Research on Potential Network Markers and Signaling Pathways in Type 2 Diabetes Based on Conditional Cell-Specific Network
Abstract
:1. Introduction
- We build a cell-specific direct correlation network from the perspective of gene association based on CCSN, which fundamentally reduces large noise and instability.
- Based on CCSN, hub genes with high network correlation in CCSN and pathways related to T2D are identified, which can provide more reliable biomarkers for T2D prognosis and treatment.
- By CCSN method, we are able to uncover ‘dark’ genes, which are non-differential in gene expression but sensitive to the degree value. These genes cannot be found by traditional differential analysis methods and may play important roles in network regulation of T2D. GO and KEGG functional enrichment analysis show that some ‘dark’ genes are involved in T2D-related pathways and might be potential biomarkers for T2D and its complications.
2. Materials and Methods
2.1. Data Pre-Processing
2.2. Construction of CCSN
2.3. Identification of Hub Genes Based on CCSN
2.4. Cell–Cell Communication
2.5. ‘Dark’ Genes Revealed by CNDM
3. Results
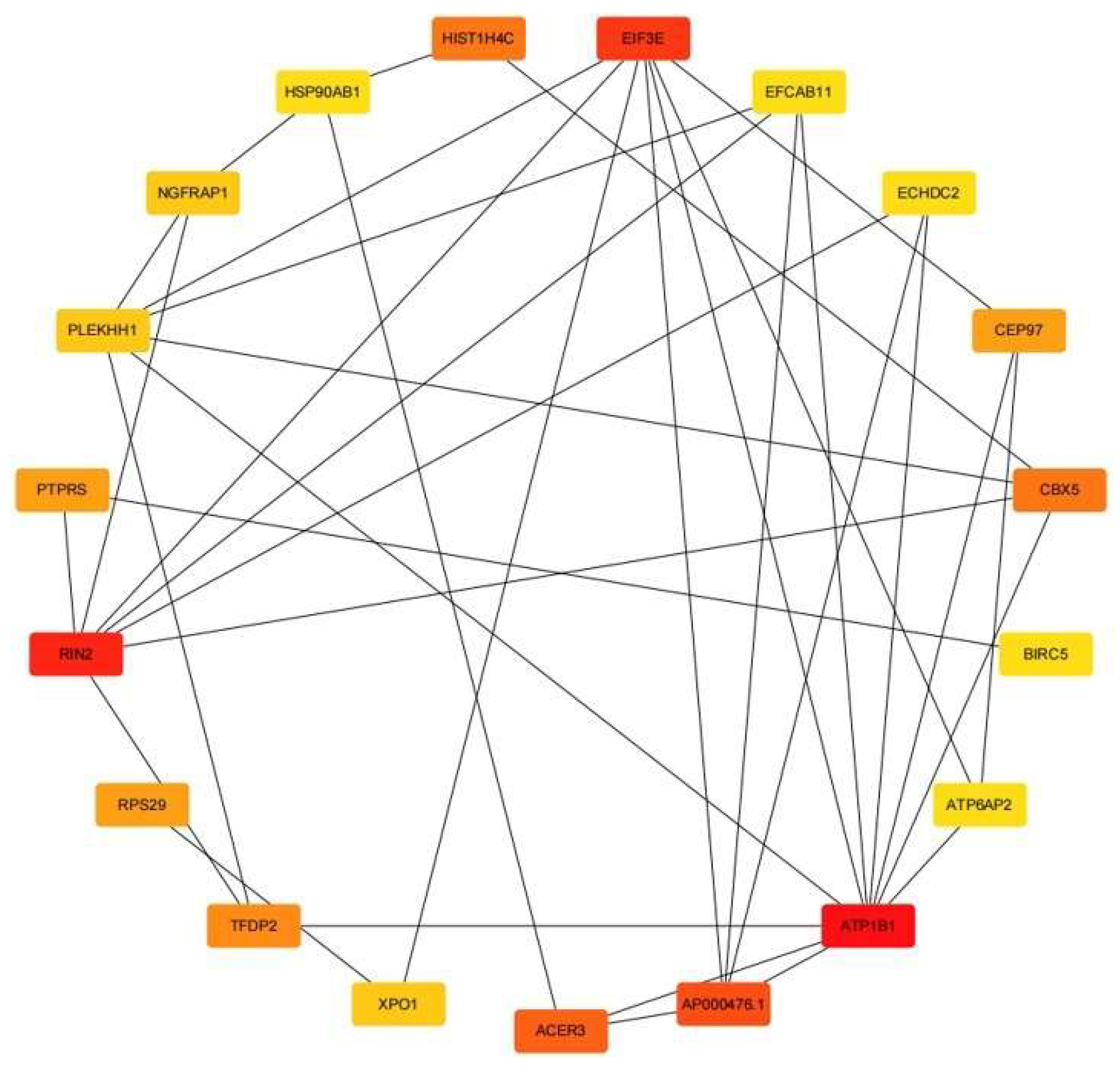
3.1. Hub Gene Analysis
3.2. Cell–Cell Communication Analysis
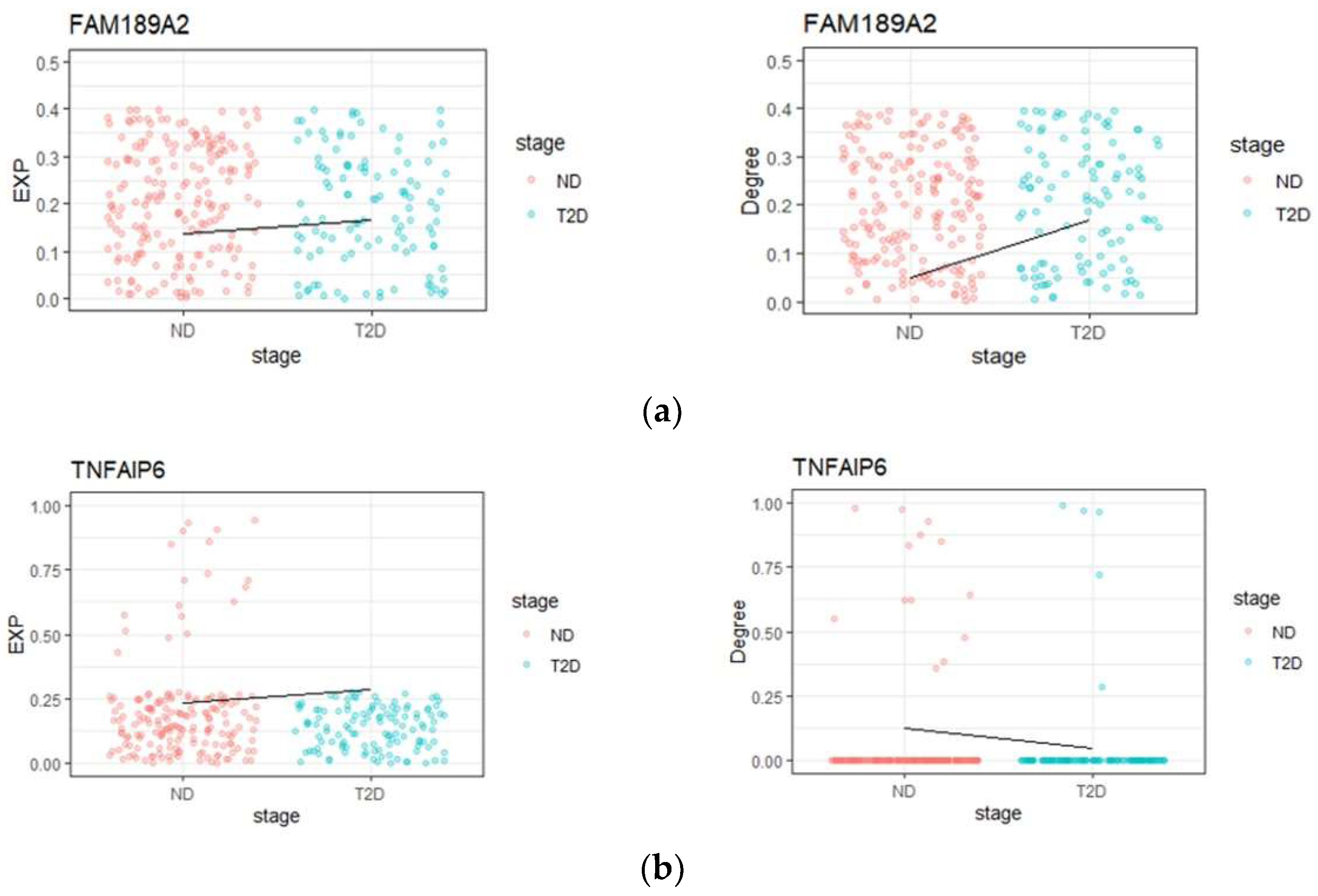
3.3. ‘Dark’ Gene Analysis
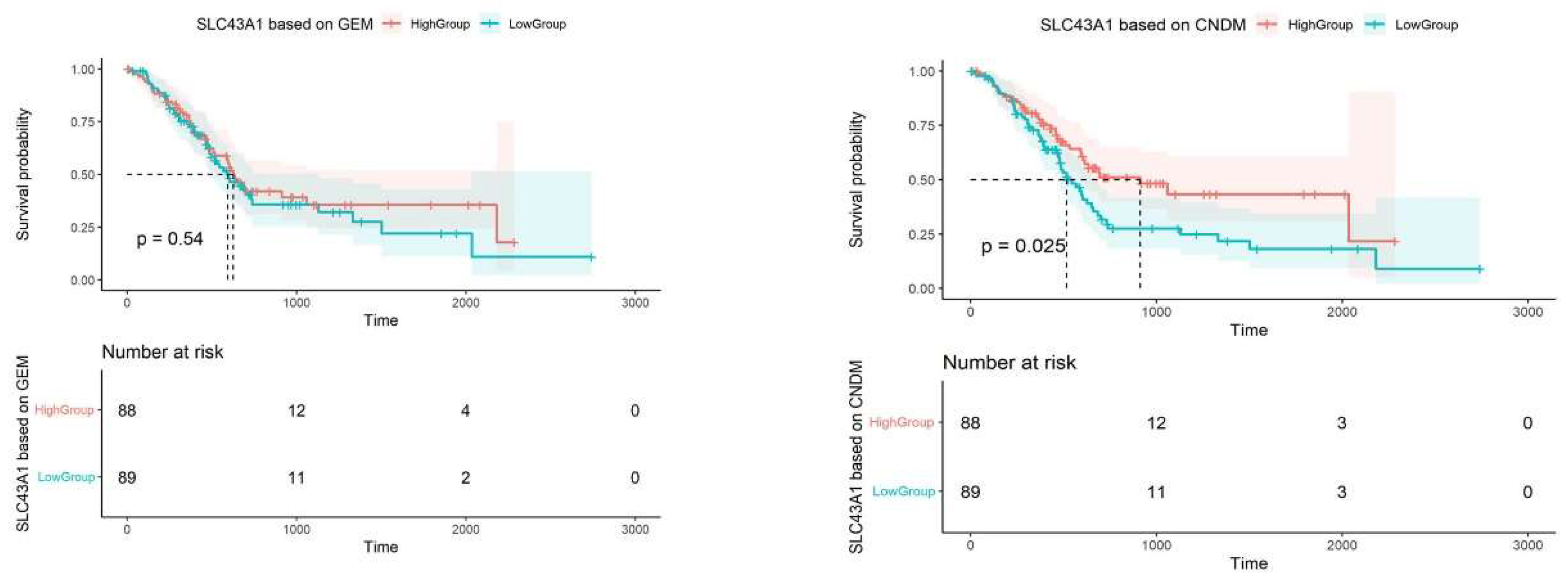
3.3.1. Prognostic Analysis of ‘Dark’ Genes
3.3.2. ‘Dark’ Gene Pathway Analysis
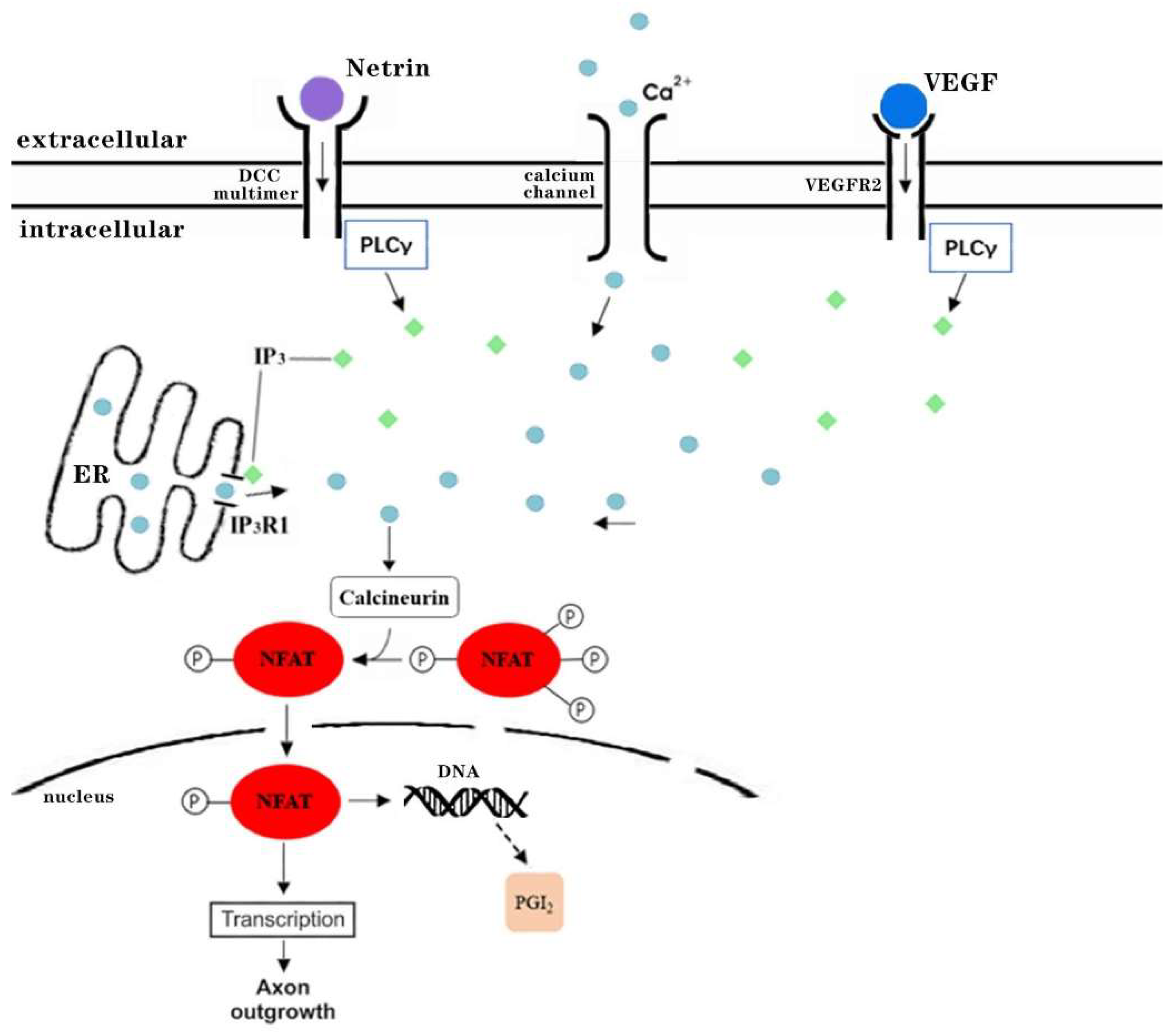
3.4. The Underlying Signaling Mechanisms Revealed by ‘Dark’ Genes NFATC2 and UNC5D
3.4.1. ‘Dark’ Gene NFATC2 Is a Key Transcription Factor of Axon Guidance and VEGF Signaling Pathway Related to Diabetes Complications
3.4.2. ‘Dark’ Gene UNC5D Is Involved in a Potential Signaling Pathway in the Development of Diabetes-Related Complications Axon Guidance
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Jin, B.; Liu, R.; Hao, S.Y.; Li, Z.; Zhu, C.Q.; Zhou, X.; Chen, P.; Fu, T.Y.; Hu, Z.K.; Wu, Q.; et al. Defining and characterizing the critical transition state prior to the type 2 diabetes disease. PLoS ONE 2017, 12, e0180937. [Google Scholar] [CrossRef] [PubMed]
