Sorbitol Reduces Sensitivity to Alternaria by Promoting Ceramide Kinases (CERK) Expression through Transcription Factor Pswrky25 in Populus (Populus simonii Carr.)
Abstract
:1. Introduction
2. Materials and Methods
2.1. Plant Material
2.2. Feeding Leaves with Exogenous Sorbitol
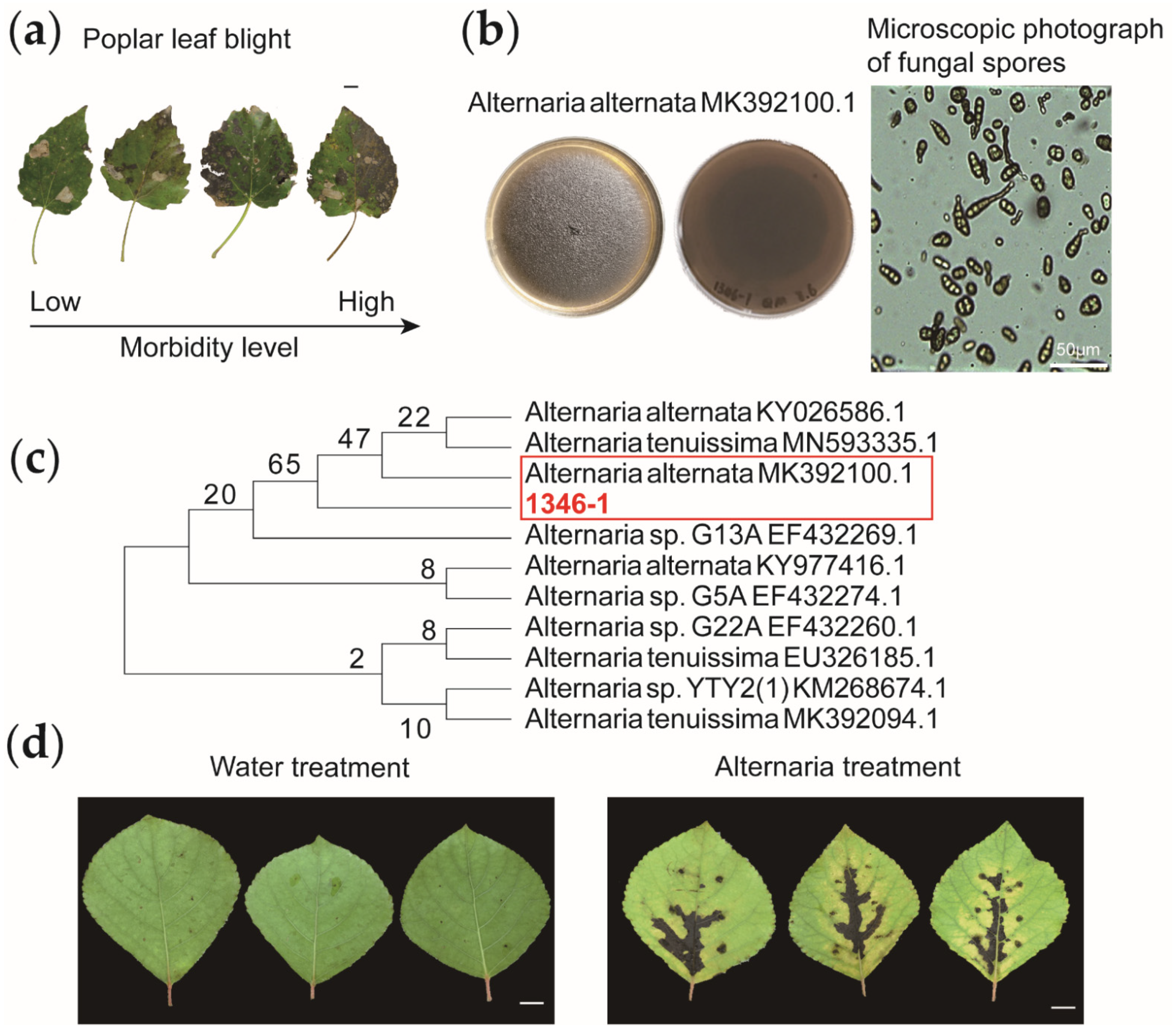
2.3. Isolation, Purification, Identification and Inoculation of Alternaria
2.4. RNA Extraction and Transcriptome Sequencing
2.5. Analysis of Differential Expression, Transcriptome Alignment and Functional Enrichment
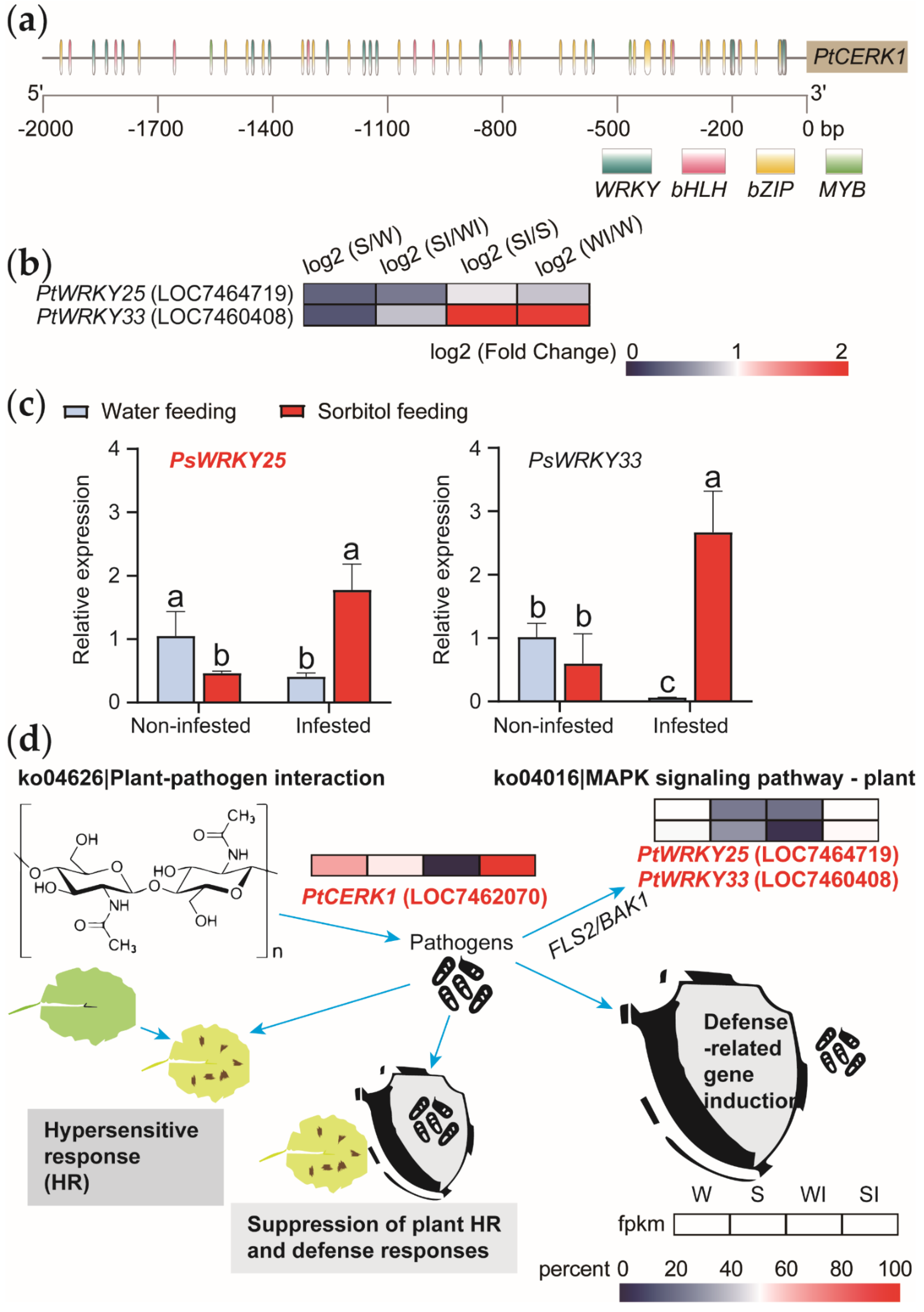
2.6. Analysis of Transcription Factor Binding Site in the Promoter Region
2.7. Construction of Transgenic Vector
2.8. Transient Transformation System of Populus simonii Carr
2.9. RNA Extraction and Real-Time Quantitative PCR (RT−qPCR) Validation
2.10. Statistical Analysis
3. Results
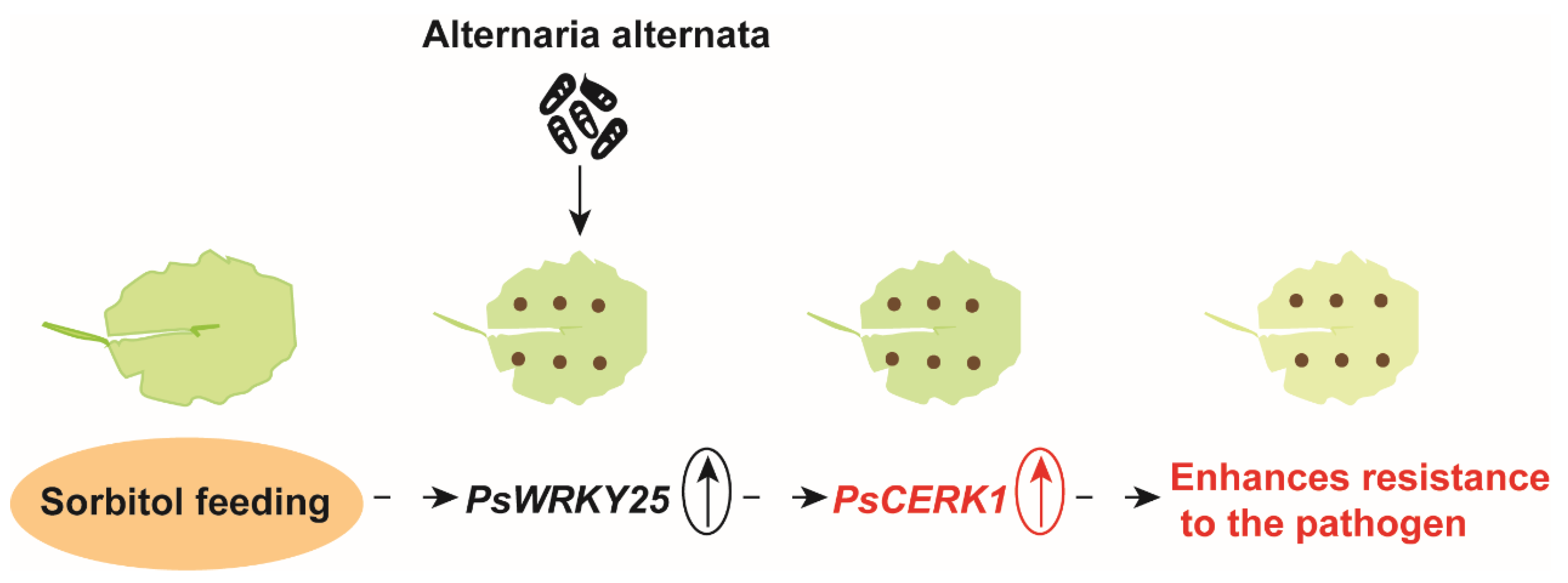
3.1. Sorbitol Feeding Improves Populus simonii Carr Resistance to Alternaria
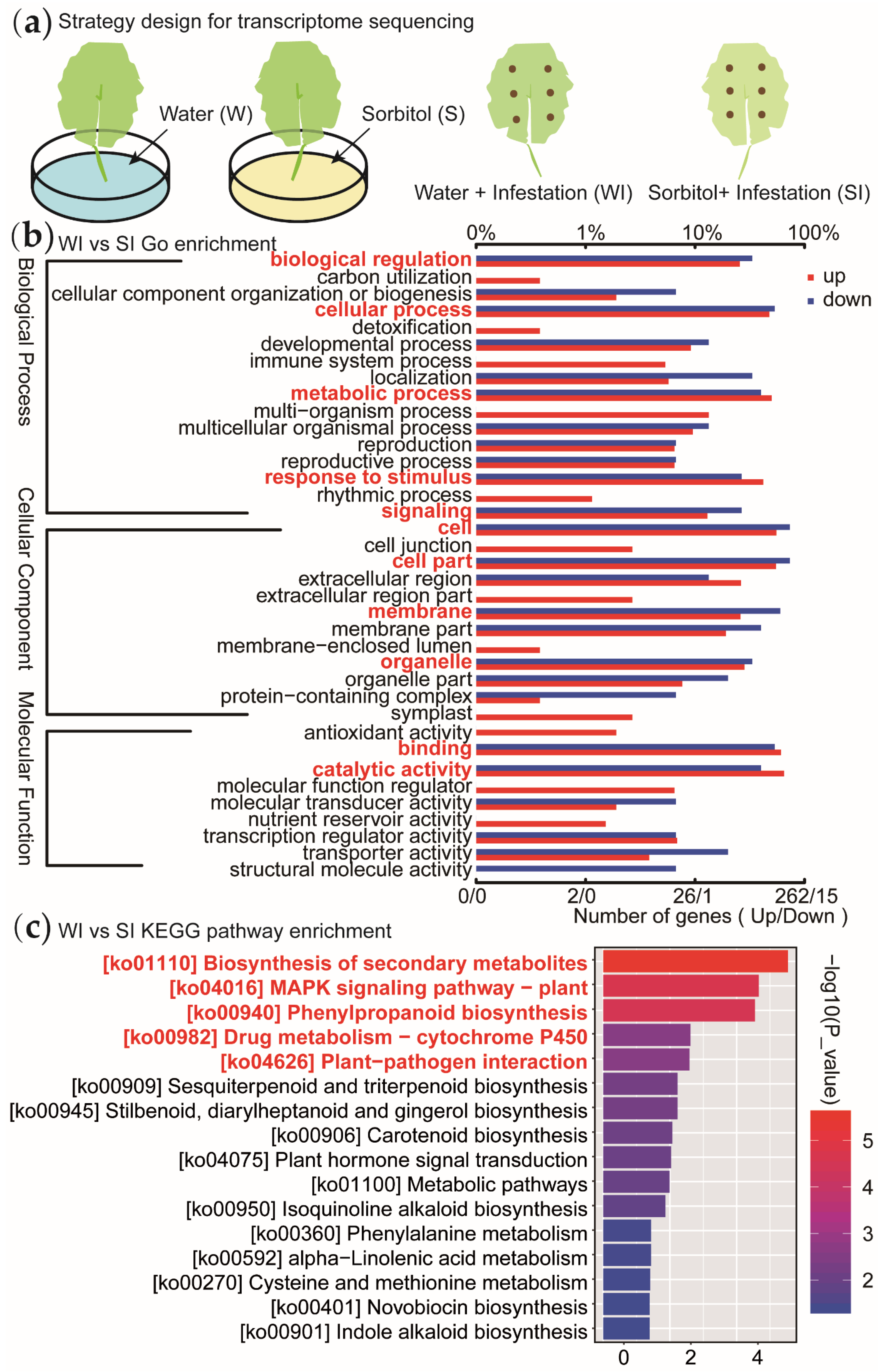
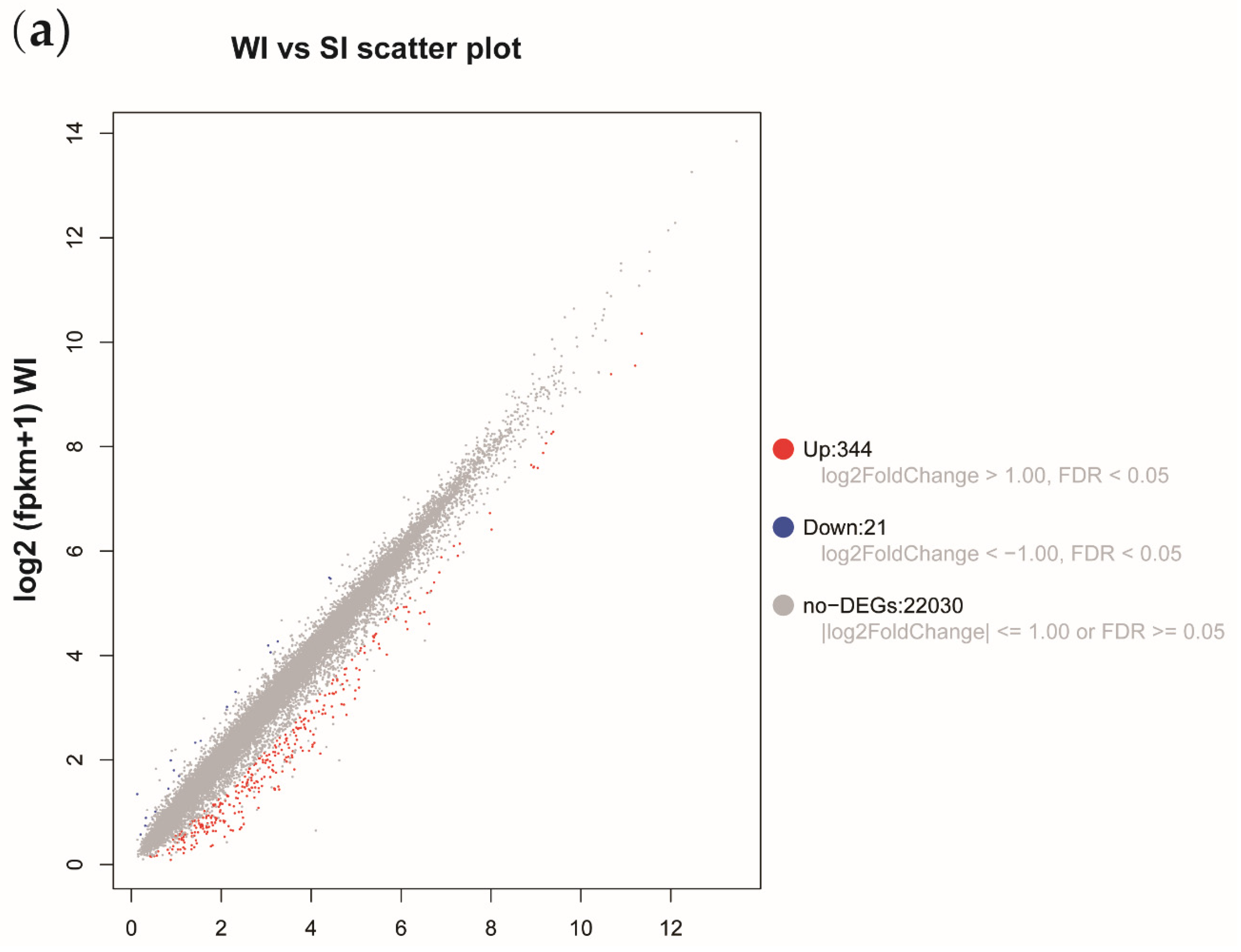
3.2. Transcriptome Sequencing Reveals Significant Differences between Sorbitol-Feeding and Water Feeding
3.3. Screening and Validation of Relevant Functional Genes in Disease-Resistance Related Pathways
3.4. Screening for Disease-Resistance-Associated Transcription Factors by Promoter Analysis
3.5. Overexpression of PsWRKY25 Increases the Expression of PsCERK1
3.6. Disease Resistance Mechanism of Poplar
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Stobrawa, K. Poplars (Populus spp.): Ecological role, applications and scientific perspectives in the 21st century. Balt. For. 2014, 20, 204–213. [Google Scholar]
