Integrated Tissue and Blood miRNA Expression Profiles Identify Novel Biomarkers for Accurate Non-Invasive Diagnosis of Breast Cancer: Preliminary Results and Future Clinical Implications
Abstract
1. Introduction
2. Materials and Methods
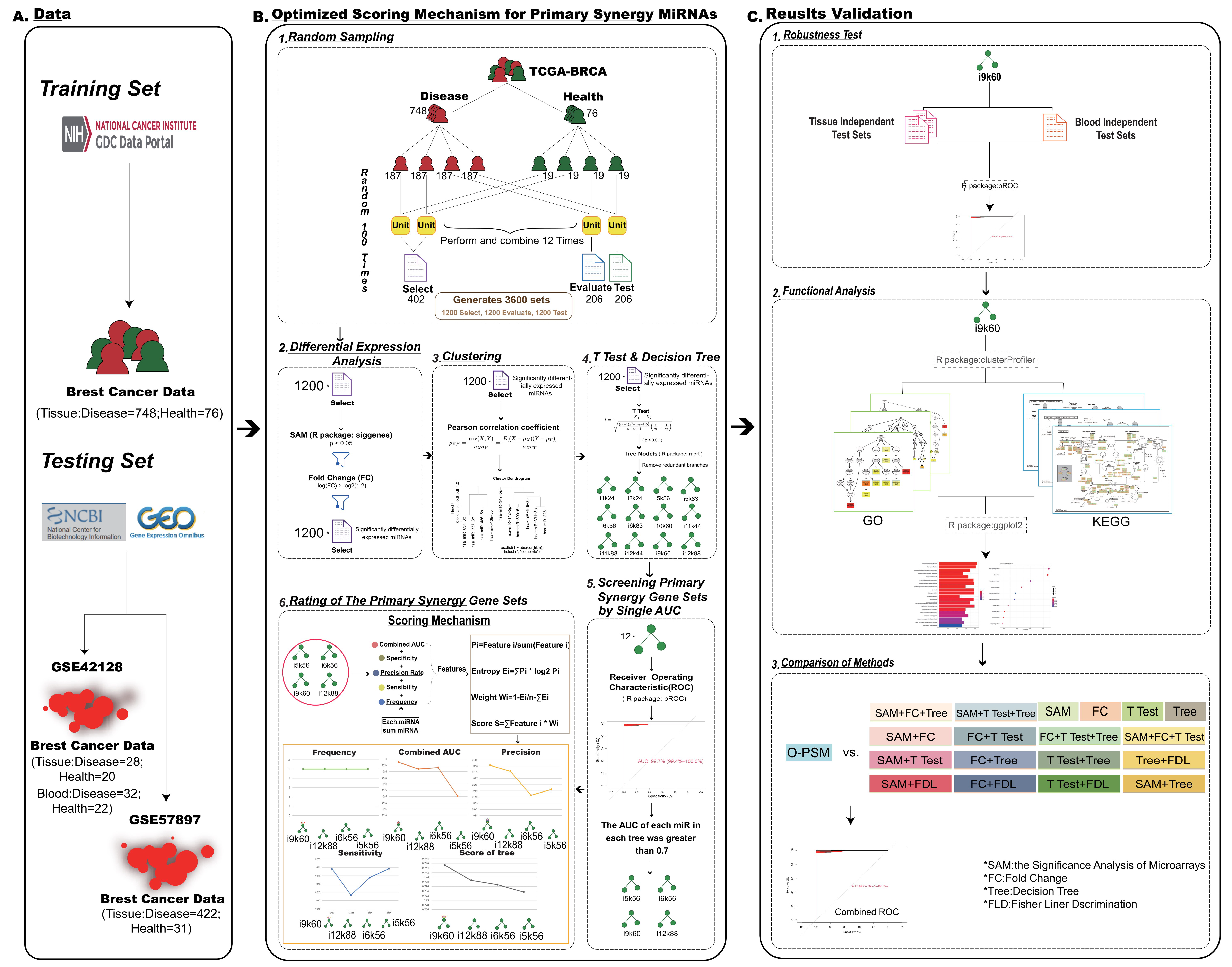
2.1. Data
2.2. Random Sampling
2.3. Differential Expression Analyses
2.4. Clustering
2.5. The T Test and Decision Tree
2.6. Screening Primary Synergy miRNA Sets by Single AUCs
2.7. Ratings of the Primary Synergy miRNA Sets
2.8. O-PSM for Primary Synergy miRNAs
2.9. Comparison of Methods
2.10. Functional Annotation of miRNAs
2.11. Construction of PPI Network and In-Depth Network Analysis
2.12. Survival Analysis
3. Results
3.1. Identification of miRNAs with Primary Synergy Using O-PSM of Tissues
3.2. Significant Categorization of Tissues
3.3. Significant Categorization of Serum
3.4. Categorization Performed Remarkably Well in Typing and Staging of BRCA
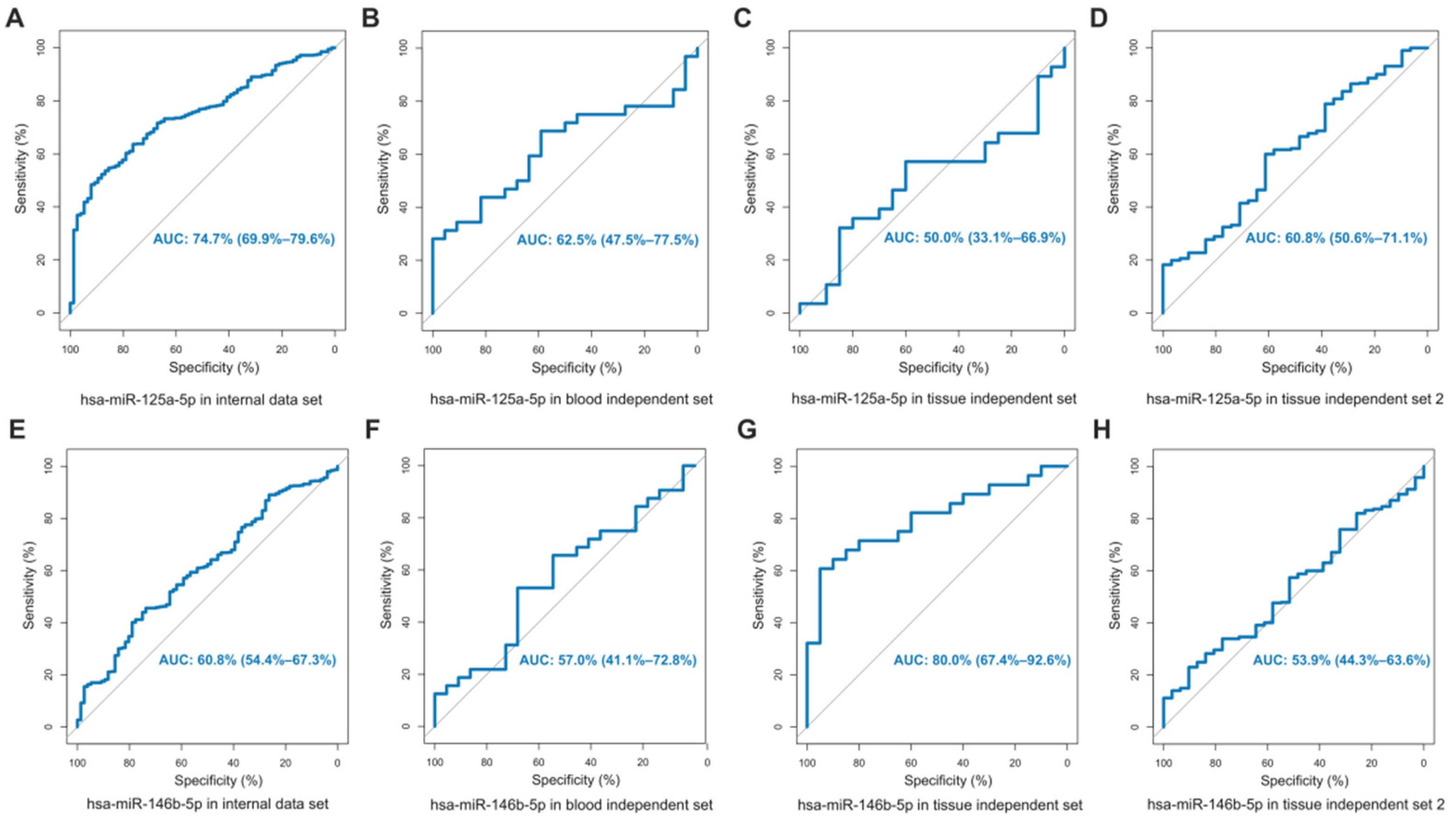
3.5. Comparison with miRNAs That Had Been Experimentally Confirmed
3.6. Comparison of Methods
3.7. Functional Annotation
3.8. PPI Network and Mechanism Analysis of Target Genes
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- De Smet, E.G.; Mestdagh, P.; Vandesompele, J.; Brusselle, G.G.; Bracke, K.R. Non-coding RNAs in the pathogenesis of COPD. Thorax 2015, 70, 782–791. [Google Scholar] [CrossRef] [PubMed]
- Harbeck, N.; Gnant, M. Breast cancer. Lancet 2017, 389, 1134–1150. [Google Scholar] [CrossRef]
- Fan, L.; Strasser-Weippl, K.; Li, J.J.; St Louis, J.; Finkelstein, D.M.; Yu, K.D.; Chen, W.Q.; Shao, Z.M.; Goss, P.E. Breast cancer in China. Lancet Oncol. 2014, 15, e279–e289. [Google Scholar] [CrossRef]
- Brook, N.; Brook, E.; Dharmarajan, A.; Dass, C.R.; Chan, A. Breast cancer bone metastases: Pathogenesis and therapeutic targets. Int. J. Biochem. Cell Biol. 2018, 96, 63–78. [Google Scholar] [CrossRef] [PubMed]
- Duijf, P.H.G.; Nanayakkara, D.; Nones, K.; Srihari, S.; Kalimutho, M.; Khanna, K.K. Mechanisms of genomic instability in breast cancer. Trends Mol. Med. 2019, 25, 595–611. [Google Scholar] [CrossRef] [PubMed]
- Merino Bonilla, J.A.; Torres Tabanera, M.; Ros Mendoza, L.H. Breast cancer in the 21st century: From early detection to new therapies. Cáncer Mama Siglo XXI Detección Precoz A Los Nuevos Tratamientos. Radiol. 2017, 59, 368–379. [Google Scholar] [CrossRef]
- Gomez, S.L.; Von Behren, J.; McKinley, M.; Clarke, C.A.; Shariff-Marco, S.; Cheng, I.; Reynolds, P.; Glaser, S.L. Breast cancer in Asian Americans in California, 1988–2013: Increasing incidence trends and recent data on breast cancer subtypes. Breast Cancer Res. Treat. 2017, 164, 139–147. [Google Scholar] [CrossRef]
- Berry, D.A.; Cronin, K.A.; Plevritis, S.K.; Fryback, D.G.; Clarke, L.; Zelen, M.; Mandelblatt, J.S.; Yakovlev, A.Y.; Habbema, J.D.; Feuer, E.J. Effect of screening and adjuvant therapy on mortality from breast cancer. N. Engl J. Med. 2005, 353, 1784–1792. [Google Scholar] [CrossRef]
