HIV Protease and Integrase Empirical Substitution Models of Evolution: Protein-Specific Models Outperform Generalist Models
Abstract
1. Introduction
2. Materials and Methods
2.1. Study Data of HIV-1 and General Virus Protease and Integrase
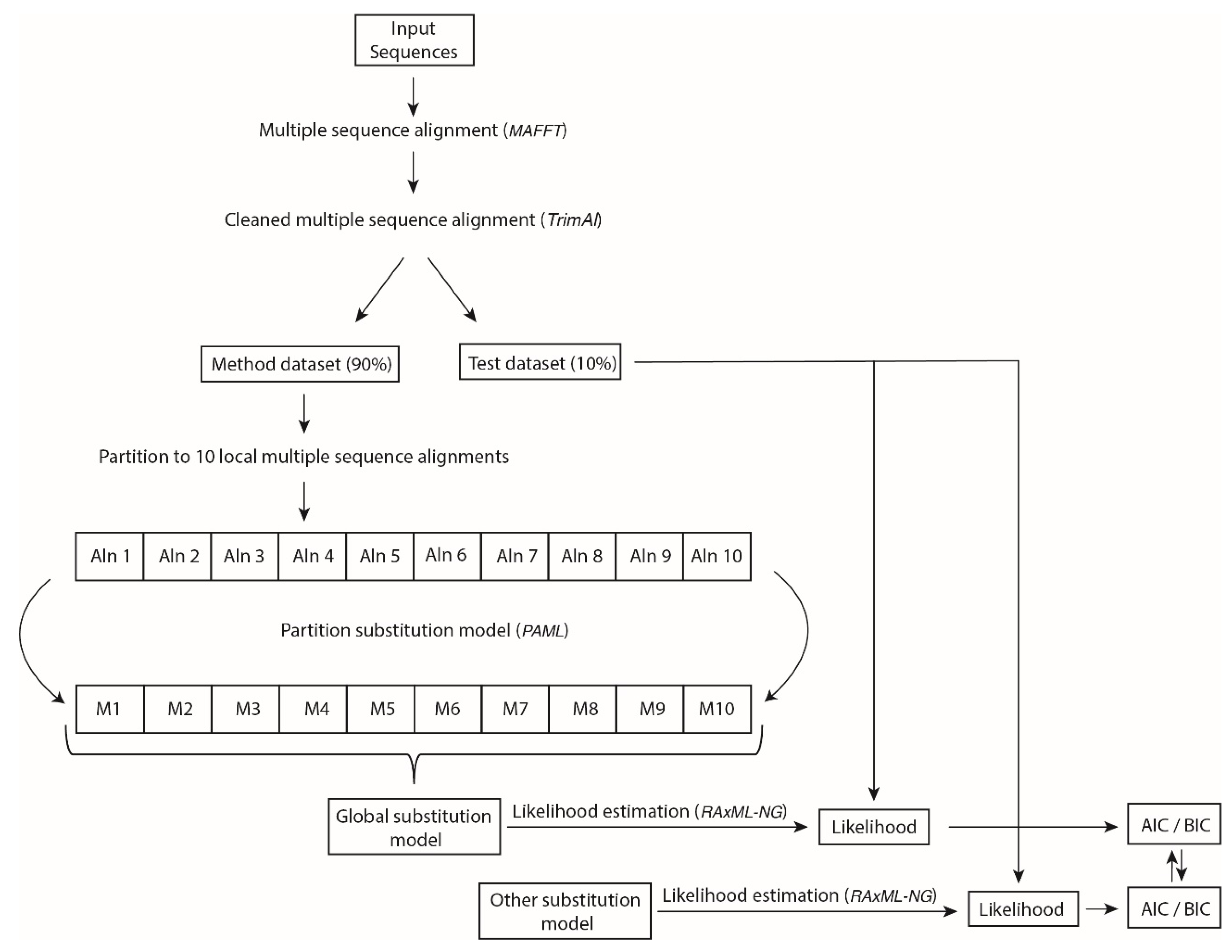
2.2. Inference of Novel Empirical Substitution Models for HIV and General Virus Protease and Integrase
2.3. Evaluation of the Novel Empirical Substitution Models of HIV-1 and General Virus Protease and Integrase
3. Results and Discussion
3.1. Novel Empirical Substitution Models for HIV and General Virus Protease and Integrase
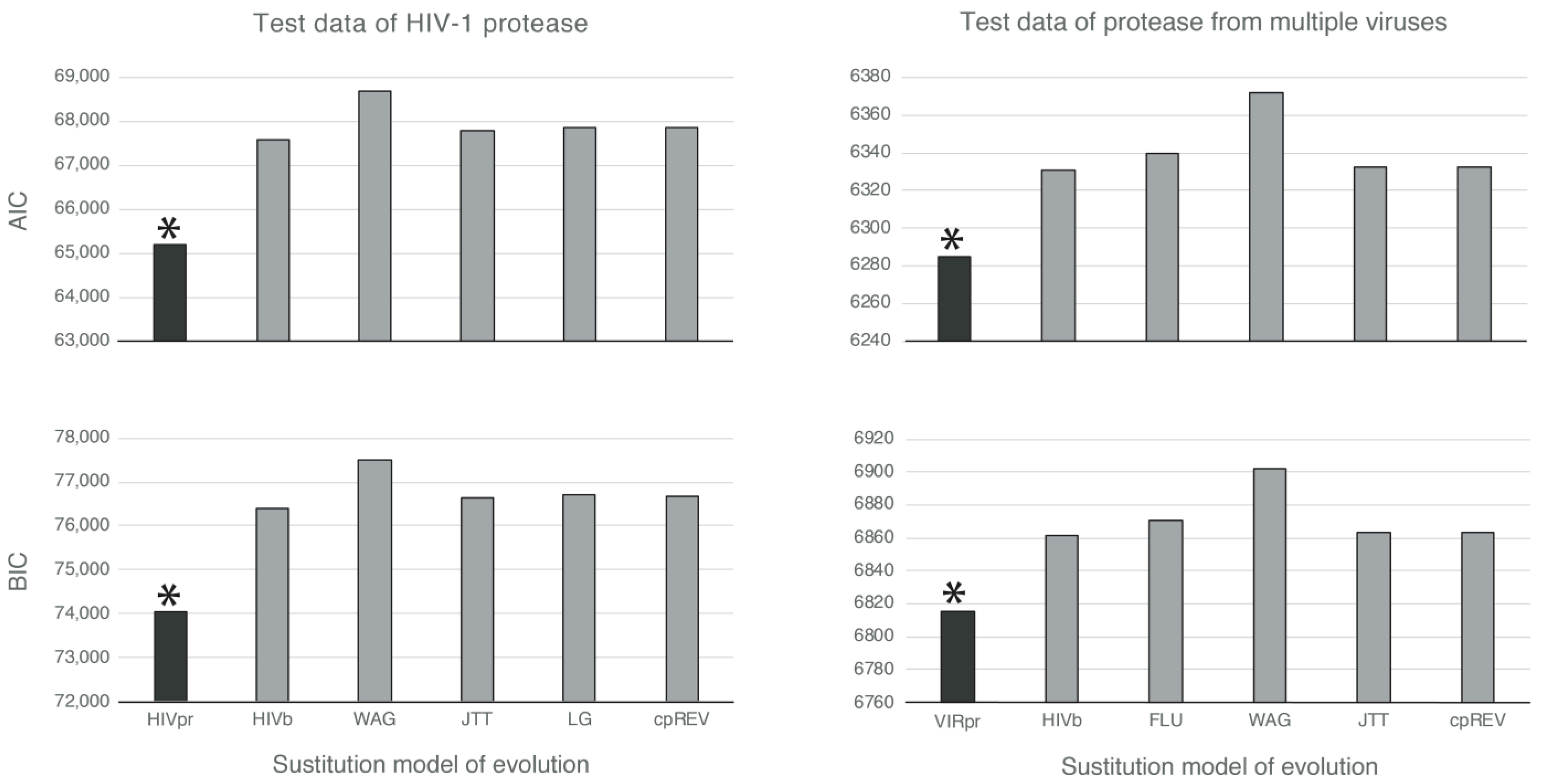
3.2. Likelihood-Based Comparisons Indicate That the Novel Empirical Substitution Models Outperform the Currently Available Empirical Substitution Models
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Conflicts of Interest
References
- Arenas, M. Trends in Substitution Models of Molecular Evolution. Front Genet 2015, 6, 319. [Google Scholar] [CrossRef] [PubMed]
- Yutin, N.; Puigbò, P.; Koonin, E.V.; Wolf, Y.I. Phylogenomics of Prokaryotic Ribosomal Proteins. PLoS ONE 2012, 7, e36972. [Google Scholar] [CrossRef]
- Shi, M.; Lin, X.-D.; Chen, X.; Tian, J.-H.; Chen, L.-J.; Li, K.; Wang, W.; Eden, J.-S.; Shen, J.-J.; Liu, L.; et al. The Evolutionary History of Vertebrate RNA Viruses. Nature 2018, 556, 197–202. [Google Scholar] [CrossRef] [PubMed]
- Furukawa, R.; Toma, W.; Yamazaki, K.; Akanuma, S. Ancestral Sequence Reconstruction Produces Thermally Stable Enzymes with Mesophilic Enzyme-like Catalytic Properties. Sci. Rep. 2020, 10, 15493. [Google Scholar] [CrossRef]
- Arenas, M.; Bastolla, U. ProtASR2: Ancestral Reconstruction of Protein Sequences Accounting for Folding Stability. Methods Ecol. Evol. 2020, 11, 248–257. [Google Scholar] [CrossRef]
- Koshi, J.M.; Mindell, D.P.; Goldstein, R.A. Using Physical-Chemistry-Based Substitution Models in Phylogenetic Analyses of HIV-1 Subtypes. Mol. Biol. Evol. 1999, 165, 173–179. [Google Scholar] [CrossRef]