- Wang, B.; Tian, C.; Liang, Y. Mixed effects of landscape structure, tree diversity and stand’s relative position on insect and pathogen damage in riparian poplar forests. For. Ecol. Manag. 2021, 479, 118555. [Google Scholar] [CrossRef]
- Ren, J.-H.; Li, H.; Wang, Y.-F.; Ye, J.-R.; Yan, A.-Q.; Wu, X.-Q. Biocontrol potential of an endophytic Bacillus pumilus JK-SX001 against poplar canker. Biol. Control. 2013, 67, 421–430. [Google Scholar] [CrossRef]
- Xhaard, C.; Barres, B.; Andrieux, A.; Bousset, L.; Halkett, F.; Frey, P. Disentangling the genetic origins of a plant pathogen during disease spread using an original molecular epidemiology approach. Mol. Ecol. 2012, 21, 2383–2398. [Google Scholar] [CrossRef] [PubMed]
- Osdaghi, E.; Kakavandi, N.R.; Nejati, M. First report of Alternaria alternata causing leaf spot on Populus euphratica in Iran. Iran. J. Plant Pathol. 2014, 50, 309–310. [Google Scholar]
- Guo, C.; Wang, C.; Zhou, T.; Jin, S.; Duan, C. First report of leaf blight caused by Alternaria brassicicola on Orychophragmus violaceus in China. Plant Dis. 2019, 103, 1031. [Google Scholar] [CrossRef]
- Hekimhan, H.; Eğerci, Y.; Uysal-Morca, A. First report of leaf blight caused by Alternaria longipes on oat in Turkey. New Dis. Rep. 2020, 42, 3. [Google Scholar] [CrossRef]
- Wang, T.; Zhao, J.; Ma, G.; Bao, S.; Wu, X. Leaf blight of sunflower caused by Alternaria tenuissima and A. alternata in Beijing, China. Can. J. Plant Pathol. 2019, 41, 372–378. [Google Scholar] [CrossRef]