- Independent UK Panel on Breast Cancer Screening. The benefits and harms of breast cancer screening: An independent review. Lancet 2012, 380, 1778–1786. [Google Scholar] [CrossRef]
- Acunzo, M.; Romano, G.; Wernicke, D.; Croce, C.M. MicroRNA and cancer–a brief overview. Adv. Biol. Regul. 2015, 57, 1–9. [Google Scholar] [CrossRef]
- Chen, C.; Tan, R.; Wong, L.; Fekete, R.; Halsey, J. Quantitation of microRNAs by real-time RT-qPCR. Methods Mol. Biol. 2011, 687, 113–134. [Google Scholar] [CrossRef] [PubMed]
- Mellis, D.; Caporali, A. MicroRNA-based therapeutics in cardiovascular disease: Screening and delivery to the target. Biochem. Soc. Trans. 2018, 46, 11–21. [Google Scholar] [CrossRef] [PubMed]
- Turchinovich, A.; Weiz, L.; Langheinz, A.; Burwinkel, B. Characterization of extracellular circulating microRNA. Nucleic Acids Res. 2011, 39, 7223–7233. [Google Scholar] [CrossRef] [PubMed]
- Fricke, A.; Cimniak, A.F.V.; Ullrich, P.V.; Becherer, C.; Bickert, C.; Pfeifer, D.; Heinz, J.; Stark, G.B.; Bannasch, H.; Braig, D.; et al. Whole blood miRNA expression analysis reveals miR-3613-3p as a potential biomarker for dedifferentiated liposarcoma. Cancer Biomark. 2018, 22, 199–207. [Google Scholar] [CrossRef] [PubMed]
- Lagos-Quintana, M.; Rauhut, R.; Yalcin, A.; Meyer, J.; Lendeckel, W.; Tuschl, T. Identification of tissue-specific microRNAs from mouse. Curr. Biol. 2002, 12, 735–739. [Google Scholar] [CrossRef]
- Osumi, H.; Shinozaki, E.; Yamaguchi, K.; Zembutsu, H. Clinical utility of circulating tumor DNA for colorectal cancer. Cancer Sci. 2019, 110, 1148–1155. [Google Scholar] [CrossRef] [PubMed]
- Castillo, J.; Bernard, V.; San Lucas, F.A.; Allenson, K.; Capello, M.; Kim, D.U.; Gascoyne, P.; Mulu, F.C.; Stephens, B.M.; Huang, J.; et al. Surfaceome profiling enables isolation of cancer-specific exosomal cargo in liquid biopsies from pancreatic cancer patients. Ann. Oncol. 2018, 29, 223–229. [Google Scholar] [CrossRef] [PubMed]
- Ko, N.Y.; Chen, L.R.; Chen, K.H. The role of micro RNA and long-non-coding RNA in osteoporosis. Int. J. Mol. Sci. 2020, 21, 4886. [Google Scholar] [CrossRef]
- Huang, H.Y.; Lin, Y.C.; Li, J.; Huang, K.Y.; Shrestha, S.; Hong, H.C.; Tang, Y.; Chen, Y.G.; Jin, C.N.; Yu, Y.; et al. miRTarBase 2020: Updates to the experimentally validated microRNA-target interaction database. Nucleic Acids Res. 2020, 48, D148–D154. [Google Scholar] [CrossRef]
- Suzuki, H.I. MicroRNA control of TGF-β signaling. Int. J. Mol. Sci. 2018, 19, 1901. [Google Scholar] [CrossRef]
- Zhang, Y.; Wang, Z.; Gemeinhart, R.A. Progress in microRNA delivery. J. Control. Release 2013, 172, 962–974. [Google Scholar] [CrossRef] [PubMed]
- Humphries, B.; Wang, Z.; Yang, C. The role of microRNAs in metal carcinogen-induced cell malignant transformation and tumorigenesis. Food Chem. Toxicol. 2016, 98, 58–65. [Google Scholar] [CrossRef] [PubMed]
- Kunz, M.; Göttlich, C.; Walles, T.; Nietzer, S.; Dandekar, G.; Dandekar, T. MicroRNA-21 versus microRNA-34: Lung cancer promoting and inhibitory microRNAs analysed in silico and in vitro and their clinical impact. Tumour Biol. 2017, 39, 1010428317706430. [Google Scholar] [CrossRef] [PubMed]
