GReNaDIne: A Data-Driven Python Library to Infer Gene Regulatory Networks from Gene Expression Data
Abstract
1. Introduction
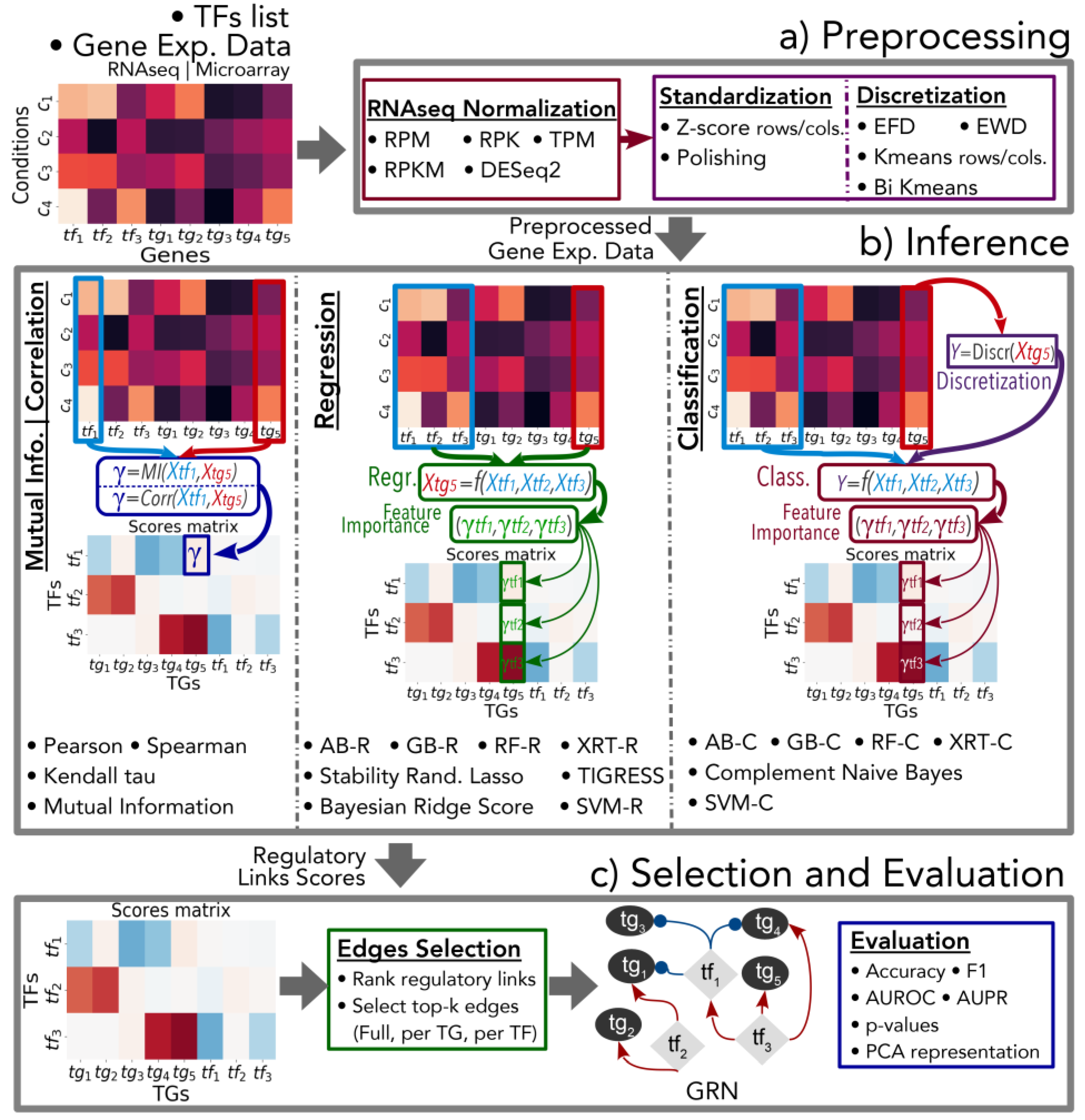
2. Implementation
2.1. Module 1: Preprocessing
2.2. Module 2: Gene Regulatory Network Inference
2.3. Module 3: Links Selection and Evaluation
3. Results and Discussion
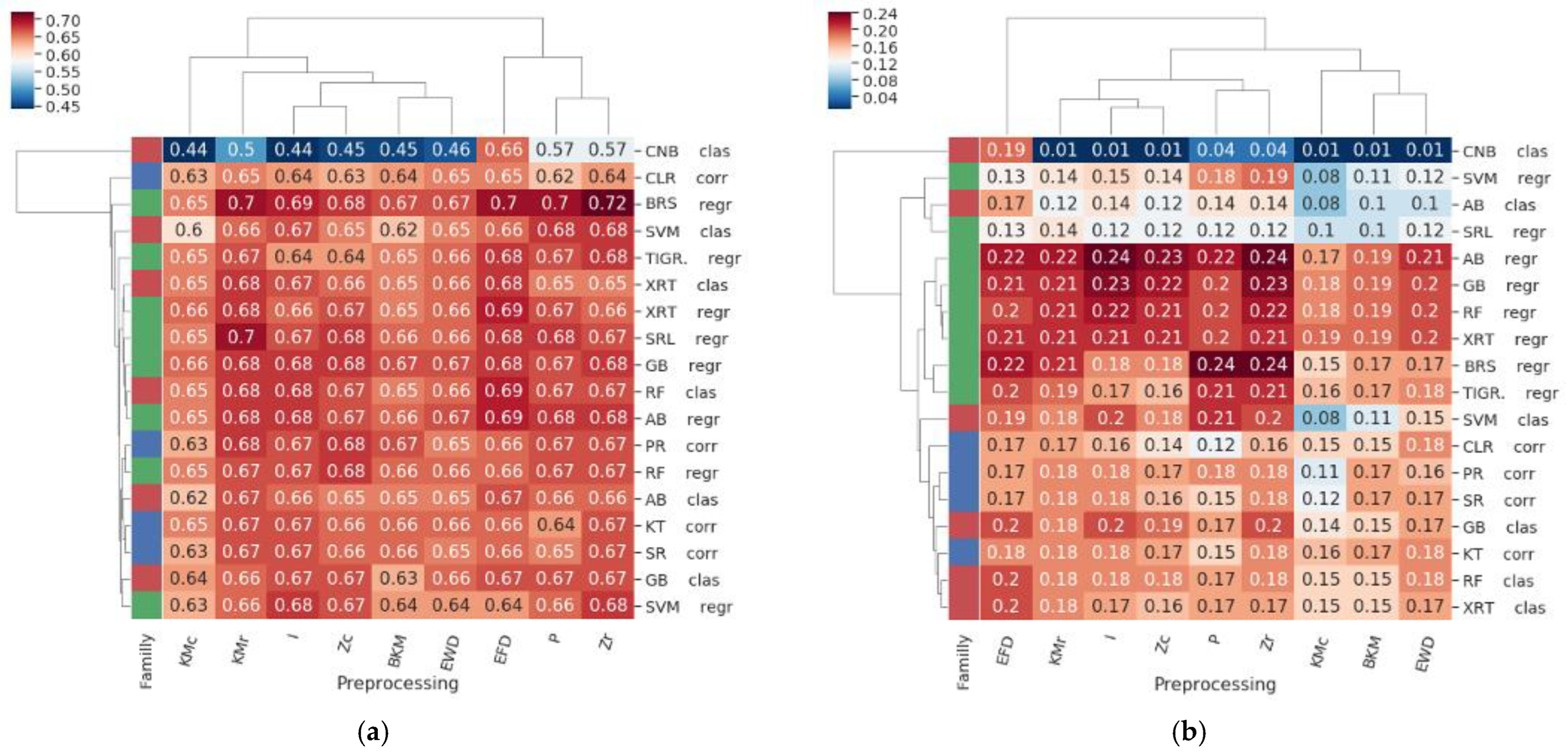
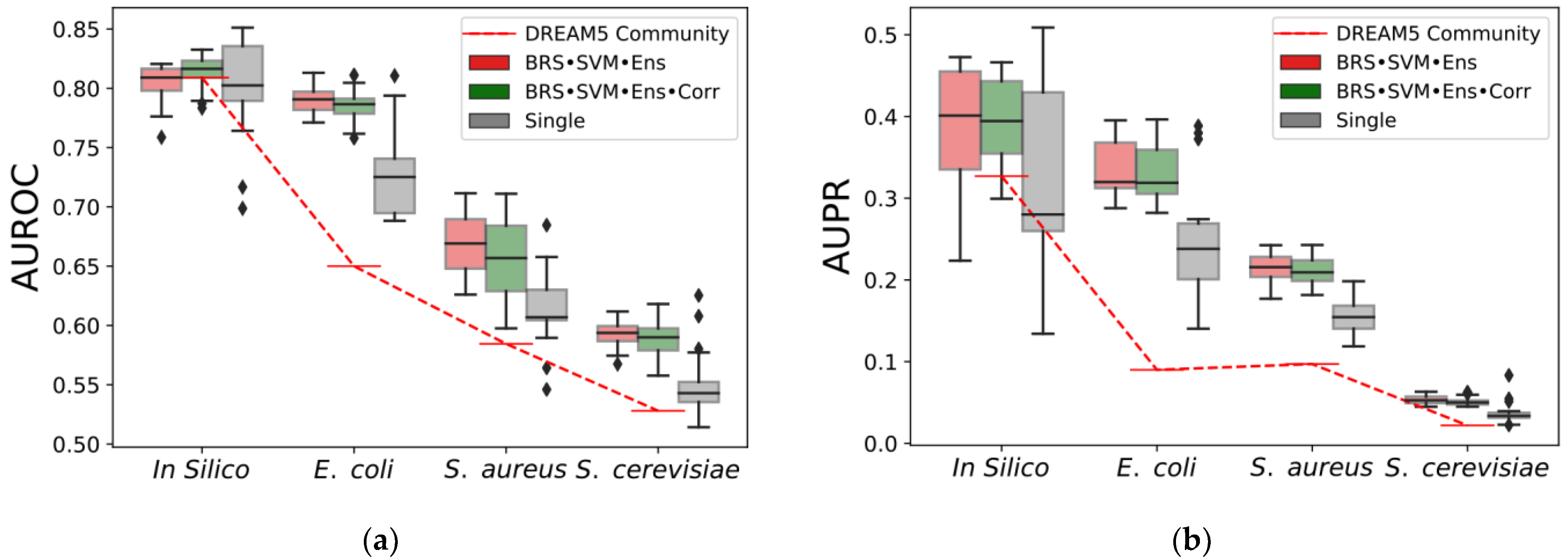
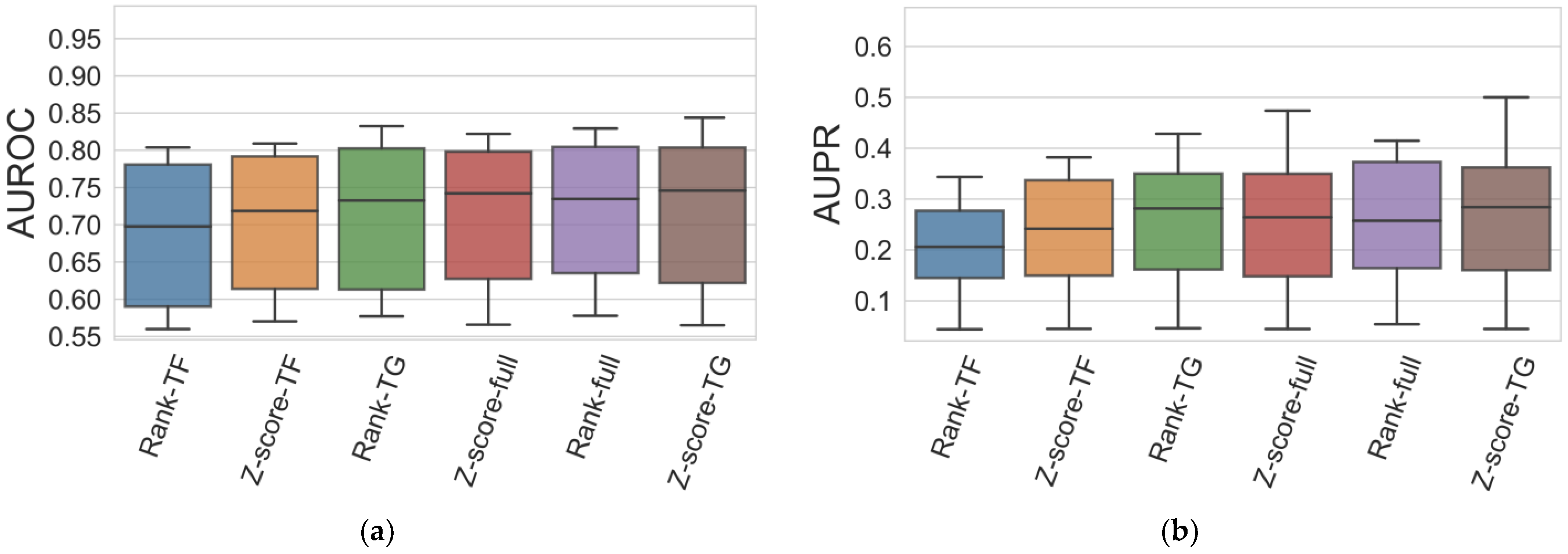
3.1. Evaluation Protocol
3.2. Results
3.3. Availability and Requirements
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| GRN | Gene Regulatory Network |
| TF | Transcription Factor |
| TG | Target Gene |
| RPM | Reads per Million |
| RPK | Reads per Kilobase |
| RPKM | Reads per Kilobase Million |
| TPM | Transcripts Per Kilobase |
| EFD | Equal Frequency Discretization |
| EWD | Equal Width Discretization |
| KMr | K-Means Row Discretization |
| KMc | K-Means Column Discretization |
| BKM | Bidirectional K-Means |
| Zc | Column-Wise Z-Score Standardization |
| Zr | Row-Wise Z-Score Standardization |
| P | Z-Score Polishing Standardization |
| I | Identity |
| MI | Mutual Information |
| SVMs | Support Vector Machines |
| AB | AdaBoost |
| GB | Gradient Boosting |
| RF | Random Forest |
| XRT | eXtreme Randomized Trees |
| c | Classifier |
| r | Regressor |
| BRSr | Bayesian Ridge Regressor |
| CNBc | Complement Naive Bayes Classifier |
| PR | Pearson Correlation Coefficient |
| SR | Spearman Correlation Coefficient |