- Pogány, M.; von Rad, U.; Grun, S.; Dongó, A.; Pintye, A.; Simoneau, P.; Bahnweg, G.; Kiss, L.; Barna, B.; Durner, J. Dual roles of reactive oxygen species and NADPH oxidase RBOHD in an Arabidopsis-Alternaria pathosystem. Plant Physiol. 2009, 151, 1459–1475. [Google Scholar] [CrossRef] [Green Version]
- Grodetskaia, T.; Fedorova, O.; Evlakov, P.; Baranov, O.Y.; Zakharova, O.; Gusev, A. Effect of Copper Oxide and Silver Nanoparticles on the Development of Tolerance to Alternaria alternata in Poplar in Vitro Clones. Nanobiotechnol. Rep. 2021, 16, 231–238. [Google Scholar] [CrossRef]
- Nešović, M.; Gašić, U.; Tosti, T.; Horvacki, N.; Nedić, N.; Sredojević, M.; Blagojević, S.; Ignjatović, L.; Tešić, Ž. Distribution of polyphenolic and sugar compounds in different buckwheat plant parts. RSC Adv. 2021, 11, 25816–25829. [Google Scholar] [CrossRef]
- Dominguez, P.G.; Niittylä, T. Mobile forms of carbon in trees: Metabolism and transport. Tree Physiol. 2021, 1–30. [Google Scholar] [CrossRef]
- Yang, X.; Chen, L.-S.; Cheng, L. Leaf Photosynthesis and Carbon Metabolism Adapt to Crop Load in ‘Gala’Apple Trees. Horticulturae 2021, 7, 47. [Google Scholar] [CrossRef]
- Kuo, T.M.; Doehlert, D.C.; Crawford, C.G. Sugar metabolism in germinating soybean seeds: Evidence for the sorbitol pathway in soybean axes. Plant Physiol. 1990, 93, 1514–1520. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, C.; Meng, D.; Piñeros, M.A.; Mao, Y.; Dandekar, A.M.; Cheng, L. A sugar transporter takes up both hexose and sucrose for sorbitol-modulated in vitro pollen tube growth in apple. Plant Cell 2020, 32, 449–469. [Google Scholar] [CrossRef] [Green Version]
