SUMO-Based Regulation of Nuclear Positioning to Spatially Regulate Homologous Recombination Activities at Replication Stress Sites
Abstract
1. Replication Stressed Forks and Homologous Recombination
2. Replication Stress Sites Move to the Nuclear Periphery
3. SUMOylation in DNA Repair
4. NPCs Anchor DNA Lesions in a SUMO-Dependent Manner to Promote DNA Repair
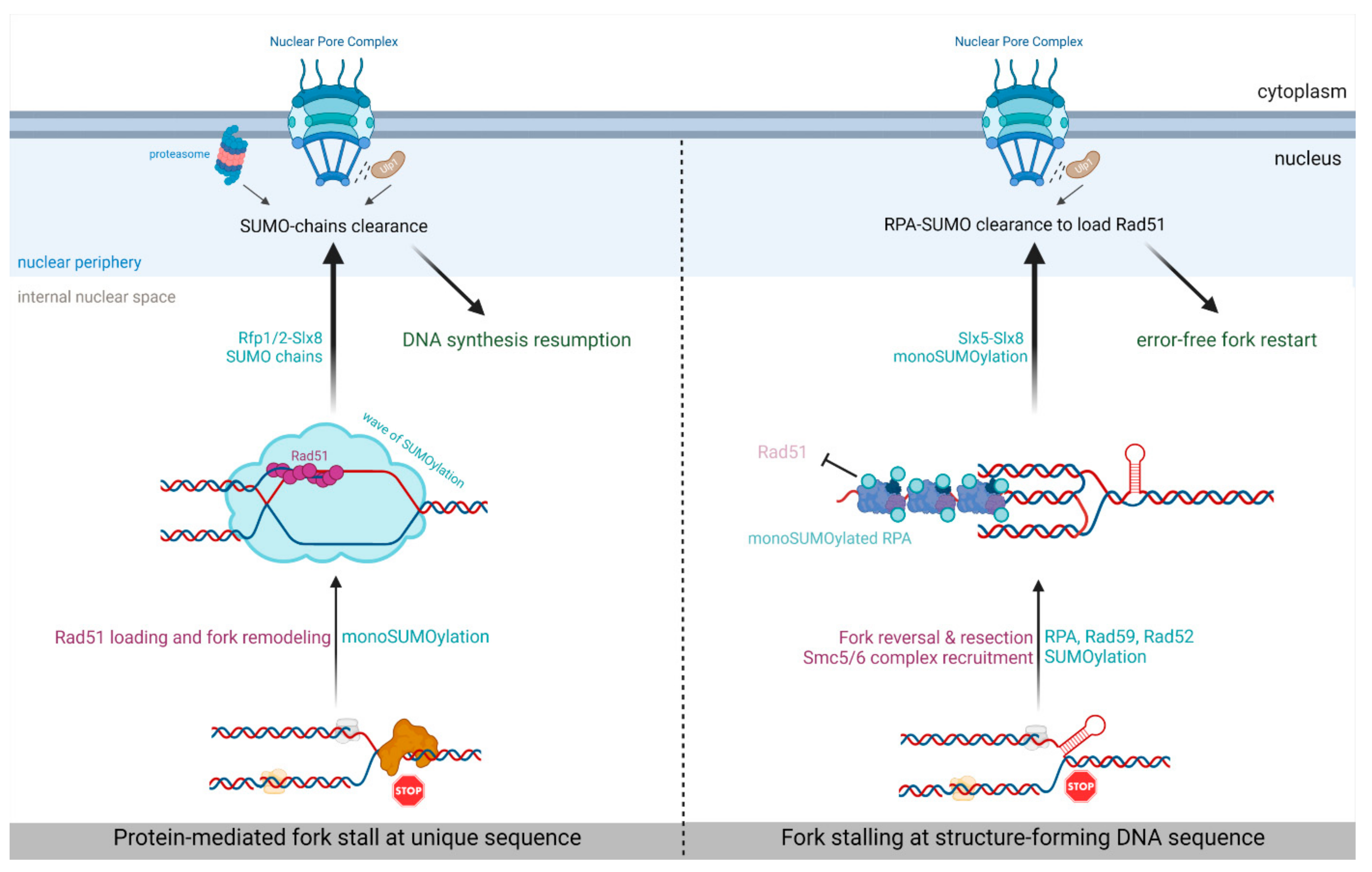
5. SUMO-Based Regulation of Nuclear Positioning Regulates Replication Fork Repair
6. Concluding Remarks
Author Contributions
Funding
Conflicts of Interest
References
- Zeman, M.K.; Cimprich, K.A. Causes and consequences of replication stress. Nat. Cell Biol. 2014, 16, 2–9. [Google Scholar] [CrossRef] [PubMed]
- Willaume, S.; Rass, E.; Paulafontanilla-Ramirez, P.; Moussa, A.; Wanschoor, P.; Bertrand, P. A link between replicative stress, lamin proteins, and inflammation. Genes 2021, 12, 552. [Google Scholar] [CrossRef] [PubMed]
- Magdalou, I.; Lopez, B.S.; Pasero, P.; Lambert, S.A.E. The causes of replication stress and their consequences on genome stability and cell fate. Semin. Cell Dev. Biol. 2014, 30, 154–164. [Google Scholar] [CrossRef] [PubMed]
- Lin, Y.L.; Pasero, P. Replication stress: From chromatin to immunity and beyond. Curr. Opin. Genet. Dev. 2021, 71, 136–142. [Google Scholar] [CrossRef] [PubMed]
- Gaillard, H.; García-Muse, T.; Aguilera, A. Replication stress and cancer. Nat. Rev. Cancer 2015, 15, 276–280. [Google Scholar] [CrossRef]
- Tomasetti, C.; Vogelstein, B. Variation in cancer risk among tissues can be explained by the number of stem cell divisions. Science 2015, 347, 78–81. [Google Scholar] [CrossRef] [PubMed]
- Berti, M.; Cortez, D.; Lopes, M. The plasticity of DNA replication forks in response to clinically relevant genotoxic stress. Nat. Rev. Mol. Cell Biol. 2020, 21, 633–651. [Google Scholar] [CrossRef] [PubMed]
- Ait Saada, A.; Lambert, S.A.E.; Carr, A.M. Preserving replication fork integrity and competence via the homologous recombination pathway. DNA Repair 2018, 71, 135–147. [Google Scholar] [CrossRef] [PubMed]
- Scully, R.; Elango, R.; Panday, A.; Willis, N.A. Recombination and restart at blocked replication forks. Curr. Opin. Genet. Dev. 2021, 71, 154–162. [Google Scholar] [CrossRef]
- Sakofsky, C.J.; Malkova, A. Break induced replication in eukaryotes: Mechanisms, functions, and consequences. Crit. Rev. Biochem. Mol. Biol. 2017, 52, 395–413. [Google Scholar] [CrossRef] [PubMed]
- Hashimoto, Y.; Puddu, F.; Costanzo, V. RAD51-and MRE11-dependent reassembly of uncoupled CMG helicase complex at collapsed replication forks. Nat. Struct. Mol. Biol. 2012, 19, 17–24. [Google Scholar] [CrossRef] [PubMed]
- Carr, A.; Lambert, S. Recombination-dependent replication: New perspectives from site-specific fork barriers. Curr. Opin. Genet. Dev. 2021, 71, 129–135. [Google Scholar] [CrossRef] [PubMed]
- Neelsen, K.J.; Lopes, M. Replication fork reversal in eukaryotes: From dead end to dynamic response. Nat. Rev. Mol. Cell Biol. 2015, 16, 207–220. [Google Scholar] [CrossRef] [PubMed]
- Quinet, A.; Lemaçon, D.; Vindigni, A. Replication Fork Reversal: Players and Guardians. Mol. Cell 2017, 68, 830–833. [Google Scholar] [CrossRef]
- Lemaçon, D.; Jackson, J.; Quinet, A.; Brickner, J.R.; Li, S.; Yazinski, S.; You, Z.; Ira, G.; Zou, L.; Mosammaparast, N.; et al. MRE11 and EXO1 nucleases degrade reversed forks and elicit MUS81-dependent fork rescue in BRCA2-deficient cells. Nat. Commun. 2017, 8, 860. [Google Scholar] [CrossRef]
