Recently Integrated Alu Elements in Capuchin Monkeys: A Resource for Cebus/Sapajus Genomics" Genes 13, no. 4: 572. https://doi.org/10.3390/genes13040572
APA StyleStorer, J. M., Walker, J. A., Rockwell, C. E., Mores, G., Beckstrom, T. O., Orkin, J. D., Melin, A. D., Phillips, K. A., Roos, C., & Batzer, M. A. (2022). Recently Integrated Alu Elements in Capuchin Monkeys: A Resource for Cebus/Sapajus Genomics. Genes, 13(4), 572. https://doi.org/10.3390/genes13040572