- Meng, D.; Li, C.; Park, H.-J.; González, J.; Wang, J.; Dandekar, A.M.; Turgeon, B.G.; Cheng, L. Sorbitol modulates resistance to Alternaria alternata by regulating the expression of an NLR resistance gene in apple. Plant Cell 2018, 30, 1562–1581. [Google Scholar] [CrossRef] [Green Version]
- Wang, L.; Zheng, J.; Pathak, J.L.; Chen, Y.; Liang, D.; Yang, L.; Sun, H.; Zhong, M.; Wu, L.; Li, L. SLIT2 overexpression in periodontitis intensifies inflammation and alveolar bone loss, possibly via the activation of MAPK pathway. Front. Cell Dev. Biol. 2020, 8, 593. [Google Scholar] [CrossRef]
- Simmons, C.W.; VanderGheynst, J.S.; Upadhyaya, S.K. A model of Agrobacterium tumefaciens vacuum infiltration into harvested leaf tissue and subsequent in planta transgene transient expression. Biotechnol. Bioe. 2009, 102, 965–970. [Google Scholar] [CrossRef]
- VanderGheynst, J.; Guo, H.-Y.; Simmons, C. Response surface studies that elucidate the role of infiltration conditions on Agrobacterium tumefaciens-mediated transient transgene expression in harvested switchgrass (Panicum virgatum). Biomass Bioenergy 2008, 32, 372–379. [Google Scholar] [CrossRef]
- Pandey, S.P.; Somssich, I.E. The role of WRKY transcription factors in plant immunity. Plant Physiol. 2009, 150, 1648–1655. [Google Scholar] [CrossRef] [Green Version]
- Huang, Z.; LePore, K.; Elkin, G.; Thanavala, Y.; Mason, H.S. High-yield rapid production of hepatitis B surface antigen in plant leaf by a viral expression system. Plant Biotechnol. J. 2008, 6, 202–209. [Google Scholar] [CrossRef]
