Association of TNF-α 308G/A and LEPR Gln223Arg Polymorphisms with the Risk of Type 2 Diabetes Mellitus
Abstract
:1. Introduction
2. Materials and Methods
2.1. Collection of Clinical Samples
2.2. Questionnaire
2.3. Genomic DNA Extraction
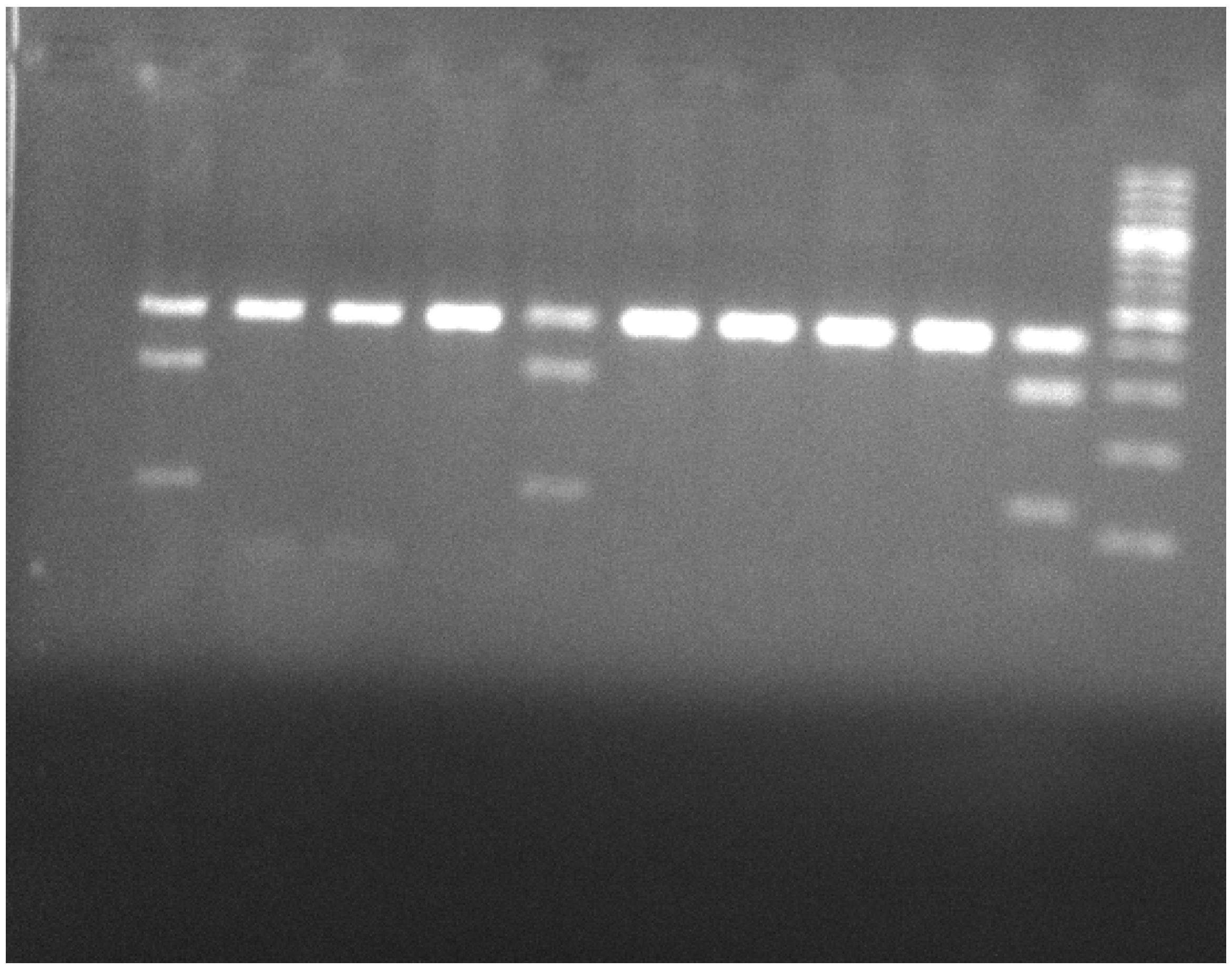
2.4. Polymerase Chain Reaction–Restricted Fragment Length Polymorphism (PCR-RFLP) for the Genomic Region of TNF-α -308G/A
2.5. Polymerase Chain Reaction–Restricted Fragment Length Polymorphism (PCR-RFLP) for the Genomic Region of LEPR Gln223Arg
2.6. Biomedical Ethics Issues
2.7. Statistical Analysis
3. Results
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Conflicts of Interest
References
- Sherwani, S.I.; Khan, H.A.; Ekhzaimy, A.; Masood, A.; Sakharkar, M.K. Significance of HbA1c Test in Diagnosis and Prognosis of Diabetic Patients. Biomark. Insights 2016, 11, S38440. [Google Scholar] [CrossRef]
- Burant, C.F. Medical Management of Type 2 Diabetes. Clin. Diabetes 2008, 26, S3. [Google Scholar]
- Coelho, M.; Oliveira, T.; Fernandes, R. State of the Art Paper Biochemistry of Adipose Tissue: An Endocrine Organ. Arch. Med. Sci. 2013, 2, 191–200. [Google Scholar] [CrossRef] [Green Version]
- Jaganathan, R.; Ravindran, R.; Dhanasekaran, S. Emerging Role of Adipocytokines in Type 2 Diabetes as Mediators of Insulin Resistance and Cardiovascular Disease. Can. J. Diabetes 2018, 42, 446–456.e1. [Google Scholar] [CrossRef]
- Feng, R.; Li, Y.; Zhao, D.; Wang, C.; Niu, Y.; Sun, C. Lack of Association between TNF 238 G/A Polymorphism and Type 2 Diabetes: A Meta-Analysis. Acta Diabetol. 2009, 46, 339–343. [Google Scholar] [CrossRef]
- Luna, G.I.; da Silva, I.C.R.; Sanchez, M.N. Association between -308G/A TNFA Polymorphism and Susceptibility to Type 2 Diabetes Mellitus: A Systematic Review. J. Diabetes Res. 2016, 2016, 6309484. [Google Scholar] [CrossRef] [Green Version]
- Thiem, K.; Stienstra, R.; Riksen, N.P.; Keating, S.T. Trained Immunity and Diabetic Vascular Disease. Clin. Sci. 2019, 133, 195–203. [Google Scholar] [CrossRef]
- Jamil, K.; Jayaraman, A.; Ahmad, J.; Joshi, S.; Yerra, S.K. TNF-α- 308G/A and −238G/A Polymorphisms and Its Protein Network Associated with Type 2 Diabetes Mellitus. Saudi J. Biol. Sci. 2017, 24, 1195–1203. [Google Scholar] [CrossRef] [Green Version]
