Chitinase Chi 2 Positively Regulates Cucumber Resistance against Fusarium oxysporum f. sp. cucumerinum
Abstract
:1. Introduction
2. Materials and Methods
2.1. Plant Materials and Treatments
2.2. Identification of the Chitinase Genes in Sequenced Cucumber
2.3. Transcriptome Analysis and Candidate Gene Identification
2.4. Quantitative Reverse-Transcription PCR (qRT-PCR) Analysis
2.5. Chi2 and Chi14 Protein Purification
2.6. Functional Characterization of Chi2 via Virus-Induced Gene Silencing (VIGS)
2.7. Histochemical Staining and Microscopic Observations
2.8. Statistical Analysis
3. Results
3.1. Expression Profiles of Chitinase Genes in Different Cucumber Tissues
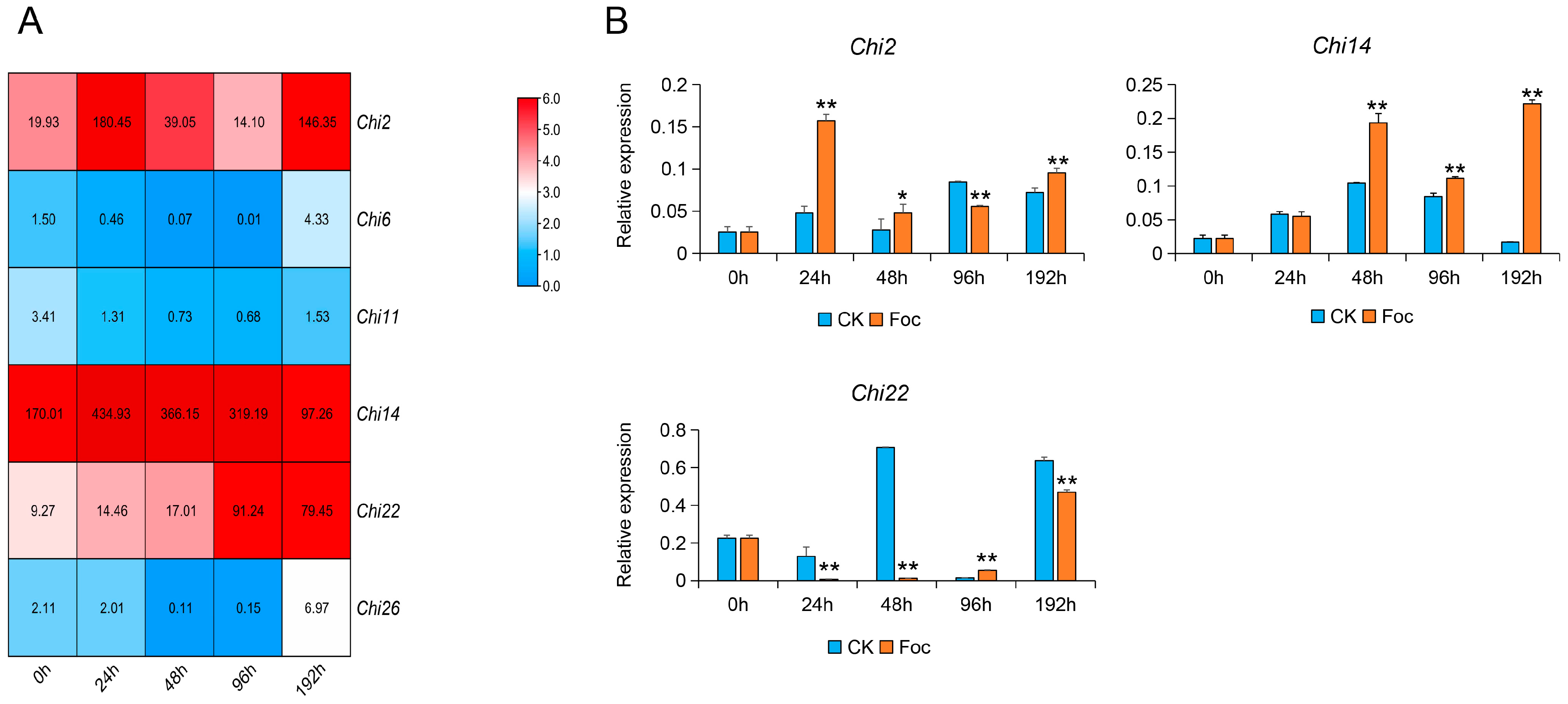
3.2. Transcriptome Analysis and Expression Verification of Chitinase Genes in Response to Foc
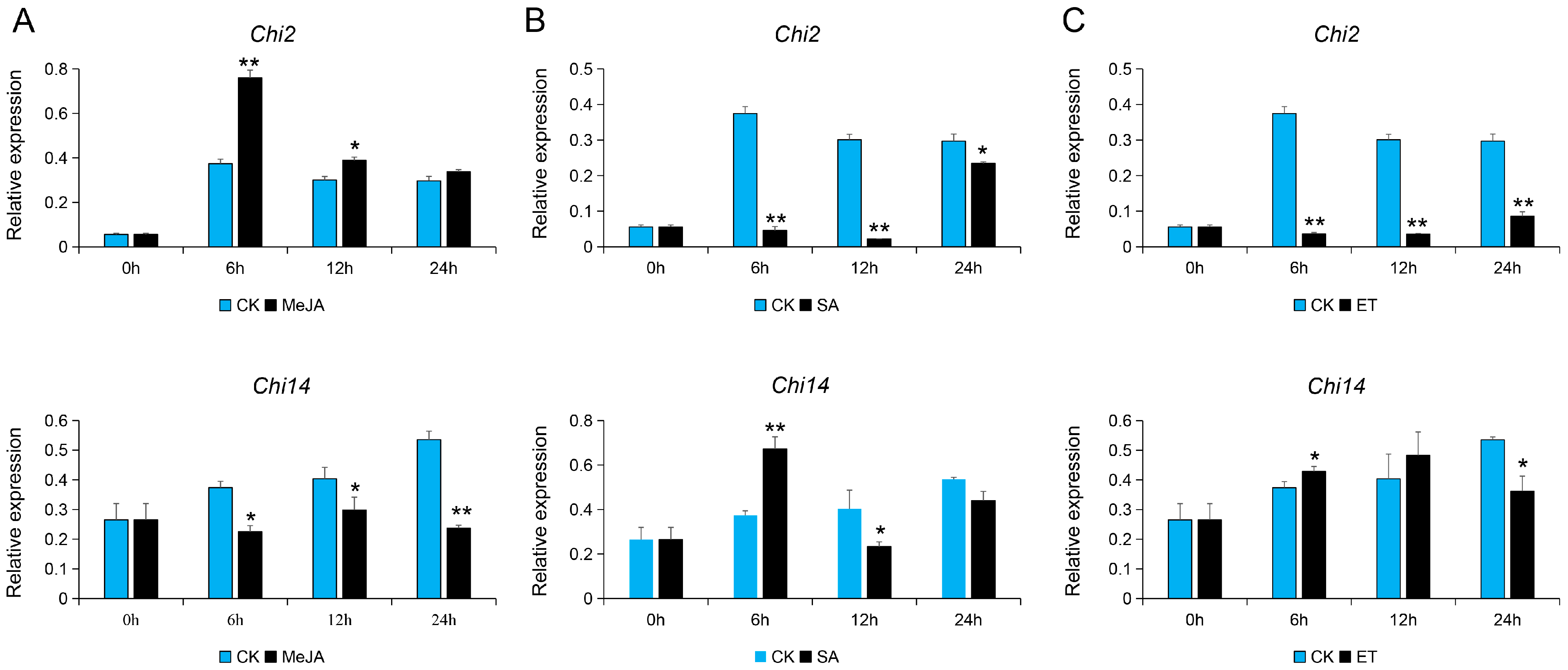
3.3. Expression Analysis of Chi2 and Chi14 in Response to Hormone Signals
3.4. Chi2 and Chi14 Suppresses Growth of Foc In Vitro
3.5. Silencing Chi2 Impairs Cucumber Tolerance to Foc
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Huang, S.W.; Li, R.Q.; Zhang, Z.H.; Li, L.; Gu, X.F.; Fan, W.; Lucas, W.J.; Wang, X.W.; Xie, B.Y.; Ni, P.X.; et al. The genome of the cucumber, Cucumis sativus L. Nat. Genet. 2009, 41, 1275–1281. [Google Scholar] [CrossRef] [Green Version]
- Hu, J.L.; Lin, X.G.; Wang, J.H.; Shen, W.S.; Wu, S.; Peng, S.P.; Mao, T.T. Arbuscular mycorrhizal fungal inoculation enhances suppression of cucumber Fusarium wilt in greenhouse soils. Pedosphere 2010, 20, 586–593. [Google Scholar] [CrossRef]
- Ahn, I.P.; Chung, H.S.; Lee, Y.H. Vegetative compatibility groups and pathogenicity among isolates of Fusarium oxysporum f. sp. cucumerinum. Plant Dis. 1998, 82, 244–246. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhou, X.G.; Wu, F.Z. Dynamics of the diversity of fungal and Fusarium communities during continuous cropping of cucumber in the greenhouse. FEMS Microbiol. Ecol. 2012, 80, 469–478. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sun, Y.M.; Li, Y.R.; Li, Y.; Wang, M.; Mur, L.A.J.; Shen, Q.R.; Guo, S.W. Nitrate mediated resistance against Fusarium infection in cucumber plants acts via photorespiration. Plant Cell Environ. 2021, 44, 3412–3431. [Google Scholar] [CrossRef]