- Teixeira-Silva, A.; Ait Saada, A.; Hardy, J.; Iraqui, I.; Nocente, M.C.; Fréon, K.; Lambert, S.A.E. The end-joining factor Ku acts in the end-resection of double strand break-free arrested replication forks. Nat. Commun. 2017, 8, 1982. [Google Scholar] [CrossRef] [PubMed]
- Kalousi, A.; Soutoglou, E. Nuclear compartmentalization of DNA repair. Curr. Opin. Genet. Dev. 2016, 37, 148–157. [Google Scholar] [CrossRef] [PubMed]
- Aten, J.A.; Stap, J.; Krawczyk, P.M.; Van Oven, C.H.; Hoebe, R.A.; Essers, J.; Kanaar, R. Dynamics of DNA Double-Strand Breaks Revealed by Clustering of Damaged Chromosome Domains. Science 2004, 303, 92–95. [Google Scholar] [CrossRef] [PubMed]
- Seeber, A.; Gasser, S.M. Chromatin organization and dynamics in double-strand break repair. Curr. Opin. Genet. Dev. 2017, 43, 9–16. [Google Scholar] [CrossRef] [PubMed]
- Nagai, S.; Dubrana, K.; Tsai-Pflugfelder, M.; Davidson, M.B.; Roberts, T.M.; Brown, G.W.; Varela, E.; Hediger, F.; Gasser, S.M.; Krogan, N.J. Functional targeting of DNA damage to a nuclear pore-associated SUMO-dependent ubiquitin ligase. Science 2008, 322, 597–602. [Google Scholar] [CrossRef] [PubMed]
- Ryu, T.; Spatola, B.; Delabaere, L.; Bowlin, K.; Hopp, H.; Kunitake, R.; Karpen, G.H.; Chiolo, I. Heterochromatic breaks move to the nuclear periphery to continue recombinational repair. Nat. Cell Biol. 2015, 17, 1401–1411. [Google Scholar] [CrossRef] [PubMed]
- Chiolo, I.; Minoda, A.; Colmenares, S.U.; Polyzos, A.; Costes, S.V.; Karpen, G.H. Double-strand breaks in heterochromatin move outside of a dynamic HP1a domain to complete recombinational repair. Cell 2011, 144, 732–744. [Google Scholar] [CrossRef] [PubMed]
- Horigome, C.; Unozawa, E.; Ooki, T.; Kobayashi, T. Ribosomal RNA gene repeats associate with the nuclear pore complex for maintenance after DNA damage. PLoS Genet. 2019, 15, e1008103. [Google Scholar] [CrossRef]
- Marnef, A.; Finoux, A.L.; Arnould, C.; Guillou, E.; Daburon, V.; Rocher, V.; Mangeat, T.; Mangeot, P.E.; Ricci, E.P.; Legube, G. A cohesin/HUSH- And LINC-dependent pathway controls ribosomal DNA double-strand break repair. Genes Dev. 2019, 33, 1175–1190. [Google Scholar] [CrossRef] [PubMed]
- Tsouroula, K.; Furst, A.; Rogier, M.; Heyer, V.; Maglott-Roth, A.; Ferrand, A.; Reina-San-Martin, B.; Soutoglou, E. Temporal and Spatial Uncoupling of DNA Double Strand Break Repair Pathways within Mammalian Heterochromatin. Mol. Cell 2016, 63, 293–305. [Google Scholar] [CrossRef] [PubMed]
- Lemaître, C.; Grabarz, A.; Tsouroula, K.; Andronov, L.; Furst, A.; Pankotai, T.; Heyer, V.; Rogier, M.; Attwood, K.M.; Kessler, P.; et al. Nuclear position dictates DNA repair pathway choice. Genes Dev. 2014, 28, 2450–2463. [Google Scholar] [CrossRef]
- Kalocsay, M.; Hiller, N.J.; Jentsch, S. Chromosome-wide Rad51 spreading and SUMO-H2A.Z-dependent chromosome fixation in response to a persistent DNA double-strand break. Mol. Cell 2009, 33, 335–343. [Google Scholar] [CrossRef] [PubMed]
- Oza, P.; Jaspersen, S.L.; Miele, A.; Dekker, J.; Peterson, C.L. Mechanisms that regulate localization of a DNA double-strand break to the nuclear periphery. Genes Dev. 2009, 23, 912–927. [Google Scholar] [CrossRef]