- Grün, D.; Lyubimova, A.; Kester, L.; Wiebrands, K.; Basak, O.; Sasaki, N.; Clevers, H.; van Oudenaarden, A. Single-cell messenger RNA sequencing reveals rare intestinal cell types. Nature 2015, 525, 251–255. [Google Scholar] [CrossRef] [PubMed]
- Shalek, A.K.; Satija, R.; Adiconis, X.; Gertner, R.S.; Gaublomme, J.T.; Raychowdhury, R.; Schwartz, S.; Yosef, N.; Malboeuf, C.; Lu, D.; et al. Single-cell transcriptomics reveals bimodality in expression and splicing in immune cells. Nature 2013, 498, 236–240. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, J.; Klughammer, J.; Farlik, M.; Penz, T.; Spittler, A.; Barbieux, C.; Berishvili, E.; Bock, C.; Kubicek, S. Single-cell transcriptomes reveal characteristic features of human pancreatic islet cell types. EMBO Rep. 2015, 17, 178–187. [Google Scholar] [CrossRef]
- Bao, K.X.; Cui, Z.C.; Wang, H.; Xiao, H.; Li, T.; Kong, X.X.; Liu, T.M. Pseudotime Ordering Single-Cell Transcriptomic of β Cells Pancreatic Islets in Health and Type 2 Diabetes. Phenomics 2021, 1, 199–210. [Google Scholar] [CrossRef]
- Lawlor, N.; George, J.; Bolisetty, M.; Kursawe, R.; Sun, L.; Sivakamasundari, V.; Kycia, I.; Robson, P.; Stitzel, M.L. Single-cell transcriptomes identify human islet cell signatures and reveal cell-type–specific expression changes in type 2 diabetes. Genome Res. 2017, 27, 208–222. [Google Scholar] [CrossRef]
- Segerstolpe, Å.; Palasantza, A.; Eliasson, P.; Andersson, E.M.; Andréasson, A.C.; Sun, X.; Picelli, S.; Sabirsh, A.; Clausen, M.; Bjursell, M.K.; et al. Single-Cell Transcriptome Profiling of Human Pancreatic Islets in Health and Type 2 Diabetes. Cell Metab. 2016, 24, 593–607. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, L.; Dai, H.; Fang, Z.; Chen, L. c-CSN: Single-cell RNA Sequencing Data Analysis by Conditional Cell-specific Network. Genom. Proteom. Bioinform. 2021, 19, 319–329. [Google Scholar] [CrossRef]
- Yang, Y.; Huh, R.; Culpepper, H.W.; Lin, Y.; Love, M.I.; Li, Y. SAFE-clustering: Single-cell Aggregated (from Ensemble) clustering for single-cell RNA-seq data. Bioinformatics 2019, 35, 1269–1277. [Google Scholar] [CrossRef] [PubMed]