- Dong, J.P.; Wang, Y.A.; Xian, Q.Q.; Chen, X.H. Transcriptome analysis reveals ethylene-mediated defense responses to Fusarium oxysporum f. sp. cucumerinum infection in Cucumis sativus L. BMC Plant Biol. 2020, 20, 334. [Google Scholar] [CrossRef]
- Du, N.S.; Shi, L.; Du, L.T.; Yuan, Y.H.; Li, B.; Sang, T.; Sun, J.; Shu, S.; Guo, S.R. Effect of vinegar residue compost amendments on cucumber growth and Fusarium wilt. Environ. Sci. Pollut. Res. Int. 2015, 22, 19133–19141. [Google Scholar] [CrossRef]
- Moreno, A.; Alférez, A.; Avilés, M.; Diánez, F.; Blanco, R.; Santos, M.; Tello, J.C. First report of Fusarium wilt of cucumber caused by Fusarium oxysporum in Spain. Plant Pathol. 2003, 52, 410. [Google Scholar]
- Chen, L.H.; Huang, X.Q.; Zhang, F.G.; Zhao, D.K.; Yang, X.M.; Shen, Q.R. Application of Trichoderma harzianum SQR-T037 bioorganic fertiliser significantly controls Fusarium wilt and affects the microbial communities of continuously cropped soil of cucumber. J. Sci. Food Agric. 2012, 92, 2465–2470. [Google Scholar] [CrossRef]
- Huang, X.; Shi, D.; Sun, F.; Lu, H.; Liu, J.; Wu, W. Efficacy of sludge and manure compost amendments against Fusarium wilt of cucumber. Environ. Sci. Pollut. Res. Int. 2012, 19, 3895–3905. [Google Scholar] [CrossRef]
- Willie, L.; Anuratha, C.S.; Datta, K.; Potrykus, I.; Muthukrishnan, S. Genetic engineering of rice for resistance to sheath blight. Nat. Biotechnol. 1995, 13, 686–691. [Google Scholar]
- Xu, J.; Xu, X.; Tian, L.; Wang, G.; Zhang, X.; Wang, X.; Guo, W. Discovery and identification of candidate genes from the chitinase gene family for Verticillium dahliae resistance in cotton. Sci. Rep. 2016, 6, 29022. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Greene, L.H. Sequence and structural analysis of the chitinase insertion domain reveals two conserved motifs involved in chitin-binding. PLoS ONE 2010, 5, e8654. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Collinge, D.B.; Kragh, K.M.; Mikkelsen, J.D.; Nielsen, K.K.; Rasmussen, U.; Vad, K. Plant chitinases. Plant J. 1993, 3, 31–40. [Google Scholar] [CrossRef]
- Punja, Z.K.; Zhang, Y.Y. Plant chitinases and their roles in resistance to fungal diseases. J. Nematol. 1993, 25, 526–540. [Google Scholar]