- Horigome, C.; Oma, Y.; Konishi, T.; Schmid, R.; Marcomini, I.; Hauer, M.H.; Dion, V.; Harata, M.; Gasser, S.M. SWR1 and INO80 chromatin remodelers contribute to DNA double-strand break perinuclear anchorage site choice. Mol. Cell 2014, 55, 626–639. [Google Scholar] [CrossRef]
- Khadaroo, B.; Teixeira, M.T.; Luciano, P.; Eckert-Boulet, N.; Germann, S.M.; Simon, M.N.; Gallina, I.; Abdallah, P.; Gilson, E.; Géli, V.; et al. The DNA damage response at eroded telomeres and tethering to the nuclear pore complex. Nat. Cell Biol. 2009, 11, 980–987. [Google Scholar] [CrossRef] [PubMed]
- Churikov, D.; Charifi, F.; Eckert-Boulet, N.; Silva, S.; Simon, M.-N.; Lisby, M.; Géli, V. SUMO-Dependent Relocalization of Eroded Telomeres to Nuclear Pore Complexes Controls Telomere Recombination. Cell Rep. 2016, 15, 1242–1253. [Google Scholar] [CrossRef]
- Su, X.A.; Dion, V.; Gasser, S.M.; Freudenreich, C.H. Regulation of recombination at yeast nuclear pores controls repair and triplet repeat stability. Genes Dev. 2015, 29, 1006–1017. [Google Scholar] [CrossRef] [PubMed]
- Kramarz, K.; Schirmeisen, K.; Boucherit, V.; Ait Saada, A.; Lovo, C.; Palancade, B.; Freudenreich, C.; Lambert, S.A.E. The nuclear pore primes recombination-dependent DNA synthesis at arrested forks by promoting SUMO removal. Nat. Commun. 2020, 11, 5643. [Google Scholar] [CrossRef]
- Aguilera, P.; Whalen, J.; Minguet, C.; Churikov, D.; Freudenreich, C.; Simon, M.N.; Géli, V. The nuclear pore complex prevents sister chromatid recombination during replicative senescence. Nat. Commun. 2020, 11, 160. [Google Scholar] [CrossRef]
- Lamm, N.; Read, M.N.; Nobis, M.; Van Ly, D.; Page, S.G.; Masamsetti, V.P.; Timpson, P.; Biro, M.; Cesare, A.J. Nuclear F-actin counteracts nuclear deformation and promotes fork repair during replication stress. Nat. Cell Biol. 2020, 22, 1460–1470. [Google Scholar] [CrossRef]
- Pinzaru, A.M.; Kareh, M.; Lamm, N.; Lazzerini-Denchi, E.; Cesare, A.J.; Sfeir, A. Replication stress conferred by POT1 dysfunction promotes telomere relocalization to the nuclear pore. Genes Dev. 2020, 34, 1619–1636. [Google Scholar] [CrossRef] [PubMed]
- Lamm, N.; Rogers, S.; Cesare, A.J. Chromatin mobility and relocation in DNA repair. Trends Cell Biol. 2021, 31, 843–855. [Google Scholar] [CrossRef]
- Horigome, C.; Bustard, D.E.; Marcomini, I.; Delgoshaie, N.; Tsai-Pflugfelder, M.; Cobb, J.A.; Gasser, S.M. PolySUMOylation by Siz2 and Mms21 triggers relocation of DNA breaks to nuclear pores through the Slx5/Slx8 STUbL. Genes Dev. 2016, 30, 931–945. [Google Scholar] [CrossRef] [PubMed]
- Lambert, S.; Mizuno, K.; Blaisonneau, J.; Martineau, S.; Chanet, R.; Fréon, K.; Murray, J.M.; Carr, A.M.; Baldacci, G. Homologous recombination restarts blocked replication forks at the expense of genome rearrangements by template exchange. Mol. Cell 2010, 39, 346–359. [Google Scholar] [CrossRef]
- Ait Saada, A.; Teixeira-Silva, A.; Iraqui, I.; Costes, A.; Hardy, J.; Paoletti, G.; Fréon, K.; Lambert, S.A.E. Unprotected Replication Forks Are Converted into Mitotic Sister Chromatid Bridges. Mol. Cell 2017, 66, 398–410.e4. [Google Scholar] [CrossRef] [PubMed]
- Tsang, E.; Miyabe, I.; Iraqui, I.; Zheng, J.; Lambert, S.A.E.; Carr, A.M. The extent of error-prone replication restart by homologous recombination is controlled by Exo1 and checkpoint proteins. J. Cell Sci. 2014, 127, 2983–2994. [Google Scholar] [CrossRef] [PubMed]