- Obradovic, M.; Sudar-Milovanovic, E.; Soskic, S.; Essack, M.; Arya, S.; Stewart, A.J.; Gojobori, T.; Isenovic, E.R. Leptin and Obesity: Role and Clinical Implication. Front. Endocrinol. 2021, 12, 585887. [Google Scholar] [CrossRef]
- Poeggeler, B.; Schulz, C.; Pappolla, M.A.; Bodó, E.; Tiede, S.; Lehnert, H.; Paus, R. Leptin and the Skin: A New Frontier: Leptin and the Skin: A New Frontier. Exp. Dermatol. 2010, 19, 12–18. [Google Scholar] [CrossRef]
- Do Carmo Martins, M.; Faleiro, L.L.; Fonseca, A. Relação entre a leptina, a massa corporal e a síndrome metabólica numa amostra da população adulta. Port. J. Cardiol. 2012, 31, 711–719. [Google Scholar] [CrossRef]
- Duarte, S.F.P.; Francischetti, E.A.; Genelhu-Abreu, V.; Barroso, S.G.; Braga, J.U.; Cabello, P.H.; Pimentel, M.M.G. p. Q223R Leptin Receptor Polymorphism Associated with Obesity in Brazilian Multiethnic Subjects. Am. J. Hum. Biol. 2006, 18, 448–453. [Google Scholar] [CrossRef]
- Chiu, K.; Chu, A.; Chuang, L.; Saad, M. Association of Leptin Receptor Polymorphism with Insulin Resistance. Eur. J. Endocrinol. 2004, 725–729. [Google Scholar] [CrossRef] [Green Version]
- Han, H.; Ryu, H.-J.; Cha, H.; Go, M.; Ahn, Y.; Koo, B.; Cho, Y.; Lee, H.; Cho, N.; Shin, C.; et al. Genetic Variations in the Leptin and Leptin Receptor Genes Are Associated with Type 2 Diabetes Mellitus and Metabolic Traits in the Korean Female Population. Clin. Genet. 2008, 74, 105–115. [Google Scholar] [CrossRef]
- Suriyaprom, K.; Tungtrongchitr, R.; Thawnasom, K. Measurement of the Levels of Leptin, BDNF Associated with Polymorphisms LEP G2548A, LEPR Gln223Arg and BDNF Val66Met in Thai with Metabolic Syndrome. Diabetol. Metab. Syndr. 2014, 6, 6. [Google Scholar] [CrossRef] [Green Version]
- Liu, Y.; Chen, S.-Q.; Jing, Z.-H.; Hou, X.; Chen, Y.; Song, X.-J.; Lv, W.-S.; Wang, R.; Wang, Y.-G. Association of LEPR Gln223Arg Polymorphism with T2DM: A Meta-Analysis. Diabetes Res. Clin. Pract. 2015, 109, e21–e26. [Google Scholar] [CrossRef]
- Su, S.; Zhang, C.; Zhang, F.; Li, H.; Yang, X.; Tang, X. The Association between Leptin Receptor Gene Polymorphisms and Type 2 Diabetes Mellitus: A Systematic Review and Meta-Analysis. Diabetes Res. Clin. Pract. 2016, 121, 49–58. [Google Scholar] [CrossRef]
