The Neglected Contribution of Streptomycin to the Tuberculosis Drug Resistance Problem
Abstract
1. Introduction
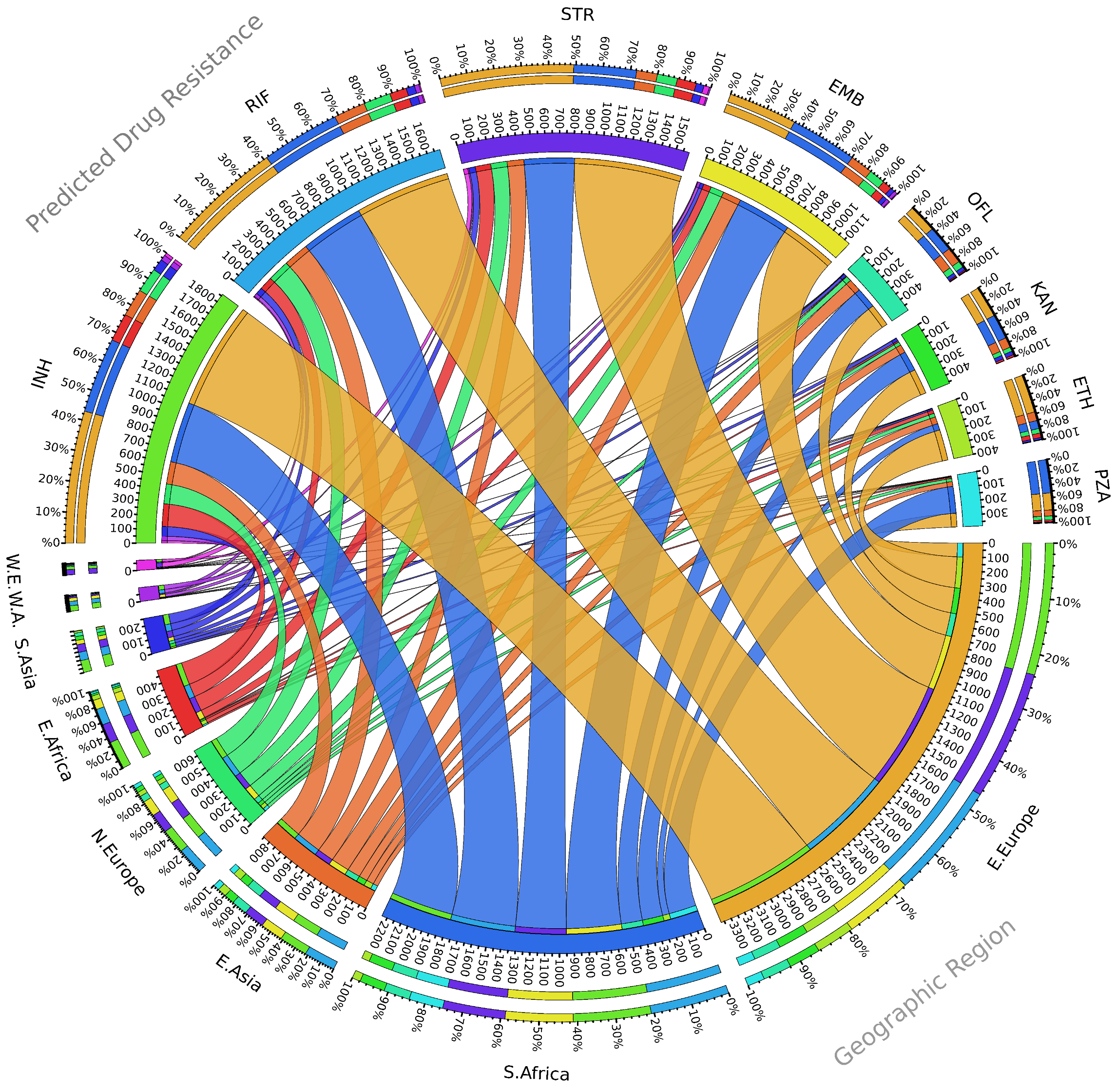
2. Seventy Years of Streptomycin Resistance in Tuberculosis, Where Do We Stand?
3. Emergence of Streptomycin-Resistant Strains
3.1. Mode of Action of Streptomycin
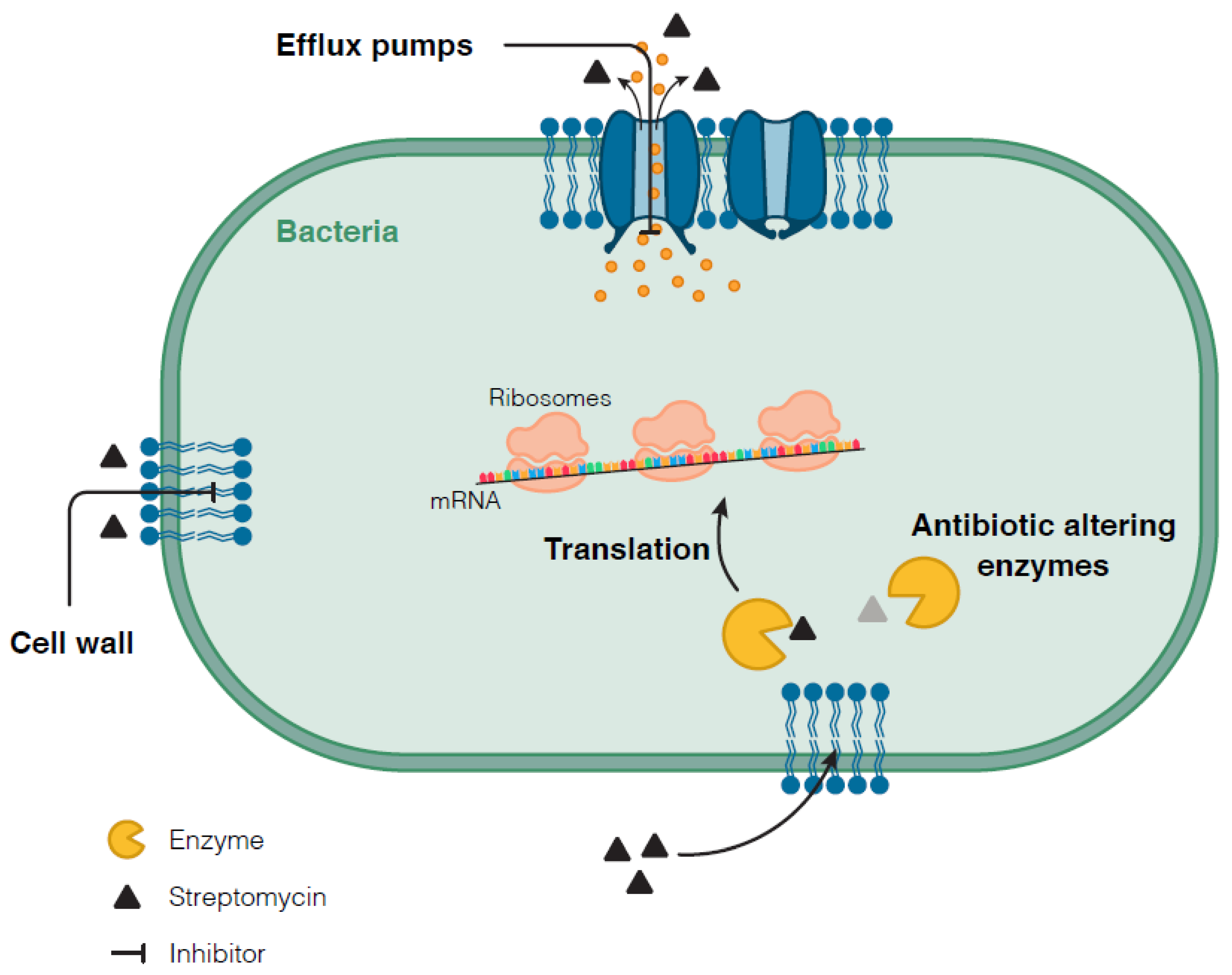
3.2. Molecular Mechanisms of Streptomycin Resistance
3.2.1. Streptomycin Resistance Mutations in rpsL, rrs, and gid
3.2.2. Other Mechanisms Conferring Resistance to Streptomycin
3.3. M. tuberculosis Mechanisms Associated with Tolerance to Streptomycin
4. Streptomycin-Resistant Mutations and Fitness Costs
4.1. Does Streptomycin Predisposes M. tuberculosis to Other Drug Resistances?
4.2. Fitness of M. tuberculosis Lineages with Streptomycin-Resistant Mutations
5. Conclusions and Future Perspectives
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- World Health Organization. Global Tuberculosis Report 2021; WHO: Geneva, Switzerland, 2021. [Google Scholar]
- Prasanna, A.; Niranjan, V. Classification of Mycobacterium Tuberculosis DR, MDR, XDR Isolates and Identification of Signature Mutation Pattern of Drug Resistance. Bioinformation 2019, 15, 261–268. [Google Scholar] [CrossRef] [PubMed]
- Khawbung, J.L.; Nath, D.; Chakraborty, S. Drug Resistant Tuberculosis: A Review. Comp. Immunol. Microbiol. Infect. Dis. 2021, 74, 101574. [Google Scholar] [CrossRef]
- Dheda, K.; Gumbo, T.; Maartens, G.; Dooley, K.E.; McNerney, R.; Murray, M.; Furin, J.; Nardell, E.A.; London, L.; Lessem, E.; et al. The Epidemiology, Pathogenesis, Transmission, Diagnosis, and Management of Multidrug-Resistant, Extensively Drug-Resistant, and Incurable Tuberculosis. Lancet Respir. Med. 2017, 5, 291–360. [Google Scholar] [CrossRef]
- World Health Organization. Global Tuberculosis Report; WHO: Geneva, Switzerland, 2019. [Google Scholar]
- Falzon, D.; Schünemann, H.J.; Harausz, E.; González-Angulo, L.; Lienhardt, C.; Jaramillo, E.; Weyer, K. World Health Organization Treatment Guidelines for Drug-Resistant Tuberculosis, 2016 Update. Eur. Respir. J. 2017, 49, 1602308. [Google Scholar] [CrossRef]
- Kempker, R.R.; Vashakidze, S.; Solomonia, N.; Dzidzikashvili, N.; Blumberg, H.M. Surgical Treatment of Drug-Resistant Tuberculosis. Lancet Infect. Dis. 2012, 12, 157–166. [Google Scholar] [CrossRef]
- Vashakidze, S.; Despuig, A.; Gogishvili, S.; Nikolaishvili, K.; Shubladze, N.; Avaliani, Z.; Tukvadze, N.; Casals, M.; Caylà, J.A.; Cardona, P.J.; et al. Retrospective Study of Clinical and Lesion Characteristics of Patients Undergoing Surgical Treatment for Pulmonary Tuberculosis in Georgia. Int. J. Infect. Dis. 2017, 56, 200–207. [Google Scholar] [CrossRef] [PubMed]
- Benito, P.; Vashakidze, S.; Gogishvili, S.; Nikolaishvili, K.; Despuig, A.; Tukvadze, N.; Shubladze, N.; Avaliani, Z.; Vilaplana, C. Impact of Adjuvant Therapeutic Surgery on the Health-Related Quality of Life of Pulmonary Tuberculosis Patients. ERJ Open Res. 2020, 6, 00083–02020. [Google Scholar] [CrossRef] [PubMed]
- World Health Organization. Operational Handbook on Tuberculosis. Module 4: Treatment—Drug-Resistant Tuberculosis Treatment; World Health Organization: Geneva, Switzerland, 2020. [Google Scholar]
- Cohen, K.A.; Stott, K.E.; Munsamy, V.; Manson, A.L.; Earl, A.M.; Pym, A.S. Evidence for Expanding the Role of Streptomycin in the Management of Drug-Resistant Mycobacterium Tuberculosis. Antimicrob. Agents Chemother. 2020, 64, e00860-20. [Google Scholar] [CrossRef] [PubMed]
- Gajdács, M. The Concept of an Ideal Antibiotic: Implications for Drug Design. Molecules 2019, 24, 892. [Google Scholar] [CrossRef] [PubMed]
- Serio, A.W.; Keepers, T.; Andrews, L.; Krause, K.M. Aminoglycoside Revival: Review of a Historically Important Class of Antimicrobials Undergoing Rejuvenation. EcoSal Plus 2018, 8. [Google Scholar] [CrossRef]
- Murray, J.F.; Schraufnagel, D.E.; Hopewell, P.C. Treatment of Tuberculosis: A Historical Perspective. Ann. Am. Thorac. Soc. 2015, 12, 1749–1759. [Google Scholar] [CrossRef]
- Zwick, E.D.; Pepperell, C.S. Tuberculosis Sanatorium Treatment at the Advent of the Chemotherapy Era. BMC Infect. Dis. 2020, 20, 1–11. [Google Scholar] [CrossRef]
- Davies, P.D.O.; Yew, W.W. Recent Developments in the Treatment of Tuberculosis. Expert Opin. Investig. Drugs 2003, 12, 1297–1312. [Google Scholar] [CrossRef]