- Meng, D.; Li, Y.; Bai, Y.; Li, M.; Cheng, L. Genome-wide identification and characterization of WRKY transcriptional factor family in apple and analysis of their responses to waterlogging and drought stress. Plant Physiol. Biochem. 2016, 103, 71–83. [Google Scholar] [CrossRef] [PubMed]
- Wallaart, R.A. Distribution of sorbitol in Rosaceae. Phytochemistry 1980, 19, 2603–2610. [Google Scholar] [CrossRef]
- Greene, D.A.; Lattimer, S.A.; Sima, A.A. Sorbitol, phosphoinositides, and sodium-potassium-ATPase in the pathogenesis of diabetic complications. N. Engl. J. Med. 1987, 316, 599–606. [Google Scholar] [PubMed]
- Akhtar, K.P.; Ullah, N.; Saleem, M.Y.; Iqbal, Q.; Asghar, M.; Khan, A.R. Evaluation of tomato genotypes for early blight disease resistance caused by Alternaria solani in Pakistan. J. Plant Pathol. 2019, 101, 1159–1170. [Google Scholar] [CrossRef]
- Rushton, P.J.; Somssich, I.E.; Ringler, P.; Shen, Q.J. WRKY transcription factors. Trends Plant Sci. 2010, 15, 247–258. [Google Scholar] [CrossRef]
- Chen, L.; Song, Y.; Li, S.; Zhang, L.; Zou, C.; Yu, D. The role of WRKY transcription factors in plant abiotic stresses. Biochim. Et Biophys. Acta (BBA)-Gene Regul. Mech. 2012, 1819, 120–128. [Google Scholar] [CrossRef]
- Eulgem, T.; Rushton, P.J.; Robatzek, S.; Somssich, I.E. The WRKY superfamily of plant transcription factors. Trends Plant Sci. 2000, 5, 199–206. [Google Scholar] [CrossRef]