- Miyabe, I.; Mizuno, K.; Keszthelyi, A.; Daigaku, Y.; Skouteri, M.; Mohebi, S.; Kunkel, T.A.; Murray, J.M.; Carr, A.M. Polymerase δ replicates both strands after homologous recombination–dependent fork restart. Nat. Struct. Mol. Biol. 2015, 22, 932–938. [Google Scholar] [CrossRef]
- Nguyen, M.O.; Jalan, M.; Morrow, C.A.; Osman, F.; Whitby, M.C. Recombination occurs within minutes of replication blockage by RTS1 producing restarted forks that are prone to collapse. Elife 2015, 4, e04539. [Google Scholar] [CrossRef]
- Pichler, A.; Fatouros, C.; Lee, H.; Eisenhardt, N. SUMO conjugation—A mechanistic view. Biomol. Concepts 2017, 8, 13–36. [Google Scholar] [CrossRef]
- Ulrich, H.D. The Fast-Growing Business of SUMO Chains. Mol. Cell 2008, 32, 301–305. [Google Scholar] [CrossRef]
- Flotho, A.; Melchior, F. Sumoylation: A regulatory protein modification in health and disease. Annu. Rev. Biochem. 2013, 82, 357–385. [Google Scholar] [CrossRef] [PubMed]
- Cappadocia, L.; Lima, C.D. Ubiquitin-like Protein Conjugation: Structures, Chemistry, and Mechanism. Chem. Rev. 2018, 118, 889–918. [Google Scholar] [CrossRef] [PubMed]
- Sriramachandran, A.M.; Meyer-Teschendorf, K.; Pabst, S.; Ulrich, H.D.; Gehring, N.H.; Hofmann, K.; Praefcke, G.J.K.; Dohmen, R.J. Arkadia/RNF111 is a SUMO-targeted ubiquitin ligase with preference for substrates marked with SUMO1-capped SUMO2/3 chain. Nat. Commun. 2019, 10, 3678. [Google Scholar] [CrossRef]
- Liang, Y.C.; Lee, C.C.; Yao, Y.L.; Lai, C.C.; Schmitz, M.L.; Yang, W.M. SUMO5, a novel poly-SUMO isoform, regulates PML nuclear bodies. Sci. Rep. 2016, 6, 26509. [Google Scholar] [CrossRef] [PubMed]
- Watts, F.Z.; Skilton, A.; Ho, J.C.Y.; Boyd, L.K.; Trickey, M.A.M.; Gardner, L.; Ogi, F.X.; Outwin, E.A. The role of Schizosaccharomyces pombe SUMO ligases in genome stability. In Proceedings of the Biochemical Society Transactions. Biochem. Soc. Trans. 2007, 35, 1379–1384. [Google Scholar] [CrossRef] [PubMed]
- Sacher, M.; Pfander, B.; Jentsch, S. Identification of SUMO-protein conjugates. Methods Enzymol. 2005, 399, 392–404. [Google Scholar] [CrossRef] [PubMed]
- Jentsch, S.; Psakhye, I. Control of nuclear activities by substrate-selective and protein-group SUMOylation. Annu. Rev. Genet. 2013, 47, 167–186. [Google Scholar] [CrossRef] [PubMed]
- Soria-Bretones, I.; Cepeda-García, C.; Checa-Rodriguez, C.; Heyer, V.; Reina-San-Martin, B.; Soutoglou, E.; Huertas, P. DNA end resection requires constitutive sumoylation of CtIP by CBX4. Nat. Commun. 2017, 8, 113. [Google Scholar] [CrossRef] [PubMed]
- Locke, A.J.; Hossain, L.; McCrostie, G.; Ronato, D.A.; Fitieh, A.; Rafique, T.A.; Mashayekhi, F.; Motamedi, M.; Masson, J.Y.; Ismail, I.H. SUMOylation mediates CtIP’s functions in DNA end resection and replication fork protection. Nucleic Acids Res. 2021, 49, 928–953. [Google Scholar] [CrossRef]
- Lopez-Contreras, A.J.; Ruppen, I.; Nieto-Soler, M.; Murga, M.; Rodriguez-Acebes, S.; Remeseiro, S.; Rodrigo-Perez, S.; Rojas, A.M.; Mendez, J.; Muñoz, J.; et al. A Proteomic Characterization of Factors Enriched at Nascent DNA Molecules. Cell Rep. 2013, 3, 1105–1116. [Google Scholar] [CrossRef]
- Cremona, C.A.; Sarangi, P.; Yang, Y.; Hang, L.E.; Rahman, S.; Zhao, X. Extensive DNA damage-induced sumoylation contributes to replication and repair and acts in addition to the mec1 checkpoint. Mol. Cell 2012, 45, 422–432. [Google Scholar] [CrossRef]