- Kim, J.K.; Jang, I.C.; Wu, R.; Zuo, W.N.; Boston, R.S.; Lee, Y.H.; Ahn, I.P.; Nahm, B.H. Co-expression of a modified maize ribosome-inactivating protein and a rice basic chitinase gene in transgenic rice plants confers enhanced resistance to sheath blight. Transgenic Res. 2003, 12, 475–484. [Google Scholar] [CrossRef]
- Nishizawa, Y.; Nishio, Z.; Nakazono, K.; Soma, M.; Nakajima, E.; Ugaki, M.; Hibi, T. Enhanced resistance to blast (Magnaporthe grisea) in transgenic Japonica rice by constitutive expression of rice chitinase. Theor. Appl. Genet. 1999, 99, 383–390. [Google Scholar] [CrossRef]
- Kovács, G.; Sági, L.; Jacon, G.; Arinaitwe, G.; Busogoro, J.P.; Thiry, E.; Strosse, H.; Swennen, R.; Remy, S. Expression of a rice chitinase gene in transgenic banana (‘Gros Michel’, AAA genome group) confers resistance to black leaf streak disease. Transgenic Res. 2013, 22, 117–130. [Google Scholar] [CrossRef]
- Shah, M.R.; Mukherjee, P.K.; Eapen, S. Expression of a fungal endochitinase gene in transgenic tomato and tobacco results in enhanced tolerance to fungal pathogens. Physiol. Mol. Biol. Plants 2010, 16, 39–51. [Google Scholar] [CrossRef] [Green Version]
- Pétriacq, P.; López, A.; Luna, E. Fruit Decay to Diseases: Can Induced Resistance and Priming Help? Plants 2018, 7, 77. [Google Scholar] [CrossRef] [Green Version]
- Qi, J.J.; Liu, X.; Shen, D.; Miao, H.; Xie, B.; Li, X.; Zeng, P.; Wang, S.; Shang, Y.; Gu, X.F.; et al. A genomic variation map provides insights into the genetic basis of cucumber domestication and diversity. Nat. Genet. 2013, 45, 1510–1515. [Google Scholar] [CrossRef] [PubMed]
- Li, Q.; Li, H.; Huang, W.; Xu, Y.; Zhou, Q.; Wang, S.; Jue, R.; Huang, S.; Zhang, Z. A chromosome-scale genome assembly of cucumber (Cucumis sativus L.). Gigascience 2019, 8, giz072. [Google Scholar] [CrossRef] [Green Version]
- Finn, R.D.; Bateman, A.; Clements, J.; Coggill, P.; Eberhardt, R.Y.; Eddy, S.R.; Heger, A.; Hetherington, K.; Holm, L.; Mistry, J.; et al. Pfam: The protein families database. Nucleic Acids Res. 2014, 42, D222–D230. [Google Scholar] [CrossRef] [Green Version]