- Schito, M.; Migliori, G.B.; Fletcher, H.A.; McNerney, R.; Centis, R.; D’Ambrosio, L.; Bates, M.; Kibiki, G.; Kapata, N.; Corrah, T.; et al. Perspectives on Advances in Tuberculosis Diagnostics, Drugs, and Vaccines. Clin. Infect. Dis. 2015, 61 (Suppl. 3), S102–S118. [Google Scholar] [CrossRef]
- Dai, R.; He, J.; Zha, X.; Wang, Y.; Zhang, X.; Gao, H.; Yang, X.; Li, J.; Xin, Y.; Wang, Y.; et al. A Novel Mechanism of Streptomycin Resistance in Yersinia Pestis: Mutation in the Rpsl Gene. PLoS Negl. Trop. Dis. 2021, 15, e0009324. [Google Scholar] [CrossRef]
- Manson, A.L.; Cohen, K.A.; Abeel, T.; Desjardins, C.A.; Armstrong, D.T.; Barry, C.E.; Brand, J.; Brand, J.; Jureen, P.; Malinga, L.; et al. Genomic Analysis of Globally Diverse Mycobacterium Tuberculosis Strains Provides Insights into the Emergence and Spread of Multidrug Resistance. Nat. Genet. 2017, 49, 395–402. [Google Scholar] [CrossRef]
- Glasauer, S.; Altmann, D.; Hauer, B.; Brodhun, B.; Haas, W.; Perumal, N. First-Line Tuberculosis Drug Resistance Patterns and Associated Risk Factors in Germany, 2008–2017. PLoS ONE 2019, 14, e0217597. [Google Scholar] [CrossRef]
- Perdigão, J.; Macedo, R.; Machado, D.; Silva, C.; Jordão, L.; Couto, I.; Viveiros, M.; Portugal, I. GidB Mutation as a Phylogenetic Marker for Q1 Cluster Mycobacterium Tuberculosis Isolates and Intermediate-Level Streptomycin Resistance Determinant in Lisbon, Portugal. Clin. Microbiol. Infect. 2014, 20, O278–O284. [Google Scholar] [CrossRef]
- Perdigão, J.; Macedo, R.; Silva, C.; Machado, D.; Couto, I.; Viveiros, M.; Jordao, L.; Portugal, I. From Multidrug-Resistant to Extensively Drug-Resistant Tuberculosis in Lisbon, Portugal: The Stepwise Mode of Resistance Acquisition. J. Antimicrob. Chemother. 2013, 68, 27–33. [Google Scholar] [CrossRef]
- Rocha, D.M.G.C.; Magalhães, C.; Cá, B.; Ramos, A.; Carvalho, T.; Comas, I.; Guimarães, J.T.; Bastos, H.N.; Saraiva, M.; Osório, N.S. Heterogeneous Streptomycin Resistance Level Among Mycobacterium Tuberculosis Strains From the Same Transmission Cluster. Front. Microbiol. 2021, 12, 1380. [Google Scholar] [CrossRef]
- Sousa, J.; Cá, B.; Maceiras, A.R.; Costa, A.; Fonseca, K.; Fernandes, A.; Ramos, A.; Carvalho, T.; Barros, L.; Magalhães, C.; et al. Mycobacterium Tuberculosis Associated with Severe Tuberculosis Evades Cytosolic Surveillance Systems and Modulates IL-1ß Production. Nat. Commun. 2020, in press. [Google Scholar] [CrossRef]
- Bastos, H.N.; Osório, N.S.; Castro, A.G.; Ramos, A.; Carvalho, T.; Meira, L.; Araújo, D.; Almeida, L.; Boaventura, R.; Fragata, P.; et al. A Prediction Rule to Stratify Mortality Risk of Patients with Pulmonary Tuberculosis. PLoS ONE 2016, 11, e0162797. [Google Scholar] [CrossRef]
- Madrazo-Moya, C.F.; Cancino-Muñoz, I.; Cuevas-Córdoba, B.; González-Covarrubias, V.; Barbosa-Amezcua, M.; Soberón, X.; Muñiz-Salazar, R.; Martínez-Guarneros, A.; Bäcker, C.; Zarrabal-Meza, J.; et al. Whole Genomic Sequencing as a Tool for Diagnosis of Drug and Multidrug-Resistance Tuberculosis in an Endemic Region in Mexico. PLoS ONE 2019, 14, e0213046. [Google Scholar] [CrossRef]
- Wu, X.; Yang, J.; Tan, G.; Liu, H.; Liu, Y.; Guo, Y.; Gao, R.; Wan, B.; Yu, F. Drug Resistance Characteristics of Mycobacterium Tuberculosis Isolates From Patients With Tuberculosis to 12 Antituberculous Drugs in China. Front. Cell. Infect. Microbiol. 2019, 9, 345. [Google Scholar] [CrossRef]
- Rezaei, F.; Haeili, M.; Imani Fooladi, A.; Azari Garmjan, G.A.; Feizabadi, M.M. Screening for Streptomycin Resistance Conferring Mutations in Mycobacterium Tuberculosis Isolates from Iran. J. Chemother. 2017, 29, 14–18. [Google Scholar] [CrossRef]
- Khosravi, A.D.; Etemad, N.; Hashemzadeh, M.; Khandan Dezfuli, S.; Goodarzi, H. Frequency of Rrs and RpsL Mutations in Streptomycin-Resistant Mycobacterium Tuberculosis Isolates from Iranian Patients. J. Glob. Antimicrob. Resist. 2017, 9, 51–56. [Google Scholar] [CrossRef]
- Thida Oo, N.A.; San, L.L.; Thapa, J.; Aye, K.S.; Aung, W.W.; Nakajima, C.; Suzuki, Y. Characterization of Mutations Conferring Streptomycin Resistance to Multidrug-Resistant Mycobacterium Tuberculosis Isolates from Myanmar. Tuberculosis 2018, 111, 8–13. [Google Scholar] [CrossRef]