- Jiang, Y.; Deyholos, M.K. Functional characterization of Arabidopsis NaCl-inducible WRKY25 and WRKY33 transcription factors in abiotic stresses. Plant Mol. Biol. 2009, 69, 91–105. [Google Scholar] [CrossRef]
- Zheng, Z.; Mosher, S.L.; Fan, B.; Klessig, D.F.; Chen, Z. Functional analysis of Arabidopsis WRKY25 transcription factor in plant defense against Pseudomonas syringae. BMC Plant Biol. 2007, 7, 1–13. [Google Scholar] [CrossRef] [Green Version]
- Ross, C.A.; Liu, Y.; Shen, Q.J. The WRKY gene family in rice (Oryza sativa). J. Integr. Plant Biol. 2007, 49, 827–842. [Google Scholar] [CrossRef]
- Ishihama, N.; Yoshioka, H. Post-translational regulation of WRKY transcription factors in plant immunity. Curr. Opin. Plant Biol. 2012, 15, 431–437. [Google Scholar] [CrossRef] [PubMed]
- Qiu, Y.; Yu, D. Over-expression of the stress-induced OsWRKY45 enhances disease resistance and drought tolerance in Arabidopsis. Environ. Exp. Bot. 2009, 65, 35–47. [Google Scholar] [CrossRef]
- Yu, F.; Huaxia, Y.; Lu, W.; Wu, C.; Cao, X.; Guo, X. GhWRKY15, a member of the WRKY transcription factor family identified from cotton (Gossypium hirsutum L.), is involved in disease resistance and plant development. BMC Plant Biol. 2012, 12, 1–18. [Google Scholar] [CrossRef] [Green Version]
- Yang, P.; Chen, C.; Wang, Z.; Fan, B.; Chen, Z. A pathogen-and salicylic acid-induced WRKY DNA-binding activity recognizes the elicitor response element of the tobacco class I chitinase gene promoter. Plant J. 1999, 18, 141–149. [Google Scholar] [CrossRef]
- Yu, D.; Chen, C.; Chen, Z. Evidence for an important role of WRKY DNA binding proteins in the regulation of NPR1 gene expression. Plant Cell 2001, 13, 1527–1540. [Google Scholar] [CrossRef] [Green Version]