- Eddy, S.R. Accelerated profile HMM searches. PLoS Comput. Biol. 2011, 7, e1002195. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, L.K.; Feng, Z.X.; Wang, X.; Wang, X.W.; Zhang, X.G. DEGseq: An R package for identifying differentially expressed genes from RNA-seq data. Bioinformatics 2010, 26, 136–138. [Google Scholar] [CrossRef] [PubMed]
- Yakushiji, Y.; Yamanaka, K.; Ogura, T. Identification of a cysteine residue important for the ATPase activity of C. elegans fidgetin homologue. FEBS Lett. 2004, 578, 191–197. [Google Scholar] [CrossRef] [Green Version]
- Liu, M.; Liang, Z.; Aranda, M.A.; Hong, N.; Liu, L.; Kang, B.; Gu, Q. A cucumber green mottle mosaic virus vector for virus-induced gene silencing in cucurbit plants. Plant Methods 2020, 16, 9. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yoshioka, H.; Numata, N.; Nakajima, K.; Katou, S.; Kawakita, K.; Rowland, O.; Jones, J.D.; Doke, N. Nicotiana benthamiana gp91z homologs NbrbohA and NbrbohB participate in H2O2 accumulation and resistance to Phytophthora infestans. Plant Cell 2003, 15, 706–718. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, Y.Y.; Wu, F.Z.; Liu, S.W. Allelopathic effects of root exudates from wheat, oat and soybean on seed germination and growth of cucumber. Allelopath. J. 2009, 24, 103–112. [Google Scholar]
- Baayen, R.P.; O’Donnell, K.; Bonants, P.J.; Cigelnik, E.; Kroon, L.P.; Roebroeck, E.J.; Waalwijk, C. Gene Genealogies and AFLP Analyses in the Fusarium oxysporum Complex Identify Monophyletic and Nonmonophyletic Formae Speciales Causing Wilt and Rot Disease. Phytopathology 2000, 90, 891–900. [Google Scholar] [CrossRef] [Green Version]
- Bari, R.; Jones, J.D. Role of plant hormones in plant defence responses. Plant Mol. Biol. 2009, 69, 473–488. [Google Scholar] [CrossRef]
- Kasprzewska, A. Plant chitinases-regulation and function. Cell. Mol. Biol. Lett. 2003, 8, 809–824. [Google Scholar]
- Xu, F.; Fan, C.; He, Y. Chitinases in Oryza sativas sp. Japonica and Arabidopsis thaliana. J. Genet. Genom. 2007, 34, 138–150. [Google Scholar] [CrossRef]