| KT | Kendall Tau Statistic |
| CLR | Context Likelihood of Relatedness |
| TIGRESS | Trustful Inference of Gene Regulation using Stability Selection |
| SRL | Stability Randomized Lasso |
| AUROC | Area Under the Receiver Operating Characteristic Curve |
| AUPR | Area Under the Precision Recall Curve |
References
- Levine, M.; Davidson, E.H. Gene regulatory networks for development. Proc. Natl. Acad. Sci. USA 2005, 102, 4936–4949. [Google Scholar] [CrossRef] [PubMed]
- Shis, D.L.; Bennett, M.R.; Igoshin, O.A. Dynamics of bacterial gene regulatory networks. Ann. Rev. Biophys. 2018, 47, 447–467. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.-C.; Desplan, C. Gene regulatory networks during the development of the Drosophila visual system. Curr. Top. Dev. Biol. 2020, 139, 89–125. [Google Scholar] [PubMed]
- Shahbazi, M.N. Mechanisms of human embryo development: From cell fate to tissue shape and back. Development 2020, 147. [Google Scholar] [CrossRef] [PubMed]
- Aibar, S.; González-Blas, C.B.; Moerman, T.; Huynh-Thu, V.A.; Imrichova, H.; Hulselmans, G.; Rambow, F.; Marine, J.C.; Geurts, P.; Aerts, J.; et al. Scenic: Single-cell regulatory network inference and clustering. Nat. Methods 2017, 14, 1083. [Google Scholar] [CrossRef] [PubMed]
- Glass, K.; Quackenbush, J.; Kepner, J. High performance computing of gene regulatory networks using a message-passing model. In Proceedings of the 2015 IEEE High Performance Extreme Computing Conference (HPEC), Waltham, MA, USA, 15–17 September 2015; pp. 1–6. [Google Scholar]