- Lin, Y.; Ghazanfar, S.; Strbenac, D.; Wang, A.; Patrick, E.; Lin, D.M.; Speed, T.; Yang, J.Y.H.; Yang, P. Evaluating stably expressed genes in single cells. GigaScience 2019, 8, giz106. [Google Scholar] [CrossRef] [Green Version]
- Dai, H.; Li, L.; Zeng, T.; Chen, L. Cell-specific network constructed by single-cell RNA sequencing data. Nucleic Acids Res. 2019, 47, e62. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Efremova, M.; Vento-Tormo, M.; Teichmann, S.A.; Vento-Tormo, R. CellPhoneDB: Inferring cell–cell communication from combined expression of multi-subunit ligand–receptor complexes. Nat. Protoc. 2020, 15, 1484–1506. [Google Scholar] [CrossRef] [PubMed]
- Chawla, T.; Sharma, D.; Singh, A. Role of the renin angiotensin system in diabetic nephropathy. World J. Diabetes 2010, 1, 141–145. [Google Scholar] [CrossRef] [PubMed]
- Binger, K.J.; Neukam, M.; Tattikota, S.G.; Qadri, F.; Puchkov, D.; Willmes, D.M.; Wurmsee, S.; Geisberger, S.; Dechend, R.; Raile, K.; et al. Atp6ap2 deletion causes extensive vacuolation that consumes the insulin content of pancreatic β cells. Proc. Natl. Acad. Sci. USA 2019, 116, 19983–19988. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pastagia, M.; Jenkins, S.G. SalmonellaNeck Abscess as an Opportunistic Infection in Diabetes Mellitus. Case Rep. Infect. Dis. 2013, 2013, 708419. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tornatore, L.; Thotakura, A.K.; Bennett, J.; Moretti, M.; Franzoso, G. The nuclear factor kappa B signaling pathway: Integrating metabolism with inflammation. Trends Cell Biol. 2012, 22, 557–566. [Google Scholar] [CrossRef]
- Jing, E.; Sundararajan, P.; Majumdar, I.D.; Hazarika, S.; Fowler, S.; Szeto, A.; Gesta, S.; Mendez, A.J.; Vishnudas, V.K.; Sarangarajan, R.; et al. Hsp90β knockdown in DIO mice reverses insulin resistance and improves glucose tolerance. Nutr. Metab. 2018, 15, 11. [Google Scholar] [CrossRef] [Green Version]