- Bruno, W.J. Modeling Residue Usage in Aligned Protein Sequences via Maximum Likelihood. Mol. Biol. Evol. 1996, 13, 1368–1374. [Google Scholar] [CrossRef][Green Version]
- Thorne, J.L. Models of Protein Sequence Evolution and Their Applications. Curr. Opin. Genet. Dev. 2000, 10, 602–605. [Google Scholar] [CrossRef]
- Liberles, D.A.; Teichmann, S.A.; Bahar, I.; Bastolla, U.; Bloom, J.; Bornberg-Bauer, E.; Colwell, L.J.; de Koning, A.P.; Dokholyan, N.V.; Echave, J.; et al. The Interface of Protein Structure, Protein Biophysics, and Molecular Evolution. Protein Sci. 2012, 21, 769–785. [Google Scholar] [CrossRef]
- Arenas, M.; Sanchez-Cobos, A.; Bastolla, U. Maximum Likelihood Phylogenetic Inference with Selection on Protein Folding Stability. Mol. Biol. Evol. 2015, 32, 2195–2207. [Google Scholar] [CrossRef]
- Parisi, G.; Echave, J. The Structurally Constrained Protein Evolution Model Accounts for Sequence Patterns of the LbetaH Superfamily. BMC Evol. Biol. 2004, 4, 41. [Google Scholar] [CrossRef] [PubMed]
- Bordner, A.J.; Mittelmann, H.D. A New Formulation of Protein Evolutionary Models That Account for Structural Constraints. Mol. Biol. Evol. 2013, 31, 736–749. [Google Scholar] [CrossRef]
- Echave, J. Beyond Stability Constraints: A Biophysical Model of Enzyme Evolution with Selection on Stability and Activity. Mol. Biol. Evol. 2019, 36, 613–620. [Google Scholar] [CrossRef] [PubMed]
- Bastolla, U.; Arenas, M. The Influence of Protein Stability on Sequence Evolution: Applications to Phylogenetic Inference. Methods Mol. Biol. 2019, 1851, 215–231. [Google Scholar]
- Pupko, T.; Bell, R.E.; Mayrose, I.; Glaser, F.; Ben-Tal, N. Rate4Site: An Algorithmic Tool for the Identification of Functional Regions in Proteins by Surface Mapping of Evolutionary Determinants within Their Homologues. Bioinformatics 2002, 18, S71–S77. [Google Scholar] [CrossRef] [PubMed]
- Yang, Z.; Nielsen, R.; Hasegawa, M. Models of Amino Acid Substitution and Applications to Mitochondrial Protein Evolution. Mol. Biol. Evol. 1998, 15, 1600–1611. [Google Scholar] [CrossRef]