- Xiao, H. MiR-7-5p suppresses tumor metastasis of non-small cell lung cancer by targeting NOVA2. Cell Mol. Biol. Lett. 2019, 24, 60. [Google Scholar] [CrossRef] [PubMed]
- Bertoli, G.; Cava, C.; Castiglioni, I. MicroRNAs: New biomarkers for diagnosis, prognosis, therapy prediction and therapeutic tools for breast cancer. Theranostics 2015, 5, 1122–1143. [Google Scholar] [CrossRef]
- O’Bryan, S.; Dong, S.; Mathis, J.M.; Alahari, S.K. The roles of oncogenic miRNAs and their therapeutic importance in breast cancer. Eur. J. Cancer 2017, 72, 1–11. [Google Scholar] [CrossRef]
- Sauerbrei, W.; Taube, S.E.; McShane, L.M.; Cavenagh, M.M.; Altman, D.G. Reporting recommendations for tumor marker prognostic studies (REMARK): An abridged explanation and elaboration. J. Natl. Cancer Inst. 2018, 110, 803–811. [Google Scholar] [CrossRef]
- Yang, Y.; Weng, W.; Peng, J.; Hong, L.; Yang, L.; Toiyama, Y.; Gao, R.; Liu, M.; Yin, M.; Pan, C.; et al. Fusobacterium nucleatum increases proliferation of colorectal cancer cells and tumor development in mice by activating toll-like receptor 4 signaling to nuclear factor-κB, and up-regulating expression of MicroRNA-21. Gastroenterology 2017, 152, 851–866.e24. [Google Scholar] [CrossRef]
- Yu, T.; Ma, P.; Wu, D.; Shu, Y.; Gao, W. Functions and mechanisms of microRNA-31 in human cancers. Biomed. Pharmacother. 2018, 108, 1162–1169. [Google Scholar] [CrossRef]
- Mulrane, L.; McGee, S.F.; Gallagher, W.M.; O’Connor, D.P. miRNA dysregulation in breast cancer. Cancer Res. 2013, 73, 6554–6562. [Google Scholar] [CrossRef]
- Ma, X.; Guo, J.; Liu, K.; Chen, L.; Liu, D.; Dong, S.; Xia, J.; Long, Q.; Yue, Y.; Zhao, P.; et al. Identification of a distinct luminal subgroup diagnosing and stratifying early stage prostate cancer by tissue-based single-cell RNA sequencing. Mol. Cancer 2020, 19, 147. [Google Scholar] [CrossRef] [PubMed]
- Gurel, B.; Ali, T.Z.; Montgomery, E.A.; Begum, S.; Hicks, J.; Goggins, M.; Eberhart, C.G.; Clark, D.P.; Bieberich, C.J.; Epstein, J.I.; et al. NKX3.1 as a marker of prostatic origin in metastatic tumors. Am. J. Surg. Pathol. 2010, 34, 1097–1105. [Google Scholar] [CrossRef] [PubMed]
- Praharaj, P.P.; Bhutia, S.K.; Nagrath, S.; Bitting, R.L.; Deep, G. Circulating tumor cell-derived organoids: Current challenges and promises in medical research and precision medicine. Biochim. Biophys. Acta Rev. Cancer 2018, 1869, 117–127. [Google Scholar] [CrossRef] [PubMed]
- Vaidyanathan, R.; Soon, R.H.; Zhang, P.; Jiang, K.; Lim, C.T. Cancer diagnosis: From tumor to liquid biopsy and beyond. Lab Chip 2018, 19, 11–34. [Google Scholar] [CrossRef] [PubMed]
- Shan, X.; Zhang, L.; Zhu, D.X.; Zhou, X.; Zhang, H.; Liu, Q.X.; Tang, J.W.; Wen, W.; Wang, T.S.; Zhu, W.; et al. Serum microRNA expression profiling revealing potential diagnostic biomarkers for lung adenocarcinoma. Chin. Med. J. 2020, 133, 2532–2542. [Google Scholar] [CrossRef]