- Sokhi, J.; Sikka, R.; Raina, P.; Kaur, R.; Matharoo, K.; Arora, P.; Bhanwer, A. Association of genetic variants in INS (rs689), INSR (rs1799816) and PP1G.G (rs1799999) with type 2 diabetes (T2D): A case–control study in three ethnic groups from North-West India. Mol. Genet. Genom. 2016, 291, 205–216. [Google Scholar] [CrossRef]
- Fernández-de Frutos, M.; Galán-Chilet, I.; Goedeke, L.; Kim, B.; Pardo-Marqués, V.; Pérez-García, A.; Herrero, J.I.; Fernández-Hernando, C.; Kim, J.; Ramírez, C.M. MicroRNA 7 Impairs Insulin Signaling and Regulates Aβ Levels through Posttranscriptional Regulation of the Insulin Receptor Substrate 2, Insulin Receptor, Insulin-Degrading Enzyme, and Liver X Receptor Pathway. Mol. Cell. Biol. 2019, 39, e00170-19. [Google Scholar] [CrossRef]
- Ben, Q.; Xu, M.; Ning, X.; Liu, J.; Hong, S.; Huang, W.; Zhang, H.; Li, Z. Diabetes mellitus and risk of pancreatic cancer: A meta-analysis of cohort studies. Eur. J. Cancer 2011, 47, 1928–1937. [Google Scholar] [CrossRef]
- Mayers, J.R.; Wu, C.; Clish, C.B.; Kraft, P.; Torrence, M.E.; Fiske, B.P.; Yuan, C.; Bao, Y.; Townsend, M.K.; Tworoger, S.S.; et al. Elevation of circulating branched-chain amino acids is an early event in human pancreatic adenocarcinoma development. Nat. Med. 2014, 20, 1193–1198. [Google Scholar] [CrossRef] [PubMed]
- García-Jiménez, C.; Gutiérrez-Salmerón, M.; Chocarro-Calvo, A.; García-Martinez, J.M.; Castaño, A.; De la Vieja, A. From obesity to diabetes and cancer: Epidemiological links and role of therapies. Br. J. Cancer 2016, 114, 716–722. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wolpin, B.M.; Bao, Y.; Qian, Z.R.; Wu, C.; Kraft, P.; Ogino, S.; Stampfer, M.J.; Sato, K.; Ma, J.; Buring, J.E.; et al. Hyperglycemia, Insulin Resistance, Impaired Pancreatic β-Cell Function, and Risk of Pancreatic Cancer. JNCI J. Natl. Cancer Inst. 2013, 105, 1027–1035. [Google Scholar] [CrossRef] [PubMed]
- Walford, G.A.; Davis, J.; Warner, A.S.; Ackerman, R.J.; Billings, L.K.; Chamarthi, B.; Fanelli, R.R.; Hernandez, A.M.; Huang, C.; Khan, S.Q.; et al. Branched chain and aromatic amino acids change acutely following two medical therapies for type 2 diabetes mellitus. Metabolism 2013, 62, 1772–1778. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rossmeislová, L.; Gojda, J.; Smolková, K. Pancreatic cancer: Branched-chain amino acids as putative key metabolic regulators? Cancer Metastasis Rev. 2021, 40, 1115–1139. [Google Scholar] [CrossRef]