- Zhou, Q.Y.; Tian, A.G.; Zou, H.F.; Xie, Z.M.; Lei, G.; Huang, J.; Wang, C.M.; Wang, H.W.; Zhang, J.S.; Chen, S.Y. Soybean WRKY-type transcription factor genes, GmWRKY13, GmWRKY21, and GmWRKY54, confer differential tolerance to abiotic stresses in transgenic Arabidopsis plants. Plant Biotechnol. J. 2008, 6, 486–503. [Google Scholar] [CrossRef]
- Yamada, K.; Yamaguchi, K.; Shirakawa, T.; Nakagami, H.; Mine, A.; Ishikawa, K.; Fujiwara, M.; Narusaka, M.; Narusaka, Y.; Ichimura, K. The Arabidopsis CERK 1-associated kinase PBL 27 connects chitin perception to MAPK activation. EMBO J. 2016, 35, 2468–2483. [Google Scholar] [CrossRef]
- Di, T.; Zhao, L.; Chen, H.; Qian, W.; Wang, P.; Zhang, X.; Xia, T. Transcriptomic and metabolic insights into the distinctive effects of exogenous melatonin and gibberellin on terpenoid synthesis and plant hormone signal transduction pathway in Camellia sinensis. J. Agric. Food Chem. 2019, 67, 4689–4699. [Google Scholar] [CrossRef]
- Wu, T.; Wang, Y.; Zheng, Y.; Fei, Z.; Dandekar, A.M.; Xu, K.; Han, Z.; Cheng, L. Suppressing sorbitol synthesis substantially alters the global expression profile of stress response genes in apple (Malus domestica) leaves. Plant Cell Physiol. 2015, 56, 1748–1761. [Google Scholar] [CrossRef] [Green Version]
- Kumpoun, W.; Motomura, Y.; Harada, Y. Inhibition of Aspergillus rot by sorbitol in apple fruit with watercore symptoms. Postharvest Biol. Technol. 2003, 29, 121–127. [Google Scholar] [CrossRef]