- Yang, M.M.; Wang, J.; Fan, J.J.; Ng, T.K.; Sun, D.J.; Guo, X.; Teng, Y.; Li, Y.-B. Variations in the Obesity Gene “LEPR” Contribute to Risk of Type 2 Diabetes Mellitus: Evidence from a Meta-Analysis. J. Diabetes Res. 2016, 2016, 5412084. [Google Scholar] [CrossRef] [Green Version]
- Boraska, V.; Rayner, N.W.; Groves, C.J.; Frayling, T.M.; Diakite, M.; Rockett, K.A.; Kwiatkowski, D.P.; Day-Williams, A.G.; McCarthy, M.I.; Zeggini, E. Large-Scale Association Analysis of TNF/LTA Gene Region Polymorphisms in Type 2 Diabetes. BMC Med. Genet. 2010, 11, 69. [Google Scholar] [CrossRef] [Green Version]
- Zeggini, E.; Groves, C.J.; Parkinson, J.R.C.; Halford, S.; Owen, K.R.; Frayling, T.M.; Walker, M.; Hitman, G.A.; Levy, J.C.; O’Rahilly, S.; et al. Large-Scale Studies of the Association between Variation at the TNF/LTA Locus and Susceptibility to Type 2 Diabetes. Diabetologia 2005, 48, 2013–2017. [Google Scholar] [CrossRef] [Green Version]
- Liu, H.-L.; Lin, Y.-G.; Wu, J.; Sun, H.; Gong, Z.-C.; Hu, P.-C.; Yin, J.-Y.; Zhang, W.; Wang, D.; Zhou, H.-H.; et al. Impact of Genetic Polymorphisms of Leptin and TNF-α on Rosiglitazone Response in Chinese Patients with Type 2 Diabetes. Eur. J. Clin. Pharmacol. 2008, 64, 663–671. [Google Scholar] [CrossRef]
- Shiau, M.-Y.; Wu, C.-Y.; Huang, C.-N.; Hu, S.-W.; Lin, S.-J.; Chang, Y.-H. TNF-α Polymorphisms and Type 2 Diabetes Mellitus in Taiwanese Patients. Tissue Antigens 2003, 61, 393–397. [Google Scholar] [CrossRef]
- Patel, R.; Palit, S.P.; Rathwa, N.; Ramachandran, A.V.; Begum, R. Genetic Variants of Tumor Necrosis Factor-α and Its Levels: A Correlation with Dyslipidemia and Type 2 Diabetes Susceptibility. Clin. Nutr. 2019, 38, 1414–1422. [Google Scholar] [CrossRef]
- Golshani, H.; Haghani, K.; Dousti, M.; Bakhtiyari, S. Association of TNF-α 308 G/A Polymorphism With Type 2 Diabetes: A Case–Control Study in the Iranian Kurdish Ethnic Group. Osong Public Health Res. Perspect. 2015, 6, 94–99. [Google Scholar] [CrossRef] [Green Version]
- Guo, X.; Li, C.; Wu, J.; Mei, Q.; Liu, C.; Sun, W.; Xu, L.; Fu, S. The Association of TNF-α −308G/A and −238G/A Polymorphisms with Type 2 Diabetes Mellitus: A Meta-Analysis. Biosci. Rep. 2019, 39, 20191301. [Google Scholar] [CrossRef]