- Sanguinetti, G.; Huynh-Thu, V.A. Gene regulatory network inference: An introductory survey. In Gene Regulatory Networks; Springer: Berlin/Heidelberg, Germany, 2019; pp. 1–23. [Google Scholar]
- Zhang, B.; Horvath, S. A general framework for weighted gene co-expression network analysis. Stat. Appl. Genet. Mol. Biol. 2005, 4. [Google Scholar] [CrossRef] [PubMed]
- Faith, J.J.; Hayete, B.; Thaden, J.T.; Mogno, I.; Wierzbowski, J.; Cottarel, G.; Kasif, S.; Collins, J.J.; Gardner, T.S. Large-scale mapping and validation of Escherichia coli transcriptional regulation from a compendium of expression profiles. PLoS Biol. 2007, 5, e8. [Google Scholar] [CrossRef] [PubMed]
- De la Fuente, A. What are gene regulatory networks? In Handbook of Research on Computational Methodologies in Gene Regulatory Networks; IGI Global: Hershey, PA, USA, 2010; pp. 1–27. [Google Scholar]
- Huynh-Thu, V.A.; Irrthum, A.; Wehenkel, L.; Geurts, P. Inferring Regulatory Networks from expression data Using tree-based methods. PLoS ONE 2010, 5, e12776. [Google Scholar] [CrossRef] [PubMed]
- Haury, A.-C.; Mordelet, F.; Vera-Licona, P.; Vert, J.P. TIGRESS: Trustful inference of gene regulation using stability selection. BMC Syst. Biol. 2012, 6, 145. [Google Scholar] [CrossRef] [PubMed]