- Arenas, M.; Dos Santos, H.G.; Posada, D.; Bastolla, U. Protein Evolution along Phylogenetic Histories under Structurally Constrained Substitution Models. Bioinformatics 2013, 29, 3020–3028. [Google Scholar] [CrossRef]
- Kozlov, A.M.; Darriba, D.; Flouri, T.; Morel, B.; Stamatakis, A. RAxML-NG: A Fast, Scalable and User-Friendly Tool for Maximum Likelihood Phylogenetic Inference. Bioinformatics 2019, 35, 4453–4455. [Google Scholar] [CrossRef]
- Yang, Z. PAML 4: Phylogenetic Analysis by Maximum Likelihood. Mol Biol Evol 2007, 24, 1586–1591. [Google Scholar] [CrossRef]
- Kosakovsky Pond, S.L.; Frost, S.D.; Muse, S.V. HYPHY: Hypothesis Testing Using Phylogenies. Bioinformatics 2005, 21, 676–679. [Google Scholar] [CrossRef]
- Jones, D.T.; Taylor, W.R.; Thornton, J.M. The Rapid Generation of Mutation Data Matrices from Protein Sequences. Comput Appl Biosci 1992, 8, 275–282. [Google Scholar] [CrossRef]
- Whelan, S.; Goldman, N. A General Empirical Model of Protein Evolution Derived from Multiple Protein Families Using a Maximum-Likelihood Approach. Mol. Biol. Evol. 2001, 18, 691–699. [Google Scholar] [CrossRef] [PubMed]
- Adachi, J.; Hasegawa, M. Model of Amino Acid Substitution in Proteins Encoded by Mitochondrial DNA. J Mol Evol 1996, 42, 459–468. [Google Scholar] [CrossRef] [PubMed]
- Nickle, D.C.; Heath, L.; Jensen, M.A.; Gilbert, P.B.; Mullins, J.I.; Kosakovsky Pond, S.L. HIV-Specific Probabilistic Models of Protein Evolution. PLoS One 2007, 2, e503. [Google Scholar] [CrossRef] [PubMed]
- Dang, C.C.; Le, Q.S.; Gascuel, O.; Le, V.S. FLU, an Amino Acid Substitution Model for Influenza Proteins. BMC Evol Biol 2010, 10, 99. [Google Scholar] [CrossRef] [PubMed]
- Kim, T.L.; Cao, C.D.; Le, V.S. Building a Specific Amino Acid Substitution Model for Dengue Viruses. In Proceedings of the 2018 10th International Conference on Knowledge and Systems Engineering (KSE), Ho Chi Minh City, Vietnam, 1 November 2018; pp. 242–246. [Google Scholar]