- Neinast, M.D.; Jang, C.; Hui, S.; Murashige, D.S.; Chu, Q.; Morscher, R.J.; Li, X.; Zhan, L.; White, E.; Anthony, T.G.; et al. Quantitative Analysis of the Whole-Body Metabolic Fate of Branched-Chain Amino Acids. Cell Metab. 2018, 29, 417–429.e4. [Google Scholar] [CrossRef] [Green Version]
- Bodoy, S.; Fotiadis, D.; Stoeger, C.; Kanai, Y.; Palacín, M. The small SLC43 family: Facilitator system l amino acid transporters and the orphan EEG1. Mol. Asp. Med. 2013, 34, 638–645. [Google Scholar] [CrossRef]
- Ye, X.; Xu, G.; Chang, Q.; Fan, J.; Sun, Z.; Qin, Y.; Jiang, A.C. ERK1/2 signaling pathways involved in VEGF release in diabetic rat retina. Investig. Ophthalmol. Vis. Sci. 2010, 51, 5226–5233. [Google Scholar] [CrossRef]
- Restrepo, B.I.; Twahirwa, M.; Rahbar, M.H.; Schlesinger, L.S. Phagocytosis via Complement or Fc-Gamma Receptors Is Compromised in Monocytes from Type 2 Diabetes Patients with Chronic Hyperglycemia. PLoS ONE 2014, 9, e92977. [Google Scholar] [CrossRef] [Green Version]
- Keller, M.P.; Paul, P.K.; Rabaglia, M.E.; Stapleton, D.S.; Schueler, K.L.; Broman, A.T.; Ye, S.I.; Leng, N.; Brandon, C.J.; Neto, E.C.; et al. The Transcription Factor Nfatc2 Regulates β-Cell Proliferation and Genes Associated with Type 2 Diabetes in Mouse and Human Islets. PLoS Genet. 2016, 12, e1006466. [Google Scholar] [CrossRef]
- Simonett, S.P.; Shin, S.; Herring, J.A.; Bacher, R.; Smith, L.A.; Dong, C.; Rabaglia, M.E.; Stapleton, D.S.; Schueler, K.L.; Choi, J.; et al. Identification of direct transcriptional targets of NFATC2 that promote β cell proliferation. J. Clin. Investig. 2021, 131, e144833. [Google Scholar] [CrossRef] [PubMed]
- Zherebitskaya, E.; Akude, E.; Smith, D.R.; Fernyhough, P. Development of Selective Axonopathy in Adult Sensory Neurons Isolated from Diabetic Rats. Diabetes 2009, 58, 1356–1364. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sivaskandarajah, G.A.; Jeansson, M.; Maezawa, Y.; Eremina, V.; Baelde, H.J.; Quaggin, S.E. Vegfa Protects the Glomerular Microvasculature in Diabetes. Diabetes 2012, 61, 2958–2966. [Google Scholar] [CrossRef] [Green Version]