- Maung, H.M.W.; Palittapongarnpim, P.; Aung, H.L.; Surachat, K.; Nyunt, W.W.; Chongsuvivatwong, V. Geno-Spatial Distribution of Mycobacterium Tuberculosis and Drug Resistance Profiles in Myanmar⇓thai Border Area. Trop. Med. Infect. Dis. 2020, 5, 153. [Google Scholar] [CrossRef]
- Smittipat, N.; Juthayothin, T.; Billamas, P.; Jaitrong, S.; Rukseree, K.; Dokladda, K.; Chaiyasirinroje, B.; Disratthakit, A.; Chaiprasert, A.; Mahasirimongkol, S.; et al. Mutations in Rrs, RpsL and GidB in Streptomycin-Resistant Mycobacterium Tuberculosis Isolates from Thailand. J. Glob. Antimicrob. Resist. 2016, 4, 5–10. [Google Scholar] [CrossRef]
- Yasha, E.; Avika, D.; Luca Freschi, M.F. Tuberculosis Resistance Acquisition in Space and Time: An Analysis of Globally Diverse. Bioarxive 2019, 11, 837096. [Google Scholar] [CrossRef]
- Forson, A.; Kwara, A.; Kudzawu, S.; Omari, M.; Otu, J.; Gehre, F.; de Jong, B.; Antonio, M. A Cross-Sectional Study of Tuberculosis Drug Resistance among Previously Treated Patients in a Tertiary Hospital in Accra, Ghana: Public Health Implications of Standardized Regimens. BMC Infect. Dis. 2018, 18, 4–9. [Google Scholar] [CrossRef]
- Bainomugisa, A.; Lavu, E.; Hiashiri, S.; Majumdar, S.; Honjepari, A.; Moke, R.; Dakulala, P.; Hill-Cawthorne, G.A.; Pandey, S.; Marais, B.J.; et al. Multi-Clonal Evolution of Multi-Drug-Resistant/Extensively Drug-Resistant Mycobacterium Tuberculosis in a High-Prevalence Setting of Papua New Guinea for over Three Decades. Microb. Genom. 2018, 4, e000147. [Google Scholar] [CrossRef]
- Ektefaie, Y.; Dixit, A.; Freschi, L.; Farhat, M.R. Globally Diverse Mycobacterium Tuberculosis Resistance Acquisition: A Retrospective Geographical and Temporal Analysis of Whole Genome Sequences. Lancet Microbe 2021, 2, e96–e104. [Google Scholar] [CrossRef]
- Holt, K.E.; McAdam, P.; Thai, P.V.K.; Thuong, N.T.T.; Ha, D.T.M.; Lan, N.N.; Lan, N.H.; Nhu, N.T.Q.; Hai, H.T.; Ha, V.T.N.; et al. Frequent Transmission of the Mycobacterium Tuberculosis Beijing Lineage and Positive Selection for the EsxW Beijing Variant in Vietnam. Nat. Genet. 2018, 50, 849–856. [Google Scholar] [CrossRef]
- Buu, T.N.; van Soolingen, D.; Huyen, M.N.T.; Lan, N.T.N.; Quy, H.T.; Tiemersma, E.W.; Kremer, K.; Borgdorff, M.W.; Cobelens, F.G.J. Increased Transmission of Mycobacterium Tuberculosis Beijing Genotype Strains Associated with Resistance to Streptomycin: A Population-Based Study. PLoS ONE 2012, 7, e42323. [Google Scholar] [CrossRef]
- Jagielski, T.; Ignatowska, H.; Bakuła, Z.; Dziewit, Ł.; Napiórkowska, A.; Augustynowicz-Kopeć, E.; Zwolska, Z.; Bielecki, J. Screening for Streptomycin Resistance-Conferring Mutations in Mycobacterium Tuberculosis Clinical Isolates from Poland. PLoS ONE 2014, 9, e100078. [Google Scholar] [CrossRef]
- Bwalya, P.; Yamaguchi, T.; Solo, E.S.; Chizimu, J.Y.; Mbulo, G.; Nakajima, C.; Suzuki, Y. Characterization of Mutations Associated with Streptomycin Resistance in Multidrug-Resistant Mycobacterium Tuberculosis in Zambia. Antibiotics 2021, 10, 1169. [Google Scholar] [CrossRef]
- Cohen, K.A.; Abeel, T.; Manson McGuire, A.; Desjardins, C.A.; Munsamy, V.; Shea, T.P.; Walker, B.J.; Bantubani, N.; Almeida, D.V.; Alvarado, L.; et al. Evolution of Extensively Drug-Resistant Tuberculosis over Four Decades: Whole Genome Sequencing and Dating Analysis of Mycobacterium Tuberculosis Isolates from KwaZulu-Natal. PLoS Med. 2015, 12, e1001880. [Google Scholar] [CrossRef]
- Serio, A.W.; Magalhães, M.L.; Blanchard, J.S.; Connolly, L.E. Aminoglycosides: Mechanisms of Action and Resistance. In Antimicrobial Drug Resistance; Springer International Publishing: Cham, Switzerland, 2017; pp. 213–229. [Google Scholar] [CrossRef]
- Wimberly, B.T.; Brodersen, D.E.; Clemons, W.M.; Morgan-Warren, R.J.; Carter, A.P.; Vonrheln, C.; Hartsch, T.; Ramakrishnan, V. Structure of the 30S Ribosomal Subunit. Nature 2000, 407, 327–339. [Google Scholar] [CrossRef]
- Carter, A.P.; Clemons, W.M.; Brodersen, D.E.; Morgan-Warren, R.J.; Wimberly, B.T.; Ramakrishnan, V. Functional Insights from the Structure of the 30S Ribosomal Subunit and Its Interactions with Antibiotics. Nature 2000, 407, 340–348. [Google Scholar] [CrossRef]
- Finken, M.; Kirschner, P.; Meier, A.; Wrede, A.; Böttger, E.C. Molecular Basis of Streptomycin Resistance in Mycobacterium Tuberculosis: Alterations of the Ribosomal Protein S12 Gene and Point Mutations within a Functional 16S Ribosomal RNA Pseudoknot. Mol. Microbiol. 1993, 9, 1239–1246. [Google Scholar] [CrossRef]