- Zhao, Y.; Li, Z.; Zhang, L.; Zhang, Y.; Yang, Y.; Tang, Y.; Fu, P. The TNF-α- 308G/A Polymorphism Is Associated with Type 2 Diabetes Mellitus: An Updated Meta-Analysis. Mol. Biol. Rep. 2014, 41, 73–83. [Google Scholar] [CrossRef]
- Guo, Y.; Huang, Z.; Sang, D.; Gao, Q.; Li, Q. The Role of Nutrition in the Prevention and Intervention of Type 2 Diabetes. Front. Bioeng. Biotechnol. 2020, 8, 575442. [Google Scholar] [CrossRef]
- Shoily, S.S.; Ahsan, T.; Fatema, K.; Sajib, A.A. Common genetic variants and pathways in diabetes and associated complications and vulnerability of populations with different ethnic origins. Sci. Rep. 2021, 11, 7504. [Google Scholar] [CrossRef]
- Wang, D.D.; Hu, F.B. Precision Nutrition for Prevention and Management of Type 2 Diabetes. Lancet Diabetes Endocrinol. 2018, 6, 416–426. [Google Scholar] [CrossRef]
- Ballini, A.; Scacco, S.; Boccellino, M.; Santacroce, L.; Arrigoni, R. Microbiota and Obesity: Where Are We Now? Biology 2020, 9, 415. [Google Scholar] [CrossRef]
- Wu, H.; Tang, W.; Liu, C.; Xue, Y.; Hou, J. The Association of LEPR Rs1137101 G>A Polymorphism with the Risk of Type 2 Diabetes Mellitus: From a Case-Control Study to a Meta-Analysis. Int. J. Clin. Exp. Med. 2019, 12, 3798–3809. [Google Scholar]
- Cordero, P.; Campion, J.; Milagro, F.I.; Goyenechea, E.; Steemburgo, T.; Javierre, B.M.; Martinez, J.A. Leptin and TNF-α Promoter Methylation Levels Measured by MSP Could Predict the Response to a Low-Calorie Diet. J. Physiol. Biochem. 2011, 463–470. [Google Scholar] [CrossRef]
| LEPR Gln223Arg | T2DM | Control |
|---|---|---|
| Gln/Gln (AA) | 46 (42.6%) a | 34 (65.4%) b |
| Gln/Arg (AG) | 48 (44.4%) a | 14 (26.9%) c |
| Arg/Arg (GG) | 14 (13%) b | 4 (7.7%) a |
| LEPR Gln223Arg | T2DM | Control |
|---|---|---|
| G | 35.2% a | 21.2% b |
| A | 64.8% b | 78.8% a |
| TNF-α -308G/A | T2DM | Control |
|---|---|---|
| G/G | 74 (68.5%) a | 27 (51.9%) b |
| G/A | 34 (31.5%) c | 25 (48.1%) b |
| TNF-α -308G/A | T2DM | Control |
|---|---|---|
| G | 84.3% a | 76% b |
| A | 15.7% b | 24.0% a |
| Gender | TNF-α -308G/A (GA) | TNF-α -308G/A (GG) | LEPR Gln223Arg GG | LEPR Gln223Arg GA | LEPR Gln223Arg AA |
|---|---|---|---|---|---|
| Female (Control Group) | 17 (56.7%) a | 13 (43.3%) a | 3 (10%) a | 8 (26.7%) b | 19 (63.3%) c |