- Hoege, C.; Pfander, B.; Moldovan, G.L.; Pyrowolakis, G.; Jentsch, S. RAD6-dependent DNA repair is linked to modification of PCNA by ubiquitin and SUMO. Nature 2002, 419, 135–141. [Google Scholar] [CrossRef] [PubMed]
- Golebiowski, F.; Matic, I.; Tatham, M.H.; Cole, C.; Yin, Y.; Nakamura, A.; Cox, J.; Barton, G.J.; Mann, M.; Hay, R.T. System-wide changes to sumo modifications in response to heat shock. Sci. Signal. 2009, 2, ra24. [Google Scholar] [CrossRef]
- Branzei, D.; Sollier, J.; Liberi, G.; Zhao, X.; Maeda, D.; Seki, M.; Enomoto, T.; Ohta, K.; Foiani, M. Ubc9- and Mms21-Mediated Sumoylation Counteracts Recombinogenic Events at Damaged Replication Forks. Cell 2006, 127, 509–522. [Google Scholar] [CrossRef] [PubMed]
- Klein, H.L. A SUMOry of DNA Replication: Synthesis, Damage, and Repair. Cell 2006, 127, 455–457. [Google Scholar] [CrossRef][Green Version]
- Psakhye, I.; Jentsch, S. Protein group modification and synergy in the SUMO pathway as exemplified in DNA repair. Cell 2012, 151, 807–820. [Google Scholar] [CrossRef]
- Sarangi, P.; Zhao, X. SUMO-mediated regulation of DNA damage repair and responses. Trends Biochem. Sci. 2015, 40, 233–242. [Google Scholar] [CrossRef] [PubMed]
- Keiten-Schmitz, J.; Schunck, K.; Müller, S. SUMO Chains Rule on Chromatin Occupancy. Front. Cell Dev. Biol. 2020, 7, 343. [Google Scholar] [CrossRef] [PubMed]
- Prudden, J.; Perry, J.J.P.; Nie, M.; Vashisht, A.A.; Arvai, A.S.; Hitomi, C.; Guenther, G.; Wohlschlegel, J.A.; Tainer, J.A.; Boddy, M.N. DNA Repair and Global Sumoylation Are Regulated by Distinct Ubc9 Noncovalent Complexes. Mol. Cell. Biol. 2011, 31, 2299–2310. [Google Scholar] [CrossRef] [PubMed]
- Bergink, S.; Jentsch, S. Principles of ubiquitin and SUMO modifications in DNA repair. Nature 2009, 458, 461–467. [Google Scholar] [CrossRef] [PubMed]
- Jackson, S.P.; Durocher, D. Regulation of DNA Damage Responses by Ubiquitin and SUMO. Mol. Cell 2013, 49, 795–807. [Google Scholar] [CrossRef]
- Reindle, A.; Belichenko, I.; Bylebyl, G.R.; Chen, X.L.; Gandhi, N.; Johnson, E.S. Multiple domains in Siz SUMO ligases contribute to substrate selectivity. J. Cell Sci. 2006, 119, 4749–4757. [Google Scholar] [CrossRef]
- Xhemalce, B.; Riising, E.M.; Baumann, P.; Dejean, A.; Arcangioli, B.; Seeler, J.S. Role of SUMO in the dynamics of telomere maintenance in fission yeast. Proc. Natl. Acad. Sci. USA 2007, 104, 893–898. [Google Scholar] [CrossRef]
- Pichler, A.; Gast, A.; Seeler, J.S.; Dejean, A.; Melchior, F. The nucleoporin RanBP2 has SUMO1 E3 ligase activity. Cell 2002, 108, 109–120. [Google Scholar] [CrossRef]
- Yang, Q.; Tang, J.; Xu, C.; Zhao, H.; Zhou, Y.; Wang, Y.; Yang, M.; Chen, X.; Chen, J. Histone deacetylase 4 inhibits NF-κB activation by facilitating IκBα sumoylation. J. Mol. Cell Biol. 2020, 12, 933–945. [Google Scholar] [CrossRef]
- Peng, J.; Wysocka, J. It takes a PHD to SUMO. Trends Biochem. Sci. 2008, 33, 191–194. [Google Scholar] [CrossRef] [PubMed]
- Kagey, M.H.; Melhuish, T.A.; Wotton, D. The polycomb protein Pc2 is a SUMO E3. Cell 2003, 113, 127–137. [Google Scholar] [CrossRef]
- Weger, S.; Hammer, E.; Heilbronn, R. Topors acts as a SUMO-1 E3 ligase for p53 in vitro and in vivo. FEBS Lett. 2005, 579, 5007–5012. [Google Scholar] [CrossRef]
- Palancade, B.; Liu, X.; Garcia-Rubio, M.; Aguilera, A.; Zhao, X.; Doye, V. Nucleoporins prevent DNA damage accumulation by modulating Ulp1-dependent sumoylation processes. Mol. Biol. Cell 2007, 18, 2912–2923. [Google Scholar] [CrossRef] [PubMed]