- Zhang, Y.; Roth, J.A.; Yu, H.; Ye, Y.; Xie, K.; Zhao, H.; Chang, D.W.; Huang, M.; Li, H.; Qu, J.; et al. A 5-microRNA signature identified from serum microRNA profiling predicts survival in patients with advanced stage non-small cell lung cancer. Carcinogenesis 2019, 40, 643–650. [Google Scholar] [CrossRef]
- Kondrotienė, A.; Daukša, A.; Pamedytytė, D.; Kazokaitė, M.; Žvirblienė, A.; Daukšienė, D.; Simanavičienė, V.; Klimaitė, R.; Golubickaitė, I.; Stakaitis, R.; et al. Plasma-derived miRNA-222 as a candidate marker for papillary thyroid cancer. Int. J. Mol. Sci. 2020, 21, 6445. [Google Scholar] [CrossRef]
- Shan, X.; Zhang, H.; Zhang, L.; Zhou, X.; Wang, T.; Zhang, J.; Shu, Y.; Zhu, W.; Wen, W.; Liu, P. Identification of four plasma microRNAs as potential biomarkers in the diagnosis of male lung squamous cell carcinoma patients in China. Cancer Med. 2018, 7, 2370–2381. [Google Scholar] [CrossRef]
- Dittmar, R.L.; Liu, S.; Tai, M.C.; Rajapakshe, K.; Huang, Y.; Longton, G.; DeCapite, C.; Hurd, M.W.; Paris, P.L.; Kirkwood, K.S.; et al. Plasma miRNA biomarkers in limited volume samples for detection of early-stage pancreatic cancer. Cancer Prev. Res. 2021, 14, 729–740. [Google Scholar] [CrossRef]
- Lou, W.; Ding, B.; Xu, L.; Fan, W. Construction of potential glioblastoma multiforme-related miRNA-mRNA regulatory network. Front. Mol. Neurosci. 2019, 12, 66. [Google Scholar] [CrossRef]
- Cheng, Y.; Su, Y.; Wang, S.; Liu, Y.; Jin, L.; Wan, Q.; Liu, Y.; Li, C.; Sang, X.; Yang, L.; et al. Identification of circRNA-lncRNA-miRNA-mRNA competitive endogenous RNA network as novel prognostic markers for acute myeloid leukemia. Genes 2020, 11, 868. [Google Scholar] [CrossRef] [PubMed]
- Lu, Z.; Gao, Y. Screening differentially expressed genes between endometriosis and ovarian cancer to find new biomarkers for endometriosis. Ann. Med. 2021, 53, 1377–1389. [Google Scholar] [CrossRef] [PubMed]
- Wang, F.; Xu, X.; Zhang, N.; Chen, Z. Identification and integrated analysis of hepatocellular carcinoma-related circular RNA signature. Ann. Transl. Med. 2020, 8, 294. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Tao, H.; Li, J.; Zhang, E.; Liang, H.; Zhang, B. Comprehensive analysis of the competing endogenous circRNA-lncRNA-miRNA-mRNA network and identification of a novel potential biomarker for hepatocellular carcinoma. Aging 2021, 13, 15990–16008. [Google Scholar] [CrossRef] [PubMed]
- Elango, R.; Alsaleh, K.A.; Vishnubalaji, R.; Manikandan, M.; Ali, A.M.; Abd El-Aziz, N.; Altheyab, A.; Al-Rikabi, A.; Alfayez, M.; Aldahmash, A.; et al. MicroRNA expression profiling on paired primary and lymph node metastatic breast cancer revealed distinct microRNA profile associated With LNM. Front. Oncol. 2020, 10, 756. [Google Scholar] [CrossRef] [PubMed]
- Fu, S.W.; Chen, L.; Man, Y.G. miRNA biomarkers in breast cancer detection and management. J. Cancer 2011, 2, 116–122. [Google Scholar] [CrossRef]
- Li, M.; Zou, X.; Xia, T.; Wang, T.; Liu, P.; Zhou, X.; Wang, S.; Zhu, W. A five-miRNA panel in plasma was identified for breast cancer diagnosis. Cancer Med. 2019, 8, 7006–7017. [Google Scholar] [CrossRef]