| Male (Control Group) | 8 (36.4%) b | 14(63.6%) c | 1 (4.5%) a | 6 (27.3%) b | 15 (68.2%) c |
| Female T2DM | 17(34%) b | 33 (66%) c | 4 (8%) a | 28 (56%) c | 18 (36%) b |
| Male T2DM | 17 (29.3%) b | 41(70.7%) c | 10 (17.2%) a | 20 (34.5%) b | 28 (48.3%) b |
| Model | Model Fitting Criteria | Likelihood Ratio Tests | ||
|---|---|---|---|---|
| −2 Log Likelihood | Chi-Square | df | Sig. | |
| Intercept Only | 85.541 | |||
| Final | 60.156 | 25.385 | 8 | 0.001 |
| Effect | Model Fitting Criteria | Likelihood Ratio Tests | ||
|---|---|---|---|---|
| −2 Log Likelihood of Reduced Model | Chi-Square | df | Sig. | |
| Intercept | 60.156 a | 0.000 | 0 | |
| gender | 64.007 | 3.851 | 2 | 0.146 |
| agegroup | 74.445 | 14.289 | 4 | 0.006 |
| T2DM | 76.766 | 16.610 | 2 | 0.000 |
| LEPRa | B | Std. Error | Wald | df | Sig. | Exp(B) | 95% Confidence Interval for Exp(B) | ||
|---|---|---|---|---|---|---|---|---|---|
| Lower Bound | Upper Bound | ||||||||
| AA | Intercept | 2.756 | 0.808 | 11.650 | 1 | 0.001 | |||
| [gender = FEMALE] | 0.263 | 0.575 | 0.209 | 1 | 0.648 | 1.300 | 0.421 | 4.013 | |
| [gender = MALE] | 0 b | . | . | 0 | . | . | . | . | |
| [agegroup = >65] | 1.618 | 0.692 | 5.465 | 1 | 0.019 | 5.043 | 1.299 | 19.580 | |
| [agegroup = 0–45] | −1.535 | 0.790 | 3.774 | 1 | 0.052 | 0.215 | 0.046 | 1.014 | |
| [agegroup = 45–65] | 0 b | . | . | 0 | . | . | . | . | |
| [T2DM = DIABETIC] | −2.343 | 0.787 | 8.853 | 1 | 0.003 | 0.096 | 0.021 | 0.449 | |
| [T2DM = NORMAL] | 0 b | . | . | 0 | . | . | . | . | |
| GA | Intercept | 1.382 | 0.827 | 2.791 | 1 | 0.095 | |||
| [gender = FEMALE] | 0.853 | 0.574 | 2.211 | 1 | 0.137 | 2.347 | 0.762 | 7.222 | |
| [gender = MALE] | 0 b | . | . | 0 | . | . | . | . | |
| [agegroup = >65] | 1.015 | 0.686 | 2.188 | 1 | 0.139 | 2.760 | 0.719 | 10.590 | |
| [agegroup = 0–45] | −1.072 | 0.767 | 1.955 | 1 | 0.162 | 0.342 | 0.076 | 1.538 | |
| [agegroup = 45–65] | 0 b | . | . | 0 | . | . | . | . | |
| [T2DM = DIABETIC] | −0.832 | 0.781 | 1.135 | 1 | 0.287 | 0.435 | 0.094 | 2.011 | |
| [T2DM = NORMAL] | 0 b | . | . | 0 | . | . | . | . | |
| Model | Model Fitting Criteria | Likelihood Ratio Tests | ||
|---|---|---|---|---|
| −2 Log Likelihood | Chi-Square | df | Sig. | |
| Intercept Only | 42.626 | |||