- Peignier, S.; Schmitt, P.; Calevro, F. Data-driven gene regulatory network inference based on classification algorithms. In Proceedings of the 31st International Conference on Tools with Artificial Intelligence, Portland, OR, USA, 4-6 November 2019; pp. 1065–1072. [Google Scholar]
- Hastie, T.; Tibshirani, R.; Friedman, J.H.; Friedman, J.H. The Elements of Statistical Learning: Data Mining, Inference, and Prediction; Springer: New York, NY, USA, 2009; Volume 2, pp. 1–758. [Google Scholar]
- Wang, Y.; Wang, R.S.; Joshi, T.; Xu, D.; Zhang, X.S.; Chen, L. A linear programming framework for inferring gene regulatory networks by integrating heterogeneous data. In Handbook of Research on Computational Methodologies in Gene Regulatory Networks; IGI Global: Hershey, PA, USA, 2010; pp. 450–475. [Google Scholar]
- Marbach, D.; Costello, J.C.; Küffner, R.; Vega, N.M.; Prill, R.J.; Camacho, D.M.; Allison, K.R.; Kellis, M.; Collins, J.J.; Stolovitzky, G. Wisdom of crowds for robust gene network inference. Nat. Methods 2012, 9, 796. [Google Scholar] [CrossRef] [PubMed]
- Pedregosa, F.; Varoquaux, G.; Gramfort, A.; Michel, V.; Thirion, B.; Grisel, O.; Blondel, M.; Prettenhofer, P.; Weiss, R.; Dubourg, V.; et al. Scikit-learn: Machine learning in python. JMLR 2011, 12, 2825–2830. [Google Scholar]
- Oliphant, T. A Guide to NumPy; Trelgol Publishing: Spanish Fork, UT, USA, 2006. [Google Scholar]
- McKinney, W. Data structures for statistical computing in Python. In Proceedings of the 9th Python in Science Conference, Austin, TX, USA, 28 June–3 July 2010; pp. 51–56. [Google Scholar]