- Tahergorabi, Z.; Khazaei, M. Imbalance of angiogenesis in diabetic complications: The mechanisms. Int. J. Prev. Med. 2012, 3, 827–838. [Google Scholar] [CrossRef] [PubMed]
- Modi, S.J.; Kulkarni, V.M. Vascular Endothelial Growth Factor Receptor (VEGFR-2)/KDR Inhibitors: Medicinal Chemistry Perspective. Med. Drug Discov. 2019, 2, 100009. [Google Scholar] [CrossRef]
- Inoguchi, T.; Umeda, F.; Watanabe, J.; Ibayashi, H. Reduced Serum-Stimulatory Activity on Prostacyclin Production by Cultured Aortic Endothelial Cells in Diabetes mellitus. Pathophysiol. Haemost. Thromb. 1986, 16, 447–452. [Google Scholar] [CrossRef] [PubMed]
- Stitham, J.; Hwa, J. Prostacyclin, Atherothrombosis and Diabetes Mellitus: Physiologic and Clinical Considerations. Curr. Mol. Med. 2016, 16, 328–342. [Google Scholar] [CrossRef] [PubMed]
- Finci, L.; Zhang, Y.; Meijers, R.; Wang, J.-H. Signaling mechanism of the netrin-1 receptor DCC in axon guidance. Prog. Biophys. Mol. Biol. 2015, 118, 153–160. [Google Scholar] [CrossRef] [Green Version]
- Dong, D.; Zhang, L.; Bai, C.; Ma, N.; Ji, W.; Jia, L.; Zhang, A.; Zhang, P.; Ren, L.; Zhou, Y. UNC5D, suppressed by promoter hypermethylation, inhibits cell metastasis by activating death-associated protein kinase 1 in prostate cancer. Cancer Sci. 2019, 110, 1244–1255. [Google Scholar] [CrossRef]
- Liu, R.; Chen, P.; Chen, L. Single-sample landscape entropy reveals the imminent phase transition during disease progression. Bioinformatics 2019, 36, 1522–1532. [Google Scholar] [CrossRef]
- Chagnon, M.J.; Uetani, N.; Tremblay, M.L. Functional significance of the LAR receptor protein tyrosine phosphatase family in development and diseases. Biochem. Cell Biol. 2004, 82, 664–675. [Google Scholar] [CrossRef] [PubMed]
- Norris, K.; Norris, F.; Kono, D.H.; Vestergaard, H.; Pedersen, O.; Theofilopoulos, A.N.; Møller, N.P. Expression of protein-tyrosine phosphatases in the major insulin target tissues. FEBS Lett. 1997, 415, 243–248. [Google Scholar] [CrossRef] [Green Version]