- Honore, N.; Marchal, G.; Cole, S.T. Novel Mutation in 16S RRNA Associated with Streptomycin Dependence in Mycobacterium Tuberculosis. Antimicrob. Agents Chemother. 1995, 39, 769–770. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Okamoto, S.; Tamaru, A.; Nakajima, C.; Nishimura, K.; Tanaka, Y.; Tokuyama, S.; Suzuki, Y.; Ochi, K. Loss of a Conserved 7-Methylguanosine Modification in 16S RRNA Confers Low-Level Streptomycin Resistance in Bacteria. Mol. Microbiol. 2007, 63, 1096–1106. [Google Scholar] [CrossRef]
- Gygli, S.M.; Borrell, S.; Trauner, A.; Gagneux, S. Antimicrobial Resistance in Mycobacterium Tuberculosis: Mechanistic and Evolutionary Perspectives. FEMS Microbiol. Rev. 2017, 41, 354–373. [Google Scholar] [CrossRef] [PubMed]
- Spies, F.S.; Ribeiro, A.W.; Ramos, D.F.; Ribeiro, M.O.; Martin, A.; Carlos Palomino, J.; Lucia Rossetti, M.R.; Eduardo da Silva, P.A.; Zaha, A. Streptomycin Resistance and Lineage-Specific Polymorphisms in Mycobacterium Tuberculosis GidB Gene. J. Clin. Microbiol. 2011, 49, 2625–2630. [Google Scholar] [CrossRef] [PubMed]
- Mabhula, A.; Singh, V. Drug-Resistance in: Mycobacterium Tuberculosis: Where We Stand. Medchemcomm 2019, 10, 1342–1360. [Google Scholar] [CrossRef] [PubMed]
- Lai, Y.P.; Ioerger, T.R. Exploiting Homoplasy in Genome-Wide Association Studies to Enhance Identification of Antibiotic-Resistance Mutations in Bacterial Genomes. Evol. Bioinform. 2020, 16, 13–15. [Google Scholar] [CrossRef]
- Nonghanphithak, D.; Kaewprasert, O.; Chaiyachat, P.; Reechaipichitkul, W.; Chaiprasert, A.; Faksri, K. Whole-Genome Sequence Analysis and Comparisons between Drug-Resistance Mutations and Minimum Inhibitory Concentrations of Mycobacterium Tuberculosis Isolates Causing M/XDR-TB. PLoS ONE 2020, 15, e0244829. [Google Scholar] [CrossRef]
- Jnawali, H.N.; Yoo, H.; Ryoo, S.; Lee, K.J.; Kim, B.J.; Koh, W.J.; Kim, C.K.; Kim, H.J.; Park, Y.K. Molecular Genetics of Mycobacterium Tuberculosis Resistant to Aminoglycosides and Cyclic Peptide Capreomycin Antibiotics in Korea. World J. Microbiol. Biotechnol. 2013, 29, 975–982. [Google Scholar] [CrossRef]
- Feuerriegel, S.; Oberhauser, B.; George, A.G.; Dafae, F.; Richter, E.; Rüsch-Gerdes, S.; Niemann, S. Sequence Analysis for Detection of First-Line Drug Resistance in Mycobacterium Tuberculosis Strains from a High-Incidence Setting. BMC Microbiol. 2012, 12, 90. [Google Scholar] [CrossRef]
- Shrestha, D.; Maharjan, B.; Thida Oo, N.A.; Isoda, N.; Nakajima, C.; Suzuki, Y. Molecular Analysis of Streptomycin-Resistance Associating Genes in Mycobacterium Tuberculosis Isolates from Nepal. Tuberculosis 2020, 125, 101985. [Google Scholar] [CrossRef]
- Macedo, R.; Nunes, A.; Portugal, I.; Duarte, S.; Vieira, L.; Gomes, J.P. Dissecting Whole-Genome Sequencing-Based Online Tools for Predicting Resistance in Mycobacterium Tuberculosis: Can We Use Them for Clinical Decision Guidance? Tuberculosis 2018, 110, 44–51. [Google Scholar] [CrossRef]
- Miotto, P.; Tessema, B.; Tagliani, E.; Chindelevitch, L.; Starks, A.M.; Emerson, C.; Hanna, D.; Kim, P.S.; Liwski, R.; Zignol, M.; et al. A Standardised Method for Interpreting the Association between Mutations and Phenotypic Drug Resistance in Mycobacterium Tuberculosis. Eur. Respir. J. 2017, 50, 1701354. [Google Scholar] [CrossRef]
- Sun, H.; Zhang, C.; Xiang, L.; Pi, R.; Guo, Z.; Zheng, C.; Li, S.; Zhao, Y.; Tang, K.; Luo, M.; et al. Characterization of Mutations in Streptomycin-Resistant Mycobacterium Tuberculosis Isolates in Sichuan, China and the Association between Beijing-Lineage and Dual-Mutation in GidB. Tuberculosis 2016, 96, 102–106. [Google Scholar] [CrossRef] [PubMed]
- Tantivitayakul, P.; Ruangchai, W.; Juthayothin, T.; Smittipat, N.; Disratthakit, A.; Mahasirimongkol, S.; Viratyosin, W.; Tokunaga, K.; Palittapongarnpim, P. Homoplastic Single Nucleotide Polymorphisms Contributed to Phenotypic Diversity in Mycobacterium Tuberculosis. Sci. Rep. 2020, 10, 8024. [Google Scholar] [CrossRef]
- Gygli, S.M.; Keller, P.M.; Ballif, M.; Blöchliger, N.; Hömke, R.; Reinhard, M.; Loiseau, C.; Ritter, C.; Sander, P.; Borrell, S.; et al. Whole-Genome Sequencing for Drug Resistance Profile Prediction in Mycobacterium Tuberculosis. Antimicrob. Agents Chemother. 2019, 63, e02175-18. [Google Scholar] [CrossRef] [PubMed]