- Virtanen, P.; Gommers, R.; Oliphant, T.E.; Haberland, M.; Reddy, T.; Cournapeau, D.; Burovski, E.; Peterson, P.; Weckesser, W.; Bright, J.; et al. SciPy 1.0: Fundamental algorithms for scientific computing in Python. Nat. Methods 2020, 17, 261–272. [Google Scholar] [CrossRef] [PubMed]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with deseq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef] [PubMed]
- Jung, S.; Bi, Y.; Davuluri, R.V. Evaluation of data discretization methods to derive platform independent isoform expression signatures for multi-class tumor subtyping. BMC Genom. 2015, 16, S3. [Google Scholar] [CrossRef] [PubMed]
- Olshen, R.A.; Rajaratnam, B. Successive normalization of rectangular arrays. Ann. Stat. 2010, 38, 1638. [Google Scholar] [CrossRef] [PubMed]
- Friedman, J.H. Stochastic gradient boosting. Comput. Stat. Data Anal. 2002, 38, 367–378. [Google Scholar] [CrossRef]
- Neal, R.M. Bayesian Learning for Neural Networks; Springer Science & Business Media: Berlin/Heidelberg, Germany, 2012; Volume 118. [Google Scholar]
- Rennie, J.D. Tackling the poor assumptions of naive bayes text classifiers. In Proceedings of the 20th international conference on machine learning, Washington, DC, USA, 21–24 August 2003; pp. 616–623. [Google Scholar]
- Peignier, S.; Sorin, B.; Calevro, F. Ensemble Learning Based Gene Regulatory Network Inference. In Proceedings of the 2021 IEEE 33rd International Conference on Tools with Artificial Intelligence, Washington, DC, USA, 1–3 November 2021; pp. 113–120. [Google Scholar]