- Bartholomew, E.S.; Black, K.; Feng, Z.; Liu, W.; Shan, N.; Zhang, X.; Wu, L.; Bailey, L.; Zhu, N.; Qi, C.; et al. Comprehensive Analysis of the Chitinase Gene Family in Cucumber (Cucumis sativus L.): From Gene Identification and Evolution to Expression in Response to Fusarium oxysporum. Int. J. Mol. Sci. 2019, 20, 5309. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Passarinho, P.A.; Van Hengel, A.J.; Fransz, P.F.; de Vries, S.C. Expression pattern of the Arabidopsis thaliana AtEP3/AtchitIV endochitinase gene. Planta 2001, 212, 556–567. [Google Scholar] [CrossRef] [PubMed]
- Hamel, F.; Bellemare, G. Characterization of a class I chitinase gene and of wound-inducible, root and flower-specific chitinase expression in Brassica napus. Biochim. Biophys. Acta 1995, 1263, 212–220. [Google Scholar] [CrossRef]
- Cohen-Kupiec, R.; Chet, I. The molecular biology of chitin digestion. Curr. Opin. Biotechnol. 1998, 9, 270–277. [Google Scholar] [CrossRef]
- Hanfrey, C.; Fife, M.; Buchanan-Wollaston, V. Leaf senescence in Brassica naupus: Expression of genes encoding pathogenesis related proteins. Plant Mol. Biol. 1996, 30, 595–609. [Google Scholar] [CrossRef] [PubMed]
- Bishop, J.G.; Dean, A.M.; Mitchell-Olds, T. Rapid evolution in plant chitinases: Molecular targets of selection in plant-pathogen coevolution. Proc. Natl. Acad. Sci. USA 2000, 97, 5322–5327. [Google Scholar] [CrossRef] [Green Version]
- Qi, X.H.; Zhu, Y.M.; Li, S.F.; Zhou, H.X.; Xu, X.W.; Xu, Q.; Chen, X.H. Identification of genes related to mesocarp development in cucumber. Hortic. Plant J. 2020, 6, 293–300. [Google Scholar] [CrossRef]
- Xu, J.; Xian, Q.; Zhang, N.; Wang, K.; Zhou, X.; Li, Y.; Dong, J.; Chen, X. Identification of miRNA-Target Gene Pairs Responsive to Fusarium Wilt of Cucumber via an Integrated Analysis of miRNA and Transcriptome Profiles. Biomolecules 2021, 11, 1620. [Google Scholar] [CrossRef] [PubMed]
- Kim, D.S.; Kim, N.H.; Hwang, B.K. The Capsicum annuum class IV chitinase ChitIV interacts with receptor-like cytoplasmic protein kinase PIK1 to accelerate PIK1-triggered cell death and defence responses. J. Exp. Bot. 2015, 66, 1987–1999. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pieterse, C.M.; Leon-Reyes, A.; Van der Ent, S.; Van Wees, S.C. Networking by small-molecule hormones in plant immunity. Nat. Chem. Biol. 2009, 5, 308–316. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dempsey, D.A.; Klessig, D.F. SOS-toomany signals for systemic acquired resistance? Trends Plant Sci. 2012, 17, 538–545. [Google Scholar] [CrossRef] [PubMed]