- Desjardins, C.A.; Cohen, K.A.; Munsamy, V.; Abeel, T.; Maharaj, K.; Walker, B.J.; Shea, T.P.; Almeida, D.V.; Manson, A.L.; Salazar, A.; et al. Genomic and Functional Analyses of Mycobacterium Tuberculosis Strains Implicate Ald in D-Cycloserine Resistance. Nat. Genet. 2016, 48, 544–551. [Google Scholar] [CrossRef] [PubMed]
- Eldholm, V.; Monteserin, J.; Rieux, A.; Lopez, B.; Sobkowiak, B.; Ritacco, V.; Balloux, F. Four Decades of Transmission of a Multidrug-Resistant Mycobacterium Tuberculosis Outbreak Strain. Nat. Commun. 2015, 6, 7119. [Google Scholar] [CrossRef] [PubMed]
- Rodríguez-García, Á.; Mares-Alejandre, R.E.; Muñoz-Muñoz, P.L.A.; Ruvalcaba-Ruiz, S.; González-Sánchez, R.A.; Bernáldez-Sarabia, J.; Meléndez-López, S.G.; Licea-Navarro, A.F.; Ramos-Ibarra, M.A. Molecular Analysis of Streptomycin Resistance Genes in Clinical Strains of Mycobacterium Tuberculosis and Biocomputational Analysis of the MtGidB L101F Variant. Antibiotics 2021, 10, 807. [Google Scholar] [CrossRef]
- Doddam, S.N.; Peddireddy, V.; Yerra, P.; Sai Arun, P.P.; Qaria, M.A.; Baddam, R.; Sarker, N.; Ahmed, N. Mycobacterium Tuberculosis DosR Regulon Gene Rv2004c Contributes to Streptomycin Resistance and Intracellular Survival. Int. J. Med. Microbiol. 2019, 309, 151353. [Google Scholar] [CrossRef] [PubMed]
- Arriaga-Guerrero, A.L.; Hernández-Luna, C.E.; Rigal-Leal, J.; Robles-González, R.J.; González-Escalante, L.A.; Silva-Ramírez, B.; Mercado-Hernández, R.; Vargas-Villarreal, J.; Bermúdez De León, M.; Peñuelas-Urquides, K. LipF Increases Rifampicin and Streptomycin Sensitivity in a Mycobacterium Tuberculosis Surrogate. BMC Microbiol. 2020, 20, 132. [Google Scholar] [CrossRef]
- Nzungize, L.; Ali, M.K.; Wang, X.; Huang, X.; Yang, W.; Duan, X.; Yan, S.; Li, C.; Abdalla, A.E.; Jeyakkumar, P.; et al. Mycobacterium Tuberculosis MetC (Rv3340) Derived Hydrogen Sulphide Conferring Bacteria Stress Survival. J. Drug Target. 2019, 27, 1004–1016. [Google Scholar] [CrossRef] [PubMed]
- Reeves, A.Z.; Campbell, P.J.; Sultana, R.; Malik, S.; Murray, M.; Plikaytis, B.B.; Shinnick, T.M.; Posey, J.E. Aminoglycoside Cross-Resistance in Mycobacterium Tuberculosis Due to Mutations in the 5’ Untranslated Region of WhiB7. Antimicrob. Agents Chemother. 2013, 57, 1857–1865. [Google Scholar] [CrossRef]
- Geiman, D.E.; Raghunand, T.R.; Agarwal, N.; Bishai, W.R. Differential Gene Expression in Response to Exposure to Antimycobacterial Agents and Other Stress Conditions among Seven Mycobacterium Tuberculosis WhiB-like Genes. Antimicrob. Agents Chemother. 2006, 50, 2836–2841. [Google Scholar] [CrossRef] [PubMed]
- Burian, J.; Ramón-García, S.; Sweet, G.; Gómez-Velasco, A.; Av-Gay, Y.; Thompson, C.J. The Mycobacterial Transcriptional Regulator WhiB7 Gene Links Redox Homeostasis and Intrinsic Antibiotic Resistance. J. Biol. Chem. 2012, 287, 299–310. [Google Scholar] [CrossRef] [PubMed]
- Sharma, D.; Bisht, D. Secretory Proteome Analysis of Streptomycin-Resistant Mycobacterium Tuberculosis Clinical Isolates. SLAS Discov. 2017, 22, 1229–1238. [Google Scholar] [CrossRef]
- Hicks, N.D.; Yang, J.; Zhang, X.; Zhao, B.; Grad, Y.H.; Liu, L.; Ou, X.; Chang, Z.; Xia, H.; Zhou, Y.; et al. Clinically Prevalent Mutations in Mycobacterium Tuberculosis Alter Propionate Metabolism and Mediate Multidrug Tolerance. Nat. Microbiol. 2018, 3, 1032–1042. [Google Scholar] [CrossRef]
- Beltran, C.G.G.; Heunis, T.; Gallant, J.; Venter, R.; du Plessis, N.; Loxton, A.G.; Trost, M.; Winter, J.; Malherbe, S.T.; Kana, B.D.; et al. Investigating Non-Sterilizing Cure in TB Patients at the End of Successful Anti-TB Therapy. Front. Cell. Infect. Microbiol. 2020, 10, 443. [Google Scholar] [CrossRef]
- Castro, R.A.D.; Borrell, S.; Gagneux, S. The Within-Host Evolution of Antimicrobial Resistance in Mycobacterium Tuberculosis. FEMS Microbiol. Rev. 2020, 45, fuaa071. [Google Scholar] [CrossRef]
- Goossens, S.N.; Sampson, S.L.; Van Rie, A. Mechanisms of Drug-Induced Tolerance in Mycobacterium Tuberculosis. Clin. Microbiol. Rev. 2020, 34, e00141-20. [Google Scholar] [CrossRef]
- Pisu, D.; Provvedi, R.; Espinosa, D.M.; Payan, J.B.; Boldrin, F.; Palù, G.; Hernandez-Pando, R.; Manganelli, R. The Alternative Sigma Factors SigE and SigB Are Involved in Tolerance and Persistence to Antitubercular Drugs. Antimicrob. Agents Chemother. 2017, 61, e01596-17. [Google Scholar] [CrossRef] [PubMed]