- Le, T.K.; Vinh, L.S. FLAVI: An Amino Acid Substitution Model for Flaviviruses. J. Mol. Evol. 2020, 88, 445–452. [Google Scholar] [CrossRef] [PubMed]
- Lemmon, A.R.; Moriarty, E.C. The Importance of Proper Model Assumption in Bayesian Phylogenetics. Syst Biol 2004, 53, 265–277. [Google Scholar] [CrossRef]
- Minin, V.; Abdo, Z.; Joyce, P.; Sullivan, J. Performance-Based Selection of Likelihood Models for Phylogeny Estimation. Syst. Biol. 2003, 52, 674–683. [Google Scholar] [CrossRef]
- Yang, Z.; Goldman, N.; Friday, A. Comparison of Models for Nucleotide Substitution Used in Maximum-Likelihood Phylogenetic Estimation. Mol. Biol. Evol. 1994, 11, 316–324. [Google Scholar]
- Zhang, J.; Nei, M. Accuracies of Ancestral Amino Acid Sequences Inferred by the Parsimony, Likelihood, and Distance Methods. J Mol Evol 1997, 44 (Suppl. 1), S139–S146. [Google Scholar] [CrossRef]
- Zhang, J. Performance of Likelihood Ratio Tests of Evolutionary Hypotheses under Inadequate Substitution Models. Mol. Biol. Evol. 1999, 16, 868–875. [Google Scholar] [CrossRef]
- Abascal, F.; Zardoya, R.; Posada, D. ProtTest: Selection of Best-Fit Models of Protein Evolution. Bioinformatics 2005, 21, 2104–2105. [Google Scholar] [CrossRef] [PubMed]
- Keane, T.M.; Creevey, C.J.; Pentony, M.M.; Naughton, T.J.; McLnerney, J.O. Assessment of Methods for Amino Acid Matrix Selection and Their Use on Empirical Data Shows That Ad Hoc Assumptions for Choice of Matrix Are Not Justified. BMC Evol. Biol. 2006, 6, 29. [Google Scholar] [CrossRef]
- Katoh, K.; Standley, D.M. MAFFT Multiple Sequence Alignment Software Version 7: Improvements in Performance and Usability. Mol. Biol. Evol. 2013, 30, 772–780. [Google Scholar] [CrossRef] [PubMed]
- Capella-Gutierrez, S.; Silla-Martinez, J.M.; Gabaldon, T. TrimAl: A Tool for Automated Alignment Trimming in Large-Scale Phylogenetic Analyses. Bioinformatics 2009, 25, 1972–1973. [Google Scholar] [CrossRef]
- Abascal, F.; Posada, D.; Zardoya, R. MtArt: A New Model of Amino Acid Replacement for Arthropoda. Mol. Biol. Evol. 2007, 24, 1–5. [Google Scholar] [CrossRef] [PubMed]