- Chagnon, M.J.; Elchebly, M.; Uetani, N.; Dombrowski, L.; Cheng, A.; Mooney, R.A.; Marette, A.; Tremblay, M.L. Altered glucose homeostasis in mice lacking the receptor protein tyrosine phosphatase sigma. Can. J. Physiol. Pharmacol. 2006, 84, 755–763. [Google Scholar] [CrossRef] [PubMed]
- Ouni, M.; Saussenthaler, S.; Eichelmann, F.; Jähnert, M.; Stadion, M.; Wittenbecher, C.; Rönn, T.; Zellner, L.; Gottmann, P.; Ling, C.; et al. Epigenetic Changes in Islets of Langerhans Preceding the Onset of Diabetes. Diabetes 2020, 69, 2503–2517. [Google Scholar] [CrossRef]
- Altieri, D.C. The molecular basis and potential role of survivin in cancer diagnosis and therapy. Trends Mol. Med. 2001, 7, 542–547. [Google Scholar] [CrossRef]
- Xu, L.; Yu, W.; Xiao, H.; Lin, K. BIRC5 is a prognostic biomarker associated with tumor immune cell infiltration. Sci. Rep. 2021, 11, 390. [Google Scholar] [CrossRef]
- Kami, K.; Doi, R.; Koizumi, M.; Toyoda, E.; Mori, T.; Ito, D.; Fujimoto, K.; Wada, M.; Miyatake, S.I.; Imamura, M. Survivin expression is a prognostic marker in pancreatic cancer patients. Surgery 2004, 136, 443–448. [Google Scholar] [CrossRef]
- Ekeblad, S.; Lejonklou, M.H.; Stålberg, P.; Skogseid, B. Prognostic relevance of survivin in pancreatic endocrine tumors. World J. Surg. 2012, 36, 1411–1418. [Google Scholar] [CrossRef] [Green Version]





| Rank | Name | Score |
|---|---|---|
| 1 | ATP1B1 | 39 |
| 2 | RIN2 | 22 |
| 3 | EIF3E | 18 |
| 4 | AP000476.1 | 15 |
| 5 | ACER3 | 11 |
| 6 | CBX5 | 10 |
| 6 | HIST1H4C | 10 |
| 8 | TFDP2 | 9 |
| 9 | RPS29 | 8 |
| 9 | PTPRS | 8 |
| 9 | CEP97 | 8 |
| 12 | RPS3 | 7 |
| 13 | NGFRAP1 | 6 |
| 13 | XPO1 | 6 |
| 13 | PLEKHH1 | 6 |
| 16 | ECHDC2 | 5 |
| 16 | EFCAB11 | 5 |
| 16 | BIRC5 | 5 |
| 16 | ATP6AP2 | 5 |
| 16 | HSP90AB1 | 5 |
| Enriched Biological Process | Enriched p Value |
|---|---|
| macromolecular complex (GO:0032991) | 0.001387 |
| nuclear-transcribed mRNA catabolic process, nonsense-mediated decay (GO:0000184) | 0.003306 |