| Final | 36.887 | 5.739 | 4 | 0.220 |
| Effect | Model Fitting Criteria | Likelihood Ratio Tests | ||
|---|---|---|---|---|
| −2 Log Likelihood of Reduced Model | Chi-Square | df | Sig. | |
| Intercept | 36.887 a | 0.000 | 0 | . |
| gender | 38.162 | 1.275 | 1 | 0.259 |
| agegroup | 39.169 | 2.283 | 2 | 0.319 |
| T2DM | 39.962 | 3.075 | 1 | 0.080 |
| TNFa | B | Std. Error | Wald | df | Sig. | Exp(B) | 95% Confidence Interval for Exp(B) | ||
|---|---|---|---|---|---|---|---|---|---|
| Lower Bound | Upper Bound | ||||||||
| GA | Intercept | −0.124 | 0.403 | 0.094 | 1 | 0.759 | |||
| [gender = FEMALE] | 0.383 | 0.340 | 1.268 | 1 | 0.260 | 1.466 | 0.753 | 2.855 | |
| [gender = MALE] | 0 b | . | . | 0 | . | . | . | . | |
| [agegroup = >65] | −0.112 | 0.392 | 0.082 | 1 | 0.775 | 0.894 | 0.414 | 1.928 | |
| [agegroup = 0–45] | −0.764 | 0.519 | 2.173 | 1 | 0.140 | 0.466 | 0.168 | 1.286 | |
| [agegroup = 45–65] | 0 b | . | . | 0 | . | . | . | . | |
| [T2DM = DIABETIC] | −0.726 | 0.417 | 3.032 | 1 | 0.082 | 0.484 | 0.214 | 1.095 | |
| [T2DM = NORMAL] | 0 b | . | . | 0 | . | . | . | . | |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Trapali, M.; Houhoula, D.; Batrinou, A.; Kanellou, A.; Strati, I.F.; Siatelis, A.; Halvatsiotis, P. Association of TNF-α 308G/A and LEPR Gln223Arg Polymorphisms with the Risk of Type 2 Diabetes Mellitus. Genes 2022, 13, 59. https://doi.org/10.3390/genes13010059
Trapali M, Houhoula D, Batrinou A, Kanellou A, Strati IF, Siatelis A, Halvatsiotis P. Association of TNF-α 308G/A and LEPR Gln223Arg Polymorphisms with the Risk of Type 2 Diabetes Mellitus. Genes. 2022; 13(1):59. https://doi.org/10.3390/genes13010059
Chicago/Turabian StyleTrapali, Maria, Dimitra Houhoula, Anthimia Batrinou, Anastasia Kanellou, Irini F. Strati, Argyris Siatelis, and Panagiotis Halvatsiotis. 2022. "Association of TNF-α 308G/A and LEPR Gln223Arg Polymorphisms with the Risk of Type 2 Diabetes Mellitus" Genes 13, no. 1: 59. https://doi.org/10.3390/genes13010059
APA StyleTrapali, M., Houhoula, D., Batrinou, A., Kanellou, A., Strati, I. F., Siatelis, A., & Halvatsiotis, P. (2022). Association of TNF-α 308G/A and LEPR Gln223Arg Polymorphisms with the Risk of Type 2 Diabetes Mellitus. Genes, 13(1), 59. https://doi.org/10.3390/genes13010059