- Kunz, K.; Piller, T.; Müller, S. SUMO-specific proteases and isopeptidases of the SENP family at a glance. J. Cell Sci. 2018, 131, jcs211904. [Google Scholar] [CrossRef] [PubMed]
- Palancade, B.; Doye, V. Sumoylating and desumoylating enzymes at nuclear pores: Underpinning their unexpected duties? Trends Cell Biol. 2008, 18, 174–183. [Google Scholar] [CrossRef] [PubMed]
- Uzunova, K.; Göttsche, K.; Miteva, M.; Weisshaar, S.R.; Glanemann, C.; Schnellhardt, M.; Niessen, M.; Scheel, H.; Hofmann, K.; Johnson, E.S.; et al. Ubiquitin-dependent proteolytic control of SUMO conjugates. J. Biol. Chem. 2007, 282, 34167–34175. [Google Scholar] [CrossRef] [PubMed]
- Chang, Y.C.; Oram, M.K.; Bielinsky, A.K. Sumo-targeted ubiquitin ligases and their functions in maintaining genome stability. Int. J. Mol. Sci. 2021, 22, 5391. [Google Scholar] [CrossRef]
- Kramarz, K.; Litwin, I.; Cal-Bakowska, M.; Szakal, B.; Branzei, D.; Wysocki, R.; Dziadkowiec, D. Swi2/Snf2-like protein Uls1 functions in the Sgs1-dependent pathway of maintenance of rDNA stability and alleviation of replication stress. DNA Repair 2014, 21, 24–35. [Google Scholar] [CrossRef]
- Prudden, J.; Pebernard, S.; Raffa, G.; Slavin, D.A.; Perry, J.J.P.; Tainer, J.A.; McGowan, C.H.; Boddy, M.N. SUMO-targeted ubiquitin ligases in genome stability. EMBO J. 2007, 26, 4089–4101. [Google Scholar] [CrossRef] [PubMed]
- Psakhye, I.; Castellucci, F.; Branzei, D. SUMO-Chain-Regulated Proteasomal Degradation Timing Exemplified in DNA Replication Initiation. Mol. Cell 2019, 76, 632–645.e6. [Google Scholar] [CrossRef]
- Liebelt, F.; Jansen, N.S.; Kumar, S.; Gracheva, E.; Claessens, L.A.; Verlaan-de Vries, M.; Willemstein, E.; Vertegaal, A.C.O. The poly-SUMO2/3 protease SENP6 enables assembly of the constitutive centromere-associated network by group deSUMOylation. Nat. Commun. 2019, 10, 3987. [Google Scholar] [CrossRef] [PubMed]
- Mullen, J.R.; Das, M.; Brill, S.J. Genetic evidence that polysumoylation bypasses the need for a SUMO-targeted UB ligase. Genetics 2011, 187, 73–87. [Google Scholar] [CrossRef][Green Version]
- Kim, S.J.; Fernandez-Martinez, J.; Nudelman, I.; Shi, Y.; Zhang, W.; Raveh, B.; Herricks, T.; Slaughter, B.D.; Hogan, J.A.; Upla, P.; et al. Integrative structure and functional anatomy of a nuclear pore complex. Nature 2018, 555, 475–482. [Google Scholar] [CrossRef] [PubMed]
- Von Appen, A.; Beck, M. Structure Determination of the Nuclear Pore Complex with Three-Dimensional Cryo electron Microscopy. J. Mol. Biol. 2016, 428, 2001–2010. [Google Scholar] [CrossRef] [PubMed]
- Knockenhauer, K.E.; Schwartz, T.U. The Nuclear Pore Complex as a Flexible and Dynamic Gate. Cell 2016, 164, 1162–1171. [Google Scholar] [CrossRef] [PubMed]
- Hoelz, A.; Debler, E.W.; Blobel, G. The Structure of the nuclear pore complex. Annu. Rev. Biochem. 2011, 80, 613–643. [Google Scholar] [CrossRef] [PubMed]
- Schwartz, T.U. The Structure Inventory of the Nuclear Pore Complex. J. Mol. Biol. 2016, 428, 1986–2000. [Google Scholar] [CrossRef]
- Ibarra, A.; Hetzer, M.W. Nuclear pore proteins and the control of genome functions. Genes Dev. 2015, 29, 337–349. [Google Scholar] [CrossRef] [PubMed]