| extracellular exosome (GO:0070062) | 0.003644 |
| translational initiation (GO:0006413) | 0.003925 |
| Kinetochore (GO:0000776) | 0.004606 |
| ribonucleoprotein complex (GO:1990904) | 0.006289 |
| Enriched Biological Process | Enriched p Value |
|---|---|
| hsa03010: Ribosome | 0.015199 |
| hsa05207: Chemical carcinogenesis—receptor activation | 0.026454 |
| hsa04614: Renin-angiotensin system | 0.027904 |
| hsa04964: Proximal tubule bicarbonate reclamation | 0.027904 |
| hsa05171: Coronavirus disease—COVID-19 | 0.031280 |
| hsa05132: Salmonella infection | 0.035643 |
| Enriched Biological Process | Enriched p Value |
|---|---|
| carboxylic ester hydrolase activity (GO:0052689) | 0.018599 |
| filopodium (GO:0030175) | 0.046597 |
| Enriched Biological Process | Enriched p Value |
|---|---|
| hsa04360: Axon guidance | 0.002895 |
| hsa04370: VEGF signaling pathway | 0.028677 |
| hsa04662: B cell receptor signaling pathway | 0.039688 |
| hsa05235: PD-L1 expression and PD-1 checkpoint pathway in cancer | 0.043020 |
| hsa04658: Th1 and Th2 cell differentiation | 0.044445 |
| hsa04666: Fc gamma R-mediated phagocytosis | 0.046818 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Xie, Y.; Cui, Z.; Wang, N.; Li, P. Research on Potential Network Markers and Signaling Pathways in Type 2 Diabetes Based on Conditional Cell-Specific Network. Genes 2022, 13, 1155. https://doi.org/10.3390/genes13071155
Xie Y, Cui Z, Wang N, Li P. Research on Potential Network Markers and Signaling Pathways in Type 2 Diabetes Based on Conditional Cell-Specific Network. Genes. 2022; 13(7):1155. https://doi.org/10.3390/genes13071155
Chicago/Turabian StyleXie, Yuke, Zhizhong Cui, Nan Wang, and Peiluan Li. 2022. "Research on Potential Network Markers and Signaling Pathways in Type 2 Diabetes Based on Conditional Cell-Specific Network" Genes 13, no. 7: 1155. https://doi.org/10.3390/genes13071155
APA StyleXie, Y., Cui, Z., Wang, N., & Li, P. (2022). Research on Potential Network Markers and Signaling Pathways in Type 2 Diabetes Based on Conditional Cell-Specific Network. Genes, 13(7), 1155. https://doi.org/10.3390/genes13071155