- Chan, M.; Liaw, C.S.; Ji, S.M.; Tan, H.H.; Wong, C.Y.; Thike, A.A.; Tan, P.H.; Ho, G.H.; Lee, A.S. Identification of circulating microRNA signatures for breast cancer detection. Clin. Cancer Res. 2013, 19, 4477–4487. [Google Scholar] [CrossRef]
- Zhao, X.G.; Hu, J.Y.; Tang, J.; Yi, W.; Zhang, M.Y.; Deng, R.; Mai, S.J.; Weng, N.Q.; Wang, R.Q.; Liu, J.; et al. miR-665 expression predicts poor survival and promotes tumor metastasis by targeting NR4A3 in breast cancer. Cell Death Dis. 2019, 10, 479. [Google Scholar] [CrossRef]
- Aure, M.R.; Vitelli, V.; Jernström, S.; Kumar, S.; Krohn, M.; Due, E.U.; Haukaas, T.H.; Leivonen, S.K.; Vollan, H.K.; Lüders, T.; et al. Integrative clustering reveals a novel split in the luminal A subtype of breast cancer with impact on outcome. Breast Cancer Res. 2017, 19, 44. [Google Scholar] [CrossRef]
- Knutsen, E.; Lellahi, S.M.; Aure, M.R.; Nord, S.; Fismen, S.; Larsen, K.B.; Gabriel, M.T.; Hedberg, A.; Bjørklund, S.S.; Bofin, A.M.; et al. The expression of the long NEAT1_2 isoform is associated with human epidermal growth factor receptor 2-positive breast cancers. Sci. Rep. 2020, 10, 1277. [Google Scholar] [CrossRef] [PubMed]
- Pladsen, A.V.; Nilsen, G.; Rueda, O.M.; Aure, M.R.; Borgan, Ø.; Liestøl, K.; Vitelli, V.; Frigessi, A.; Langerød, A.; Mathelier, A.; et al. DNA copy number motifs are strong and independent predictors of survival in breast cancer. Commun. Biol. 2020, 3, 153. [Google Scholar] [CrossRef] [PubMed]
- Hironaka-Mitsuhashi, A.; Matsuzaki, J.; Takahashi, R.U.; Yoshida, M.; Nezu, Y.; Yamamoto, Y.; Shiino, S.; Kinoshita, T.; Ushijima, T.; Hiraoka, N.; et al. A tissue microRNA signature that predicts the prognosis of breast cancer in young women. PLoS ONE 2017, 12, e0187638. [Google Scholar] [CrossRef] [PubMed]
- Shen, J.; Cao, B.; Wang, Y.; Ma, C.; Zeng, Z.; Liu, L.; Li, X.; Tao, D.; Gong, J.; Xie, D. Hippo component YAP promotes focal adhesion and tumour aggressiveness via transcriptionally activating THBS1/FAK signalling in breast cancer. J. Exp. Clin. Cancer Res. 2018, 37, 175. [Google Scholar] [CrossRef] [PubMed]
- Shao, H.; Cheng, H.Y.; Cook, R.G.; Tweardy, D.J. Identification and characterization of signal transducer and activator of transcription 3 recruitment sites within the epidermal growth factor receptor. Cancer Res. 2003, 63, 3923–3930. [Google Scholar]
- Habib, A.A.; Chatterjee, S.; Park, S.K.; Ratan, R.R.; Lefebvre, S.; Vartanian, T. The epidermal growth factor receptor engages receptor interacting protein and nuclear factor-kappa B (NF-kappa B)-inducing kinase to activate NF-kappa B. Identification of a novel receptor-tyrosine kinase signalosome. J. Biol. Chem. 2001, 276, 8865–8874. [Google Scholar] [CrossRef]
- Ullrich, A.; Coussens, L.; Hayflick, J.S.; Dull, T.J.; Gray, A.; Tam, A.W.; Lee, J.; Yarden, Y.; Libermann, T.A.; Schlessinger, J.; et al. Human epidermal growth factor receptor cDNA sequence and aberrant expression of the amplified gene in A431 epidermoid carcinoma cells. Nature 1984, 309, 418–425. [Google Scholar] [CrossRef]