- D’Angelo, M.A.; Hetzer, M.W. Structure, dynamics and function of nuclear pore complexes. Trends Cell Biol. 2008, 18, 456–466. [Google Scholar] [CrossRef] [PubMed]
- Whalen, J.M.; Freudenreich, C.H. Location, location, location: The role of nuclear positioning in the repair of collapsed forks and protection of genome stability. Genes 2020, 11, 635. [Google Scholar] [CrossRef] [PubMed]
- Loeillet, S.; Palancade, B.; Cartron, M.; Thierry, A.; Richard, G.-F.; Dujon, B.; Doye, V.; Nicolas, A. Genetic network interactions among replication, repair and nuclear pore deficiencies in yeast. DNA Repair 2005, 4, 459–468. [Google Scholar] [CrossRef] [PubMed]
- Moudry, P.; Lukas, C.; Macurek, L.; Neumann, B.; Heriche, J.K.; Pepperkok, R.; Ellenberg, J.; Hodny, Z.; Lukas, J.; Bartek, J. Nucleoporin NUP153 guards genome integrity by promoting nuclear import of 53BP1. Cell Death Differ. 2012, 19, 798–807. [Google Scholar] [CrossRef] [PubMed]
- Lemaître, C.; Fischer, B.; Kalousi, A.; Hoffbeck, A.S.; Guirouilh-Barbat, J.; Shahar, O.D.; Genet, D.; Goldberg, M.; Betrand, P.; Lopez, B.; et al. The nucleoporin 153, a novel factor in double-strand break repair and DNA damage response. Oncogene 2012, 31, 4803–4809. [Google Scholar] [CrossRef] [PubMed]
- Gaillard, H.; Santos-Pereira, J.M.; Aguilera, A. The Nup84 complex coordinates the DNA damage response to warrant genome integrity. Nucleic Acids Res. 2019, 47, 4054–4067. [Google Scholar] [CrossRef] [PubMed]
- Amaral, N.; Ryu, T.; Li, X.; Chiolo, I. Nuclear Dynamics of Heterochromatin Repair. Trends Genet. 2017, 33, 86–100. [Google Scholar] [CrossRef]
- Whalen, J.M.; Dhingra, N.; Wei, L.; Zhao, X.; Freudenreich, C.H. Relocation of Collapsed Forks to the Nuclear Pore Complex Depends on Sumoylation of DNA Repair Proteins and Permits Rad51 Association. Cell Rep. 2020, 31, 107635. [Google Scholar] [CrossRef] [PubMed]
- Capella, M.; Mandemaker, I.K.; Martín Caballero, L.; den Brave, F.; Pfander, B.; Ladurner, A.G.; Jentsch, S.; Braun, S. Nucleolar release of rDNA repeats for repair involves SUMO-mediated untethering by the Cdc48/p97 segregase. Nat. Commun. 2021, 12, 4918. [Google Scholar] [CrossRef] [PubMed]
- Torres-Rosell, J.; Sunjevaric, I.; De Piccoli, G.; Sacher, M.; Eckert-Boulet, N.; Reid, R.; Jentsch, S.; Rothstein, R.; Aragón, L.; Lisby, M. The Smc5-Smc6 complex and SUMO modification of Rad52 regulates recombinational repair at the ribosomal gene locus. Nat. Cell Biol. 2007, 9, 923–931. [Google Scholar] [CrossRef] [PubMed]
- Géli, V.; Lisby, M. Recombinational DNA repair is regulated by compartmentalization of DNA lesions at the nuclear pore complex. Bioessays 2015, 37, 1287–1292. [Google Scholar] [CrossRef] [PubMed]
- Polleys, E.J.; Freudenreich, C.H. Homologous recombination within repetitive DNA. Curr. Opin. Genet. Dev. 2021, 71, 143–153. [Google Scholar] [CrossRef] [PubMed]
- Chung, I.; Zhao, X. DNA break-induced sumoylation is enabled by collaboration between a sumo ligase and the ssdnabinding complex rpa. Genes Dev. 2015, 29, 1593–1598. [Google Scholar] [CrossRef] [PubMed]
- Nie, M.; Boddy, M.N. Pli1 PIAS1 SUMO Ligase Protected by the Nuclear Pore-associated SUMO Protease Ulp1 SENP1/2. J. Biol. Chem. 2015, 290, 22678–22685. [Google Scholar] [CrossRef] [PubMed]
- Oshidari, R.; Huang, R.; Medghalchi, M.; Tse, E.Y.W.; Ashgriz, N.; Lee, H.O.; Wyatt, H.; Mekhail, K. DNA repair by Rad52 liquid droplets. Nat. Commun. 2020, 11, 695. [Google Scholar] [CrossRef] [PubMed]