- Lei, J.; Zhang, H.; Wu, C.; Wang, X.; Yang, Y.; Zhang, X.; Huang, Y.; Wang, H. The Influence of Mycobacterium Tuberculosis Sigma Factors on the Promotion Efficiency of PtpAt Promoter in Mycobacterium Smegmatis. Curr. Microbiol. 2005, 51, 141–147. [Google Scholar] [CrossRef]
- Demaio, J.; Zhang, Y.; Ko, C.; Young, D.B.; Bishai, W.R. A Stationary-Phase Stress-Response Sigma Factor from Mycobacterium Tuberculosis. Proc. Natl. Acad. Sci. USA 1996, 93, 2790–2794. [Google Scholar] [CrossRef]
- Keren, I.; Minami, S.; Rubin, E.; Lewis, K. Characterization and Transcriptome Analysis of Mycobacterium Tuberculosis Persisters. MBio 2011, 2, 3–12. [Google Scholar] [CrossRef]
- Sala, A.; Bordes, P.; Genevaux, P. Multiple Toxin-Antitoxin Systems in Mycobacterium Tuberculosis. Toxins 2014, 6, 2. [Google Scholar] [CrossRef] [PubMed]
- Talwar, S.; Pandey, M.; Sharma, C.; Kutum, R.; Lum, J.; Carbajo, D.; Goel, R.; Poidinger, M.; Dash, D.; Singhal, A.; et al. Role of VapBC12 Toxin-Antitoxin Locus in Cholesterol-Induced Mycobacterial Persistence. mSystems 2020, 5, e00855-20. [Google Scholar] [CrossRef] [PubMed]
- Maisonneuve, E.; Shakespeare, L.J.; Jørgensen, M.G.; Gerdes, K. Bacterial Persistence by RNA Endonucleases. Proc. Natl. Acad. Sci. USA 2011, 108, 13206–13211. [Google Scholar] [CrossRef]
- Roy, M.; Bose, M.; Bankoti, K.; Kundu, A.; Dhara, S.; Das, A.K. Biochemical Characterization of VapC46 Toxin from Mycobacterium Tuberculosis. Mol. Biotechnol. 2020, 62, 335–343. [Google Scholar] [CrossRef]
- Gupta, A.; Venkataraman, B.; Vasudevan, M.; Gopinath Bankar, K. Co-Expression Network Analysis of Toxin-Antitoxin Loci in Mycobacterium Tuberculosis Reveals Key Modulators of Cellular Stress. Sci. Rep. 2017, 7, 5868. [Google Scholar] [CrossRef] [PubMed]
- Bellerose, M.M.; Baek, S.-H.; Huang, C.-C.; Moss, C.E.; Koh, E.-I.; Proulx, M.K.; Smith, C.M.; Baker, R.E.; Lee, J.S.; Eum, S.; et al. Common Variants in the Glycerol Kinase Gene Reduce Tuberculosis Drug Efficacy. MBio 2019, 10, 1–15. [Google Scholar] [CrossRef] [PubMed]
- Safi, H.; Gopal, P.; Lingaraju, S.; Ma, S.; Levine, C.; Dartois, V.; Yee, M.; Li, L.; Blanc, L.; Liang, H.P.H.; et al. Phase Variation in Mycobacterium Tuberculosis GlpK Produces Transiently Heritable Drug Tolerance. Proc. Natl. Acad. Sci. USA 2019, 116, 19665–19674. [Google Scholar] [CrossRef] [PubMed]
- Vargas, R.; Farhat, M.R. Antibiotic Treatment and Selection for GlpK Mutations in Patients with Active Tuberculosis Disease. Proc. Natl. Acad. Sci. USA 2020, 117, 3910–3912. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.J.; Lee, S.-K.; Song, N.; Nathan, T.O.; Swarts, B.M.; Eum, S.-Y.; Ehrt, S.; Cho, S.-N.; Eoh, H. Transient Drug-Tolerance and Permanent Drug-Resistance Rely on the Trehalose-Catalytic Shift in Mycobacterium Tuberculosis. Nat. Commun. 2019, 10, 2928. [Google Scholar] [CrossRef]
- Gopal, P.; Sarathy, J.P.; Yee, M.; Ragunathan, P.; Shin, J.; Bhushan, S.; Zhu, J.; Akopian, T.; Kandror, O.; Lim, T.K.; et al. Pyrazinamide Triggers Degradation of Its Target Aspartate Decarboxylase. Nat. Commun. 2020, 11, 1661. [Google Scholar] [CrossRef]
- Safi, H.; Sherman, D.R.; Dick, T.; Alland, D. Mycobacterium Tuberculosis GlpK Mutants in Human Tuberculosis. Proc. Natl. Acad. Sci. USA 2020, 117, 3913–3914. [Google Scholar] [CrossRef]
- Bouziane, F.; Allem, R.; Sebaihia, M.; Kumanski, S.; Mougari, F.; Sougakoff, W.; Raskine, L.; Yala, D.; Cambau, E. First Genetic Characterisation of Multidrug-Resistant Mycobacterium Tuberculosis Isolates from Algeria. J. Glob. Antimicrob. Resist. 2019, 19, 301–307. [Google Scholar] [CrossRef]
- Durão, P.; Ramiro, R.S.; Pereira, C.; Jurič, J.; Pereira, D.; Gordo, I. Radial Expansion Facilitates the Maintenance of Double Antibiotic Resistances. Antimicrob. Agents Chemother. 2020, 64, e00668-20. [Google Scholar] [CrossRef]
- Moura de Sousa, J.; Balbontín, R.; Durão, P.; Gordo, I. Multidrug-Resistant Bacteria Compensate for the Epistasis between Resistances. PLoS Biol. 2017, 15, e2001741. [Google Scholar] [CrossRef]
- Wong, A. Epistasis and the Evolution of Antimicrobial Resistance. Front. Microbiol. 2017, 8, 246. [Google Scholar] [CrossRef]