- Maemondo, M.; Inoue, A.; Kobayashi, K.; Sugawara, S.; Oizumi, S.; Isobe, H.; Gemma, A.; Harada, M.; Yoshizawa, H.; Kinoshita, I.; et al. Gefitinib or chemotherapy for non-small-cell lung cancer with mutated EGFR. N. Engl. J. Med. 2010, 362, 2380–2388. [Google Scholar] [CrossRef]
- Talukdar, S.; Emdad, L.; Das, S.K.; Fisher, P.B. EGFR: An essential receptor tyrosine kinase-regulator of cancer stem cells. Adv. Cancer Res. 2020, 147, 161–188. [Google Scholar] [CrossRef]
- Zhang, Z.; Stiegler, A.L.; Boggon, T.J.; Kobayashi, S.; Halmos, B. EGFR-mutated lung cancer: A paradigm of molecular oncology. Oncotarget 2010, 1, 497–514. [Google Scholar] [CrossRef]
- Riely, G.J.; Pao, W.; Pham, D.; Li, A.R.; Rizvi, N.; Venkatraman, E.S.; Zakowski, M.F.; Kris, M.G.; Ladanyi, M.; Miller, V.A. Clinical course of patients with non-small cell lung cancer and epidermal growth factor receptor exon 19 and exon 21 mutations treated with gefitinib or erlotinib. Clin. Cancer Res. 2006, 12, 839–844. [Google Scholar] [CrossRef] [PubMed]
- Shigematsu, H.; Lin, L.; Takahashi, T.; Nomura, M.; Suzuki, M.; Wistuba, I.I.; Fong, K.M.; Lee, H.; Toyooka, S.; Shimizu, N.; et al. Clinical and biological features associated with epidermal growth factor receptor gene mutations in lung cancers. J. Natl. Cancer Inst. 2005, 97, 339–346. [Google Scholar] [CrossRef] [PubMed]
- Pao, W.; Chmielecki, J. Rational, biologically based treatment of EGFR-mutant non-small-cell lung cancer. Nat. Rev. Cancer 2010, 10, 760–774. [Google Scholar] [CrossRef] [PubMed]
- Galland, F.; Karamysheva, A.; Mattei, M.G.; Rosnet, O.; Marchetto, S.; Birnbaum, D. Chromosomal localization of FLT4, a novel receptor-type tyrosine kinase gene. Genomics 1992, 13, 475–478. [Google Scholar] [CrossRef]
- Ruzinova, M.B.; Benezra, R. Id proteins in development, cell cycle and cancer. Trends Cell Biol. 2003, 13, 410–418. [Google Scholar] [CrossRef]
- Park, M.; Dean, M.; Kaul, K.; Braun, M.J.; Gonda, M.A.; Vande Woude, G. Sequence of MET protooncogene cDNA has features characteristic of the tyrosine kinase family of growth-factor receptors. Proc. Natl. Acad. Sci. USA 1987, 84, 6379–6383. [Google Scholar] [CrossRef]
- Finkenzeller, G.; Marmé, D.; Hug, H. Sequence of human protein kinase C α. Nucleic Acids Res. 1990, 18, 2183. [Google Scholar] [CrossRef]
- Kolch, W.; Heidecker, G.; Kochs, G.; Hummel, R.; Vahidi, H.; Mischak, H.; Finkenzeller, G.; Marmé, D.; Rapp, U.R. Protein kinase C α activates RAF-1 by direct phosphorylation. Nature 1993, 364, 249–252. [Google Scholar] [CrossRef]
- Darieva, Z.; Han, N.; Warwood, S.; Doris, K.S.; Morgan, B.A.; Sharrocks, A.D. Protein kinase C regulates late cell cycle-dependent gene expression. Mol. Cell Biol. 2012, 32, 4651–4661. [Google Scholar] [CrossRef]
- Maglione, D.; Guerriero, V.; Viglietto, G.; Delli-Bovi, P.; Persico, M.G. Isolation of a human placenta cDNA coding for a protein related to the vascular permeability factor. Proc. Natl. Acad. Sci. USA 1991, 88, 9267–9271. [Google Scholar] [CrossRef]