- Gopal, J.; Iwama, K. In vitro screening of potato against water-stress mediated through sorbitol and polyethylene glycol. Plant Cell Rep. 2007, 26, 693–700. [Google Scholar] [CrossRef] [PubMed]




Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Qi, M.; Wu, R.; Song, Z.; Dong, B.; Chen, T.; Wang, M.; Cao, H.; Du, T.; Wang, S.; Li, N.; et al. Sorbitol Reduces Sensitivity to Alternaria by Promoting Ceramide Kinases (CERK) Expression through Transcription Factor Pswrky25 in Populus (Populus simonii Carr.). Genes 2022, 13, 405. https://doi.org/10.3390/genes13030405
Qi M, Wu R, Song Z, Dong B, Chen T, Wang M, Cao H, Du T, Wang S, Li N, et al. Sorbitol Reduces Sensitivity to Alternaria by Promoting Ceramide Kinases (CERK) Expression through Transcription Factor Pswrky25 in Populus (Populus simonii Carr.). Genes. 2022; 13(3):405. https://doi.org/10.3390/genes13030405
Chicago/Turabian StyleQi, Meng, Rui Wu, Zhihua Song, Biying Dong, Ting Chen, Mengying Wang, Hongyan Cao, Tingting Du, Shengjie Wang, Na Li, and et al. 2022. "Sorbitol Reduces Sensitivity to Alternaria by Promoting Ceramide Kinases (CERK) Expression through Transcription Factor Pswrky25 in Populus (Populus simonii Carr.)" Genes 13, no. 3: 405. https://doi.org/10.3390/genes13030405
APA StyleQi, M., Wu, R., Song, Z., Dong, B., Chen, T., Wang, M., Cao, H., Du, T., Wang, S., Li, N., Yang, Q., Fu, Y., & Meng, D. (2022). Sorbitol Reduces Sensitivity to Alternaria by Promoting Ceramide Kinases (CERK) Expression through Transcription Factor Pswrky25 in Populus (Populus simonii Carr.). Genes, 13(3), 405. https://doi.org/10.3390/genes13030405