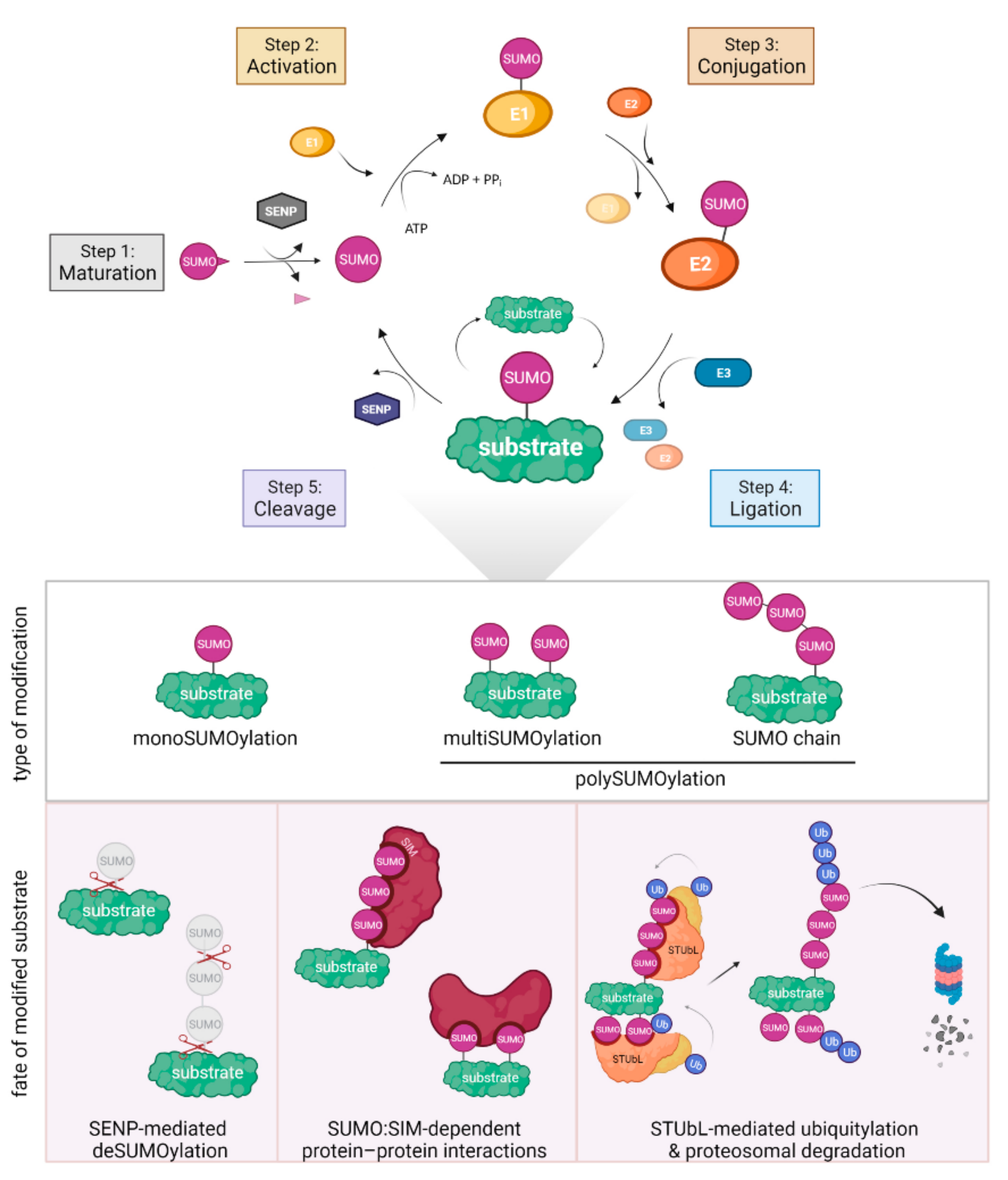
| SUMO Pathway Component | Humans | S. cerevisiae | S. pombe | |
|---|---|---|---|---|
| Small ubiquitin-like modifier (SUMO) | SUMO-1, SUMO-2, SUMO-3, SUMO-4, SUMO-5 | Smt3 | Pmt3 | |
| Activating enzyme (E1) | SAE1 SAE2 | Aos1 Uba2 | Rad31 Fub2 | |
| Conjugating enzyme (E2) | Ubc9 | Ubc9 | Hus5 | |
| SUMO ligase (E3) | SP-RING type | PIAS1, PIAS2, PIAS3, PIAS4 Mms21 | Siz1, Siz2 Mms21 Zip3 | Pli1Nse2 |
| other | RanBP2 * [69] HDAC4 [70], KPA1 [71], Pc2 [72], Topors [73] | |||
| SUMO-targeted ubiquitin ligase (STUbL) | RNF4 RNF11 | Slx5-Slx8 Uls1 | Rfp1/Rfp2-Slx8 Rrp2 (predicted) | |
| Sentrin/SUMO-specific protease (SENP) | SENP1 °,*, SENP2 °,*, SENP3, SENP5 ° SENP6, SENP7 | Ulp1 °,* Ulp2 | Ulp1 °,* Ulp2 | |
| Type of Obstacle | Protein-Mediated Fork Arrest | Structure-Forming DNA Sequence | Telomere-Specific Replication Stress | Aphidicolin Induced Replication Stress | |
|---|---|---|---|---|---|
| System description | Site-specific RFB blocking a single replisome in a polar manner | Expanded trinucleotide repeats forming hairpin structures that stall replisomes | Stalled replisomes at telomere repeats in telomerase-negative cells | Telomere-specific replication stress induced by POT1 dysfunctions | Global replication fork stalling induced |
| Organism | S. pombe | S. cerevisiae | S. cerevisiae | human cell lines | human cell lines |
| Relocation and anchorage requirements | ● Rad51-dependent fork remodeling ● Pli1 ● SUMO chain ● Rfp1-Slx8, ● Rfp2-Slx8 ● NPC-anchorage site unknown | ● Nascent DNA degradation (by Mre11, Exo1, Dna2) ● Mms21 ● SUMOylation of RPA, Rad52, Rad59 ● Slx5-SUMO interaction ● Nup1, Nup84 | ● Nup1 | ● F-actin polymerization ● ATR pathway ● Nup62, Nup153, TPR | ● F-actin polymerization |
| Relocation outcomes | Ulp1-NPCs alleviate inhibitory effect of SUMO chains on HR-mediated fork restart | Rad51 loading to promote error free fork restart and preventing CAG repeat instability | Promoting conservative fork restart pathway to avoid error-prone Rad51-dependent SCR | Preventing SCR at telomeres to promote the maintenance of repetitive DNA | Promoting replication stress response to ensure fork restart and prevent mitotic abnormalities. |
| Reference | [33] | [32,34,97] | [34] | [36] | [35] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Schirmeisen, K.; Lambert, S.A.E.; Kramarz, K. SUMO-Based Regulation of Nuclear Positioning to Spatially Regulate Homologous Recombination Activities at Replication Stress Sites. Genes 2021, 12, 2010. https://doi.org/10.3390/genes12122010
Schirmeisen K, Lambert SAE, Kramarz K. SUMO-Based Regulation of Nuclear Positioning to Spatially Regulate Homologous Recombination Activities at Replication Stress Sites. Genes. 2021; 12(12):2010. https://doi.org/10.3390/genes12122010
Chicago/Turabian StyleSchirmeisen, Kamila, Sarah A. E. Lambert, and Karol Kramarz. 2021. "SUMO-Based Regulation of Nuclear Positioning to Spatially Regulate Homologous Recombination Activities at Replication Stress Sites" Genes 12, no. 12: 2010. https://doi.org/10.3390/genes12122010
APA StyleSchirmeisen, K., Lambert, S. A. E., & Kramarz, K. (2021). SUMO-Based Regulation of Nuclear Positioning to Spatially Regulate Homologous Recombination Activities at Replication Stress Sites. Genes, 12(12), 2010. https://doi.org/10.3390/genes12122010