- Minh, B.Q.; Dang, C.C.; Vinh, L.S.; Lanfear, R. QMaker: Fast and Accurate Method to Estimate Empirical Models of Protein Evolution. Syst. Biol. 2021, 70, 1046–1060. [Google Scholar] [CrossRef]
- Arenas, M.; Villaverde, M.C.; Sussman, F. Prediction and Analysis of Binding Affinities for Chemically Diverse HIV-1 PR Inhibitors by the Modified SAFE_p Approach. J. Comput. Chem. 2009, 30, 1229–1240. [Google Scholar] [CrossRef]
- Arenas, M. Genetic Consequences of Antiviral Therapy on HIV-1. Comput. Math. Method Med. 2015, 2015, 9. [Google Scholar] [CrossRef]
- Ghosh, A.K.; Osswald, H.L.; Prato, G. Recent Progress in the Development of HIV-1 Protease Inhibitors for the Treatment of HIV/AIDS. J. Med. Chem. 2016, 59, 5172–5208. [Google Scholar] [CrossRef]
- Hazuda, D.J. HIV Integrase as a Target for Antiretroviral Therapy. Curr. Opin. HIV AIDS 2012, 7, 383–389. [Google Scholar] [CrossRef] [PubMed]
- Darriba, D.; Taboada, G.L.; Doallo, R.; Posada, D. ProtTest 3: Fast Selection of Best-Fit Models of Protein Evolution. Bioinformatics 2011, 27, 1164–1165. [Google Scholar] [CrossRef]
- Akaike, H. Information Theory and an Extension of the Maximum Likelihood Principle. In Second International Symposium on Information Theory; Petrov, B.N., Csaki, F., Eds.; Akademiai Kiado: Budapest, Hungary, 1973; pp. 267–281. [Google Scholar]
- Schwarz, G. Estimating the Dimension of a Model. Ann. Stat. 1978, 6, 461–464. [Google Scholar] [CrossRef]
- Weber, I.T.; Wang, Y.-F.; Harrison, R.W. HIV Protease: Historical Perspective and Current Research. Viruses 2021, 13, 839. [Google Scholar] [CrossRef]
- Craik, C.S.; Roczniak, S.; Largman, C.; Rutter, W.J. The Catalytic Role of the Active Site Aspartic Acid in Serine Proteases. Science 1987, 237, 909–913. [Google Scholar] [CrossRef] [PubMed]
- Engelman, A.; Craigie, R. Identification of Conserved Amino Acid Residues Critical for Human Immunodeficiency Virus Type 1 Integrase Function in Vitro. J. Virol. 1992, 66, 6361–6369. [Google Scholar] [CrossRef] [PubMed]