- Gao, W.; Long, L.; Zhu, L.F.; Xu, L.; Gao, W.H.; Sun, L.Q.; Liu, L.L.; Zhang, X.L. Proteomic and virus-induced gene silencing (VIGS) Analyses reveal that gossypol, brassinosteroids, and jasmonic acid contribute to the resistance of cotton to Verticillium dahliae. Mol. Cell. Proteom. 2013, 12, 3690–3703. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sun, L.; Zhu, L.; Xu, L.; Yuan, D.; Min, L.; Zhang, X. Cotton cytochrome P450 CYP82D regulates systemic cell death by modulating the octadecanoid pathway. Nat. Commun. 2014, 5, 5372. [Google Scholar] [CrossRef]
- Xu, Y.H.; Wang, J.W.; Wang, S.; Wang, J.Y.; Chen, X.Y. Characterization of GaWRKY1, a cotton transcription factor that regulates these squiterpene synthasegene(+)-delta-cadinenesynthase-A. Plant Physiol. 2004, 135, 507–515. [Google Scholar] [CrossRef] [Green Version]
- He, X.; Zhu, L.; Wassan, G.M.; Wang, Y.; Miao, Y.; Shaban, M.; Hu, H.; Sun, H.; Zhang, X. GhJAZ2 attenuates cotton resistance to biotic stresses via the inhibition of the transcriptional activity of GhbHLH171. Mol. Plant Pathol. 2018, 19, 896–908. [Google Scholar] [CrossRef]



Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Xu, J.; Zhang, N.; Wang, K.; Xian, Q.; Dong, J.; Qi, X.; Chen, X. Chitinase Chi 2 Positively Regulates Cucumber Resistance against Fusarium oxysporum f. sp. cucumerinum. Genes 2022, 13, 62. https://doi.org/10.3390/genes13010062
Xu J, Zhang N, Wang K, Xian Q, Dong J, Qi X, Chen X. Chitinase Chi 2 Positively Regulates Cucumber Resistance against Fusarium oxysporum f. sp. cucumerinum. Genes. 2022; 13(1):62. https://doi.org/10.3390/genes13010062
Chicago/Turabian StyleXu, Jun, Ningyuan Zhang, Ke Wang, Qianqian Xian, Jingping Dong, Xiaohua Qi, and Xuehao Chen. 2022. "Chitinase Chi 2 Positively Regulates Cucumber Resistance against Fusarium oxysporum f. sp. cucumerinum" Genes 13, no. 1: 62. https://doi.org/10.3390/genes13010062
APA StyleXu, J., Zhang, N., Wang, K., Xian, Q., Dong, J., Qi, X., & Chen, X. (2022). Chitinase Chi 2 Positively Regulates Cucumber Resistance against Fusarium oxysporum f. sp. cucumerinum. Genes, 13(1), 62. https://doi.org/10.3390/genes13010062