- Durão, P.; Balbontín, R.; Gordo, I. Evolutionary Mechanisms Shaping the Maintenance of Antibiotic Resistance. Trends Microbiol. 2018, 26, 677–691. [Google Scholar] [CrossRef] [PubMed]
- Sun, H.; Zeng, J.; Li, S.; Liang, P.; Zheng, C.; Liu, Y.; Luo, T.; Rastogi, N.; Sun, Q. Interaction between RpsL and GyrA Mutations Affects the Fitness and Dual Resistance of Mycobacterium Tuberculosis Clinical Isolates against Streptomycin and Fluoroquinolones. Infect. Drug Resist. 2018, 11, 431–440. [Google Scholar] [CrossRef] [PubMed]
- Islam, M.M.; Tan, Y.; Hameed, H.M.A.; Chhotaray, C.; Liu, Z.; Liu, Y.; Lu, Z.; Wang, S.; Cai, X.; Gao, Y.; et al. Phenotypic and Genotypic Characterization of Streptomycin-Resistant Multidrug-Resistant Mycobacterium Tuberculosis Clinical Isolates in Southern China. Microb. Drug Resist. 2020, 26, 766–775. [Google Scholar] [CrossRef]
- Cohen, K.A.; Stott, K.E.; Manson, A.L.; Munsamy, V.; Earl, A.M.; Pym, A.S. Untapped Potential for Streptomycin: Using Genomics to Optimize Aminoglycoside Selection in Drug-Resistant Mycobacterium Tuberculosis. Am. J. Respir. Crit. Care Med. 2018, 197, A1552. [Google Scholar]
- Ismail, N.A.; Mvusi, L.; Nanoo, A.; Dreyer, A.; Omar, S.V.; Babatunde, S.; Molebatsi, T.; van der Walt, M.; Adelekan, A.; Deyde, V.; et al. Prevalence of Drug-Resistant Tuberculosis and Imputed Burden in South Africa: A National and Sub-National Cross-Sectional Survey. Lancet Infect. Dis. 2018, 18, 779–787. [Google Scholar] [CrossRef]
- Mesfin, E.A.; Beyene, D.; Tesfaye, A.; Admasu, A.; Addise, D.; Amare, M.; Dagne, B.; Yaregal, Z.; Tesfaye, E.; Tessema, B. Drug-Resistance Patterns of Mycobacterium Tuberculosis Strains and Associated Risk Factors among Multi Drug-Resistant Tuberculosis Suspected Patients from Ethiopia. PLoS ONE 2018, 13, e0197737. [Google Scholar] [CrossRef]
- Knopp, M.; Andersson, D.I. Predictable Phenotypes of Antibiotic Resistance Mutations. MBio 2018, 9, e00770-18. [Google Scholar] [CrossRef]
- Zhou, Y.; Van Den Hof, S.; Wang, S.; Pang, Y.; Zhao, B.; Xia, H.; Anthony, R.; Ou, X.; Li, Q.; Zheng, Y.; et al. Association between Genotype and Drug Resistance Profiles of Mycobacterium Tuberculosis Strains Circulating in China in a National Drug Resistance Survey. PLoS ONE 2017, 12, e0174197. [Google Scholar] [CrossRef]
- Vargas, R.; Freschi, L.; Spitaleri, A.; Tahseen, S.; Barilar, I.; Niemann, S.; Miotto, P.; Cirillo, D.M.; Köser, C.U.; Farhat, M.R. The Role of Epistasis in Amikacin, Kanamycin, Bedaquiline, and Clofazimine Resistance in Mycobacterium Tuberculosis Complex. Antimicrob. Agents Chemother. 2021, 65, AAC0116421. [Google Scholar] [CrossRef]
- Georghiou, S.B.; Magana, M.; Garfein, R.S.; Catanzaro, D.G.; Catanzaro, A.; Rodwell, T.C. Evaluation of Genetic Mutations Associated with Mycobacterium Tuberculosis Resistance to Amikacin, Kanamycin and Capreomycin: A Systematic Review. PLoS ONE 2012, 7, e33275. [Google Scholar] [CrossRef]
- Alame Emane, A.K.; Guo, X.; Takiff, H.E.; Liu, S. Drug Resistance, Fitness and Compensatory Mutations in Mycobacterium Tuberculosis. Tuberculosis 2021, 129, 102091. [Google Scholar] [CrossRef]
- Cohen, T.; Sommers, B.; Murray, M. The Effect of Drug Resistance on the Fitness of Mycobacterium Tuberculosis. Lancet Infect. Dis. 2003, 3, 13–21. [Google Scholar] [CrossRef]
- Alame Emane, A.K.; Guo, X.; Takiff, H.E.; Liu, S. Highly Transmitted M. Tuberculosis Strains Are More Likely to Evolve MDR/XDR and Cause Outbreaks, but What Makes Them Highly Transmitted? Tuberculosis 2021, 129, 102092. [Google Scholar] [CrossRef] [PubMed]
- Zhan, L.; Wang, J.; Wang, L.; Qin, C. The Correlation of Drug Resistance and Virulence in Mycobacterium Tuberculosis. Biosaf. Health 2020, 2, 18–24. [Google Scholar] [CrossRef]
- Spagnolo, F.; Dykhuizen, D. Antibiotic Resistance Increases Evolvability and Maximizes Opportunities Across Fitness Landscapes. bioRxiv 2019, 750729. [Google Scholar] [CrossRef]
- De Keijzer, J.; Mulder, A.; De Beer, J.; De Ru, A.H.; Van Veelen, P.A.; Van Soolingen, D. Mechanisms of Phenotypic Rifampicin Tolerance in Mycobacterium Tuberculosis Beijing Genotype Strain B0/W148 Revealed by Proteomics. J. Proteome Res. 2016, 15, 1194–1204. [Google Scholar] [CrossRef] [PubMed]
- Muzondiwa, D.; Hlanze, H.; Reva, O.N. The Epistatic Landscape of Antibiotic Resistance of Different Clades of Mycobacterium Tuberculosis. Antibiotics 2021, 10, 857. [Google Scholar] [CrossRef]