- Taylor, A.P.; Rodriguez, M.; Adams, K.; Goldenberg, D.M.; Blumenthal, R.D. Altered tumor vessel maturation and proliferation in placenta growth factor-producing tumors: Potential relationship to post-therapy tumor angiogenesis and recurrence. Int. J. Cancer 2003, 105, 158–164. [Google Scholar] [CrossRef] [PubMed]
- Kim, B.T.; Kitagawa, H.; Tamura, J.; Saito, T.; Kusche-Gullberg, M.; Lindahl, U.; Sugahara, K. Human tumor suppressor EXT gene family members EXTL1 and EXTL3 encode α 1,4- N-acetylglucosaminyltransferases that likely are involved in heparan sulfate/ heparin biosynthesis. Proc. Natl. Acad. Sci. USA 2001, 98, 7176–7181. [Google Scholar] [CrossRef] [PubMed]
- Funderburgh, J.L. Keratan sulfate: Structure, biosynthesis, and function. Glycobiology 2000, 10, 951–958. [Google Scholar] [CrossRef] [PubMed]
- Eskelinen, E.L.; Tanaka, Y.; Saftig, P. At the acidic edge: Emerging functions for lysosomal membrane proteins. Trends Cell Biol. 2003, 13, 137–145. [Google Scholar] [CrossRef]
- Soga, T. Cancer metabolism: Key players in metabolic reprogramming. Cancer Sci. 2013, 104, 275–281. [Google Scholar] [CrossRef]
- Schink, K.O.; Raiborg, C.; Stenmark, H. Phosphatidylinositol 3-phosphate, a lipid that regulates membrane dynamics, protein sorting and cell signalling. Bioessays 2013, 35, 900–912. [Google Scholar] [CrossRef]
- Jin, H.; Tang, Y.; Yang, L.; Peng, X.; Li, B.; Fan, Q.; Wei, S.; Yang, S.; Li, X.; Wu, B.; et al. Rab GTPases: Central coordinators of membrane trafficking in cancer. Front. Cell Dev. Biol. 2021, 9, 648384. [Google Scholar] [CrossRef]
- Mancini, M.; Zazzara, M.; Zattoni, F. Stem cells, biomarkers and genetic profiling: Approaching future challenges in urology. Urologia 2016, 83, 4–13. [Google Scholar] [CrossRef]
- Bahrami, A.; Aledavood, A.; Anvari, K.; Hassanian, S.M.; Maftouh, M.; Yaghobzade, A.; Salarzaee, O.; ShahidSales, S.; Avan, A. The prognostic and therapeutic application of microRNAs in breast cancer: Tissue and circulating microRNAs. J. Cell Physiol. 2018, 233, 774–786. [Google Scholar] [CrossRef]
- Cittelly, D.M.; Das, P.M.; Spoelstra, N.S.; Edgerton, S.M.; Richer, J.K.; Thor, A.D.; Jones, F.E. Downregulation of miR-342 is associated with tamoxifen resistant breast tumors. Mol. Cancer 2010, 9, 317. [Google Scholar] [CrossRef]
- Liu, Q.; Wang, Z.; Zhou, X.; Tang, M.; Tan, W.; Sun, T.; Deng, Y. miR-342-5p inhibits osteosarcoma cell growth, migration, invasion, and sensitivity to doxorubicin through targeting Wnt7b. Cell Cycle 2019, 18, 3325–3336. [Google Scholar] [CrossRef] [PubMed]
- Du, H.; Bao, Y.; Liu, C.; Zhong, A.; Niu, Y.; Tang, X. miR-139-5p enhances cisplatin sensitivity in non-small cell lung cancer cells by inhibiting cell proliferation and promoting apoptosis via the targeting of Homeobox protein HOX-B2. Mol. Med. Rep. 2021, 23, 104. [Google Scholar] [CrossRef] [PubMed]