- Davis, J.; Goadrich, M. The relationship between precision-recall and roc curves. In Proceedings of the 23rd international conference on Machine learning, Pittsburgh, PA, USA, 25–29 June 2006; ACM: New York, NY, USA, 2006; pp. 233–240. [Google Scholar]
- Fawcett, T. An introduction to roc analysis. Pattern Recognit. Lett. 2006, 27, 861–874. [Google Scholar] [CrossRef]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Schmitt, P.; Sorin, B.; Frouté, T.; Parisot, N.; Calevro, F.; Peignier, S. GReNaDIne: A Data-Driven Python Library to Infer Gene Regulatory Networks from Gene Expression Data. Genes 2023, 14, 269. https://doi.org/10.3390/genes14020269
Schmitt P, Sorin B, Frouté T, Parisot N, Calevro F, Peignier S. GReNaDIne: A Data-Driven Python Library to Infer Gene Regulatory Networks from Gene Expression Data. Genes. 2023; 14(2):269. https://doi.org/10.3390/genes14020269
Chicago/Turabian StyleSchmitt, Pauline, Baptiste Sorin, Timothée Frouté, Nicolas Parisot, Federica Calevro, and Sergio Peignier. 2023. "GReNaDIne: A Data-Driven Python Library to Infer Gene Regulatory Networks from Gene Expression Data" Genes 14, no. 2: 269. https://doi.org/10.3390/genes14020269
APA StyleSchmitt, P., Sorin, B., Frouté, T., Parisot, N., Calevro, F., & Peignier, S. (2023). GReNaDIne: A Data-Driven Python Library to Infer Gene Regulatory Networks from Gene Expression Data. Genes, 14(2), 269. https://doi.org/10.3390/genes14020269