- Pečerska, J.; Kühnert, D.; Meehan, C.J.; Coscollá, M.; de Jong, B.C.; Gagneux, S.; Stadler, T. Quantifying Transmission Fitness Costs of Multi-Drug Resistant Tuberculosis. Epidemics 2021, 21, 100471. [Google Scholar] [CrossRef]
- Loiseau, C.; Brites, D.; Reinhard, M.; Zürcher, K.; Borrell, S.; Ballif, M.; Fenner, L.; Cox, H.; Rutaihwa, L.K.; Wilkinson, R.J.; et al. HIV Coinfection Is Associated with Low-Fitness RpoB Variants in Rifampicin-Resistant Mycobacterium Tuberculosis. Antimicrob. Agents Chemother. 2020, 64, e00782-20. [Google Scholar] [CrossRef]
- Castro, R.A.D.; Ross, A.; Kamwela, L.; Reinhard, M.; Loiseau, C.; Feldmann, J.; Borrell, S.; Trauner, A.; Gagneux, S. The Genetic Background Modulates the Evolution of Fluoroquinolone-Resistance in Mycobacterium Tuberculosis. Mol. Biol. Evol. 2020, 37, 195–207. [Google Scholar] [CrossRef]
- Degiacomi, G.; Sammartino, J.C.; Sinigiani, V.; Marra, P.; Urbani, A.; Pasca, M.R. In Vitro Study of Bedaquiline Resistance in Mycobacterium Tuberculosis Multi-Drug Resistant Clinical Isolates. Front. Microbiol. 2020, 11, 2290. [Google Scholar] [CrossRef]
- Davies, A.P.; Billington, O.J.; Bannister, B.A.; Weir, W.R.C.; McHugh, T.D.; Gillespie, S.H. Comparison of Fitness of Two Isolates of Mycobacterium Tuberculosis, One of Which Had Developed Mulit-Drug Resistance during the Course of Treatment. J. Infect. 2000, 41, 184–187. [Google Scholar] [CrossRef] [PubMed]
- Gagneux, S.; Long, C.D.; Small, P.M.; Van, T.; Schoolnik, G.K.; Bohannan, B.J.M. The Competitive Cost of Antibiotic Resistance in Mycobacterium Tuberculosis. Science 2006, 312, 1944–1946. [Google Scholar] [CrossRef] [PubMed]
- Mariam, D.H.; Mengistu, Y.; Hoffner, S.E.; Andersson, D.I. Effect of RpoB Mutations Conferring Rifampin Resistance on Fitness of Mycobacterium Tuberculosis. Antimicrob. Agents Chemother. 2004, 48, 1289–1294. [Google Scholar] [CrossRef] [PubMed]

| Gene Name | Polymorphism (Nucleotide or Amino Acid Change) | Suggested Reference |
|---|---|---|
| rpsL | Lys88Arg, Lys43Arg | [59] |
| Arg86Pro Arg86Tyr, Arg9His, Gly84Val, Lys43Thr, Lys51Asn, Lys88Gln, Lys88Met, Thr40Ile, Thr41Ser, Val52Gly, Val87Leu, Val93Met | [60] | |
| Arg86Gly | [30] | |
| Gly118Asp | [55] | |
| rrs | 190G/A, 277G/C, 427G/C, 462C/T, 513C/T, 514A/C, 516C/T, 517C/T, 628G/C, 799C/T888G/A, 905C/A, 905C/G, 906A/G, 907A/C, 907A/T, 908A/G | [60] |
| 644A/G | [55] | |
| gid | 102del, 103_104insG, 107del, 115del, 136del, 157del, 225_226insT, 294_295insAC, 297_298insA, 326del, 351del352_353insG, 366_367del, 386del, 390del, 400_401insT, 446_447insA, 450del, 452del, 455del, 471del, 519_520insA, 554_555insG, 559_572del, 58_59insT, 601del | [61] |
| Leu108Arg, Leu35fs *, Ala200Glu, Cys191Arg, Gly73Glu, Cys191Arg, Gly73Glu, Leu50Arg, Glu60fs *, Arg39fs *, Arg118fs *, Arg217Gly, Leu94Pro, Asp67Gly, Pro84Leu, Gly73Ala, Leu145Phe, Val77Gly, Val135fs *, Ser149Arg, Leu90Phe, Gly214Ala, Ala119Thr; Ala19Ser, Arg158Leu, Val66Leu, Arg137Gln, Ala134Glu, Ala138Val | [61] | |
| 259C/T | [59] | |
| Pro75Thr | ||
| Arg47Gln, Pro84Ser, Met104dup, Gly117Arg, Lys163Asn, Ile11Thr, Cys119Trp, Cys191Phe, Ser70Arg, Ala141Glu | [23] | |
| Ala80Pro, Tyr195His | [21] | |
| His168Tyr, Gly208Val | [30] | |
| Ala19Gly | [55] | |
| Met218Val | [55] | |
| Val110fs | [62] | |
| Gly13Arg, Leu101Phe | [63] | |
| 243insGC, 112delC, 254delA, 347delG, 372delG | [55] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Rocha, D.M.G.C.; Viveiros, M.; Saraiva, M.; Osório, N.S. The Neglected Contribution of Streptomycin to the Tuberculosis Drug Resistance Problem. Genes 2021, 12, 2003. https://doi.org/10.3390/genes12122003
Rocha DMGC, Viveiros M, Saraiva M, Osório NS. The Neglected Contribution of Streptomycin to the Tuberculosis Drug Resistance Problem. Genes. 2021; 12(12):2003. https://doi.org/10.3390/genes12122003
Chicago/Turabian StyleRocha, Deisy M. G. C., Miguel Viveiros, Margarida Saraiva, and Nuno S. Osório. 2021. "The Neglected Contribution of Streptomycin to the Tuberculosis Drug Resistance Problem" Genes 12, no. 12: 2003. https://doi.org/10.3390/genes12122003
APA StyleRocha, D. M. G. C., Viveiros, M., Saraiva, M., & Osório, N. S. (2021). The Neglected Contribution of Streptomycin to the Tuberculosis Drug Resistance Problem. Genes, 12(12), 2003. https://doi.org/10.3390/genes12122003