- Gong, S.; Ying, L.; Fan, Y.; Sun, Z. Fentanyl inhibits lung cancer viability and invasion via upregulation of miR-331-3p and repression of HDAC5. Onco Targets Ther. 2020, 13, 13131–13141. [Google Scholar] [CrossRef] [PubMed]
- Ji, L.; Lin, Z.; Wan, Z.; Xia, S.; Jiang, S.; Cen, D.; Cai, L.; Xu, J.; Cai, X. miR-486-3p mediates hepatocellular carcinoma sorafenib resistance by targeting FGFR4 and EGFR. Cell Death Dis. 2020, 11, 250. [Google Scholar] [CrossRef]
- Wu, H.; Li, X.; Zhang, T.; Zhang, G.; Chen, J.; Chen, L.; He, M.; Hao, B.; Wang, C. Overexpression miR-486-3p promoted by allicin enhances temozolomide sensitivity in glioblastoma via targeting MGMT. Neuromol. Med. 2020, 22, 359–369. [Google Scholar] [CrossRef]
- Niu, Y.C.; Tong, J.; Shi, X.F.; Zhang, T. MicroRNA-654-3p enhances cisplatin sensitivity by targeting QPRT and inhibiting the PI3K/AKT signaling pathway in ovarian cancer cells. Exp. Ther. Med. 2020, 20, 1467–1479. [Google Scholar] [CrossRef]
- Mancini, M.; Righetto, M.; Noessner, E. Checkpoint inhibition in bladder cancer: Clinical expectations, current evidence, and proposal of future strategies based on a tumor-specific immunobiological approach. Cancers 2021, 13, 6016. [Google Scholar] [CrossRef]




| Subtype | Combined AUC | CI |
|---|---|---|
| Basal-like | 0.840 | 78.94~89.15% |
| Normal-like | 0.876 | 83.05~92.13% |
| Luminal A | 0.680 | 62.66~73.39% |
| Luminal B | 0.686 | 62.30~75.00% |
| HER-2 (+) | 0.686 | 60.21~76.95% |
| Stage | Combined AUC | CI |
|---|---|---|
| 1 | 0.657 | 51.78~79.70% |
| 2 | 0.723 | 59.08~85.48% |
| 3 | 0.615 | 62.12~85.03% |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Su, F.; Gao, Z.; Liu, Y.; Zhou, G.; Cui, Y.; Deng, C.; Liu, Y.; Zhang, Y.; Ma, X.; Wang, Y.; et al. Integrated Tissue and Blood miRNA Expression Profiles Identify Novel Biomarkers for Accurate Non-Invasive Diagnosis of Breast Cancer: Preliminary Results and Future Clinical Implications. Genes 2022, 13, 1931. https://doi.org/10.3390/genes13111931
Su F, Gao Z, Liu Y, Zhou G, Cui Y, Deng C, Liu Y, Zhang Y, Ma X, Wang Y, et al. Integrated Tissue and Blood miRNA Expression Profiles Identify Novel Biomarkers for Accurate Non-Invasive Diagnosis of Breast Cancer: Preliminary Results and Future Clinical Implications. Genes. 2022; 13(11):1931. https://doi.org/10.3390/genes13111931
Chicago/Turabian StyleSu, Fei, Ziyu Gao, Yueyang Liu, Guiqin Zhou, Ying Cui, Chao Deng, Yuyu Liu, Yihao Zhang, Xiaoyan Ma, Yongxia Wang, and et al. 2022. "Integrated Tissue and Blood miRNA Expression Profiles Identify Novel Biomarkers for Accurate Non-Invasive Diagnosis of Breast Cancer: Preliminary Results and Future Clinical Implications" Genes 13, no. 11: 1931. https://doi.org/10.3390/genes13111931
APA StyleSu, F., Gao, Z., Liu, Y., Zhou, G., Cui, Y., Deng, C., Liu, Y., Zhang, Y., Ma, X., Wang, Y., Guan, L., Zhang, Y., & Liu, B. (2022). Integrated Tissue and Blood miRNA Expression Profiles Identify Novel Biomarkers for Accurate Non-Invasive Diagnosis of Breast Cancer: Preliminary Results and Future Clinical Implications. Genes, 13(11), 1931. https://doi.org/10.3390/genes13111931