- Kulkosky, J.; Jones, K.S.; Katz, R.A.; Mack, J.P.; Skalka, A.M. Residues Critical for Retroviral Integrative Recombination in a Region That Is Highly Conserved among Retroviral/Retrotransposon Integrases and Bacterial Insertion Sequence Transposases. Mol. Cell. Biol. 1992, 12, 2331–2338. [Google Scholar] [CrossRef] [PubMed]
- Parera, M.; Fernandez, G.; Clotet, B.; Martinez, M.A. HIV-1 Protease Catalytic Efficiency Effects Caused by Random Single Amino Acid Substitutions. Mol Biol Evol 2007, 24, 382–387. [Google Scholar] [CrossRef][Green Version]
- Ribeiro, A.J.M.; Tyzack, J.D.; Borkakoti, N.; Holliday, G.L.; Thornton, J.M. A Global Analysis of Function and Conservation of Catalytic Residues in Enzymes. J. Biol. Chem. 2020, 295, 314–324. [Google Scholar] [CrossRef] [PubMed]
- HIV Protease and Integrase Empirical Substitution Models of Evolution: Protein-Specific Models Outperform Generalist Models. Available online: https://zenodo.org/record/5763867#.YcWbnx17mjQ (accessed on 7 December 2021).

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Del Amparo, R.; Arenas, M. HIV Protease and Integrase Empirical Substitution Models of Evolution: Protein-Specific Models Outperform Generalist Models. Genes 2022, 13, 61. https://doi.org/10.3390/genes13010061
Del Amparo R, Arenas M. HIV Protease and Integrase Empirical Substitution Models of Evolution: Protein-Specific Models Outperform Generalist Models. Genes. 2022; 13(1):61. https://doi.org/10.3390/genes13010061
Chicago/Turabian StyleDel Amparo, Roberto, and Miguel Arenas. 2022. "HIV Protease and Integrase Empirical Substitution Models of Evolution: Protein-Specific Models Outperform Generalist Models" Genes 13, no. 1: 61. https://doi.org/10.3390/genes13010061
APA StyleDel Amparo, R., & Arenas, M. (2022). HIV Protease and Integrase Empirical Substitution Models of Evolution: Protein-Specific Models Outperform Generalist Models. Genes, 13(1), 61. https://doi.org/10.3390/genes13010061






